Abstract
Apolipoprotein M (ApoM) is considered a protective factor that inhibits the occurrence and development of liver cancer, but the specific underlying mechanisms require further investigation. Previous studies have demonstrated that ApoM gene knockout promotes the expression of the transcription factor sterol regulatory element-binding protein 1 (SREBP1; also known as SREBF1) in the livers of mice. SREBF1 is closely associated with factors involved in fatty acid synthesis and has a role in the promotion of tumor progression. The present study initially confirmed that the expression levels of ApoM in cancer tissues were significantly decreased compared with those in normal tissue, while the expression levels of SREBF1 were significantly increased. In addition, ApoM gene knockout significantly increased the expression levels of SREBF1 and the key glycolytic enzyme ATP-dependent 6-phosphofructokinase, liver type (PFKL). Binding site prediction and a dual-luciferase reporter gene assay indicated that SREBF1 regulates the promoter region of PFKL. To the best of our knowledge, the present study was the first to propose the regulation of glycolytic enzyme transcription levels by SREBF1. Furthermore, cell proliferation and Transwell assays demonstrated that ApoM gene knockout increased the expression levels of SREBF1 and further enhanced the activity of the promoter region of PFKL, ultimately promoting the proliferation, migration and invasion of liver cancer cells.
Keywords: liver cancer, apolipoprotein M, sterol regulatory element-binding protein 1, ATP-dependent 6-phosphofructokinase, liver type, proliferation, migration, invasion
Introduction
Liver cancer is one of the most common malignant tumors of the digestive system and exhibits characteristics of high-grade malignancy, rapid progression, high recurrence rates and a high probability of metastasis (1,2). According to statistics, liver cancer will become a global public health challenge for >1 million patients in 2025 (3). It is generally acknowledged that the Warburg effect (4), and metabolic disturbances of glutamine (5) and fatty acids (6), are able to accelerate the progression of cancer and the reprogramming of cell metabolism leads to uncontrollable proliferation activity. In primary liver cancer, metabolic diseases, such as obesity (7), non-alcoholic fatty liver disease (8) and type 2 diabetes mellitus (9) have been listed as high-risk factors. Of note, whether in the initiation or promotion stages of hepatocarcinogenesis, the stability of the metabolic environment determines the final outcome.
As a member of the apolipoprotein family, apolipoprotein M (ApoM) participates in the synthesis of high-density lipoprotein (HDL) and the reverse transport of cholesterol (10). In view of the contribution of ApoM in protecting against insulin resistance (11), exhibiting antiatherosclerotic functions (12) and reducing liver lipid accumulation (13), it is considered to be an important factor in regulating glucose and lipid metabolism. In recent years, accumulating evidence has indicated that ApoM is associated with the occurrence and development of primary liver cancer. For instance, the mRNA and protein levels of ApoM in liver cancer tissues are significantly reduced compared with those in neighboring tissues (14); hsa-microRNA (miR)-573, as a potential target of ApoM, is able to reduce ApoM expression levels with an accompanying reduction in hepatoma cell apoptosis (15). In addition, ApoM functions as a tumor suppressor to inhibit the growth and metastasis of SMMC7721 cells via vitamin D receptor signaling (16). In summary, ApoM exhibits a positive effect in suppressing tumor progression. Of note, results of a previous study by our group demonstrated that ApoM-knockout mice formed tumors faster under the induction of N-nitrosodiethylamine, which may indicate that ApoM also has an important inhibitory effect in the process of liver cancer (17).
Based on the above viewpoints, ApoM is an apolipoprotein that inhibits the occurrence and development of liver cancer, and it is closely related to the body's glucose and lipid metabolism. At present, the metabolic level and tumor metabolic microenvironment are still the focus of cancer related research. But whether ApoM affects the development of liver cancer through glycolipid metabolism remains unclear. Of note, the results of a previous study by our group demonstrated that deficiency of the ApoM gene causes damage to autophagy activity in the liver and eventually leads to lipid accumulation (18). Furthermore, the expression levels of sterol regulatory element-binding protein 1 (SREBF1; also known as SREBP1) are markedly upregulated in this process (18). ApoM has been proven to regulate various biological processes, including lipid biosynthesis (19), insulin resistance (20) and tumor growth (21). Considering that glycolysis is a typical metabolic pathway in cancer cells, the present study aimed to investigate whether ApoM regulates glycolysis through the SREBF1 pathway, thus affecting the progression of liver cancer. The present study aimed to further elucidate the potential association between ApoM, glycolysis and primary liver cancer and explored the potential mechanism by which ApoM inhibits the occurrence and development of liver cancer.
Materials and methods
Cell culture
Huh-7 cells (BeNa Culture Collection) and Mhcc97h cells (Guangzhou Saiku) were cultured in high-sugar DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Shanghai ExCell Biology, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and were incubated at 37°C in an atmosphere with 5% CO2. As for the reason for choosing Huh7 and Mhcc97h cells, it was observed that SREBF1 was highly expressed in Huh7 and Mhcc97h cells (22). Small interfering (si)RNA (Guangzhou RiboBio Co., Ltd.) was transfected into cells using Lipofectamine® 3000 reagent (cat. no. L3000150; Thermo Fisher Scientific, Inc.). The sequences were as follows: si-negative control (NC), 5′-GGCTCTAGAAAAGCCTATGC-3′ (this control was non-targeting), si-SREBF1, 5′-CGGAGAAGCTGCCTATCAA-3′ and si-ApoM, 5′-GAGCACAGATCTCAGAACT-3′.
Dual-luciferase activity detection
The Ensembl database (http://asia.ensembl.org/index.html) was used to query the promoter sequence of ATP-dependent 6-phosphofructokinase, liver type (PFKL). The JASPAR 2022 database (https://jaspar.genereg.net/) found that SREBF1 may be a transcription factor of PFKL and has a binding site. The highest scoring sequences were selected an constructed synthetically into pgl3-basic (Nanjing Qingke Co., Ltd.). The pgl3-basic and pEnCMV-SREBF1 (human)-HA plasmids (Nanjing Qingke Co., Ltd.) were then co-transfected into 293T cells. Dual-Luciferase Reporter Gene Assay kit (cat. no. E1910; Glomax; Promega Corporation) was used to detect both the firefly and Renilla luciferase gene activity.
EDU staining assay
Each group of cells (si-NC, si-SREBF1, pcDNA3.1-NC, pcDNA3.1-SREBF1, si-ApoM−/−+si-SREBF1 and si-SREBF1+pcDNA3.1-PFKL) was stained according to the instructions of the EDU Cell Proliferation kit (cat. no. C10310-1; Guangzhou RiboBio Co., Ltd.). Images were obtained using an inverted fluorescence microscope (Olympus Corporation). ImageJ software V1.8.0.112 [National Institutes of Health (NIH)] was used for data analysis.
Western blot analysis
RIPA lysis buffer (cat. no. BL651A; Biosharp Life Sciences) and PMSF (cat. no. BL507A; Biosharp Life Sciences) were used for tissue and cell protein extraction, and the protein concentration was measured using a NanoDrop® 2000 mini-spectrophotometer (Thermo Fisher Scientific, Inc.). Proteins (60 µg) were separated using SDS-PAGE (10 or 12% gel), transferred to a PVDF membrane (MilliporeSigma) and subsequently blocked with blocking solution (cat. no. P0023B; Beyotime Institute of Biotechnology) for 10 min at room temperature. Subsequently, samples were incubated with the appropriate primary antibody solution overnight at 4°C. The next day, the PVDF membranes were incubated in secondary antibody solution for 2 h at room temperature (both 1:3,000 dilution; cat. nos. BL001A and BL003A; Biosharp Life Sciences). ECL chemiluminescent fluid (cat. no. BL520A; Biosharp Life Sciences) and an imaging system (ShanghaiTanon-5200 Co., Ltd.) were used for exposure. The following antibodies were used: Anti-ApoM (cat. no. A5336; 1:1,000 dilution; ABclonal Biotech Co., Ltd.), anti-PFKL (cat. no. A7708; 1:1,000 dilution; ABclonal Biotech Co., Ltd.) and anti-SREBP1 (cat. no. ab138663; 1:1,000 dilution; Abcam) antibody were used to react with human hepatoma cells, while anti-SREBP1 (cat. no. ab28481; 1:1,000 dilution; Abcam) antibody was used to react with mouse tissue proteins and β-actin (A1978; 1:5,000 dilution; MilliporeSigma). ImageJ software V1.8.0.112 (NIH) was used for statistical analysis.
Transwell migration and invasion assays
Following cell transfection with si-NC, si-SREBF1, pcDNA3.1-NC, pcDNA3.1-SREBF1, si-ApoM−/−+si-SREBF1 or si-SREBF1+ pcDNA3.1-PFKL plasmids for 48 h at 37°C, the cell concentration was adjusted to 1×105 cells/ml using serum-free cell culture medium and 200 µl cell suspension was added to the upper chamber for the migration assay (cat. no. 3422; Corning, Inc; PC membrane, 6.5 mm; pore size, 8.0 µm). A total of 600 µl cell culture medium with 20% serum was added to the lower chamber and plates were cultured for 48 h. Cells that passed through the membrane were stained with 4% paraformaldehyde (Phygene Brotechnology Co., Ltd.) and 0.1% crystal violet solution (cat. no. C0121; Beyotime Institute of Biotechnology.) for 10 min at room temperature. Finally, use a cotton swab to wipe the cells that have not crossed the membrane. Migrated cells were counted under a microscope (Olympus Corporation). For the invasion assay, Dilute Matrigel® to 200 µg/ml with PBS, and 50 µl Matrigel® was added to the upper chamber prior to incubation at 37°C for 2 h. The remaining steps were followed in an identical manner to those of the migration assay above. ImageJ software V1.8.0.112 (National Institutes of Health) was used for data analysis.
Wound-healing assay
Following cell transfection with si-NC, si-SREBF1, pcDNA3.1-NC, pcDNA3.1-SREBF1, si-ApoM−/−+ si-SREBF1 or si-SREBF1+pcDNA3.1-PFKL plasmids for 48 h, a 200-µl pipette tip was used to make a linear scratch on the cell monolayers. Cells were subsequently washed three times with PBS and cultured in DMEM. After 48 h of incubation, the width of the gap refilled by the cells was measured and recorded, and the wound-healing rate was calculated. ImageJ software V1.8.0.112 (NIH) was used for data analysis.
Animals
The present study was approved by the Experimental Animal Welfare and Ethics Committee of Wannan Medical College (approval no. LLSC-2020-001). According to literature reports, there are more new cases of liver cancer in Chinese males than in females (23), and they are more likely to get liver cancer. Therefore, male mice were selected to establish the liver cancer induction model. C57BL/6J mice were purchased from the Shanghai Model Organisms Center. The experimental mice were reared in a specific pathogen-free area at the Experimental Animal Center of Wannan Medical College and kept at a constant temperature of 22–24°C, humidity of 38% and a 12-h light/dark cycle. The animal cage and drinking water bottle were subjected to high-temperature and high-pressure sterilization (121°C, 30 min). Mice had free access to food and water and were regularly observed and cared for every day. Furthermore, 40 8-week-old male WT mice (body weight, 20–23 g; Qinglongshan Animal Farm) were randomly divided into two groups by intraperitoneal injection of N-nitrosodiethylamine (35 mg/kg, Shanghai McLean Co., Ltd.) solution and an equal volume of normal saline once a week until liver tumors developed. The same applies to the experimental grouping of ApoM−/− mice. During this period, three mice were randomly selected from each group every month. Mice were anesthetized by intraperitoneal injection of 1% sodium pentobarbital (50 mg/kg) and blood was collected from the medial canthus. All animals were killed by cervical dislocation and a part of the liver tissue was taken out and placed in 10% neutral formalin solution (Jiangsu Siyan Biotechnology), and another part was stored in a −80°C refrigerator. The experimental end-point was the appearance of relevant indications of clinical symptoms (such as the appearance of liver tumor). Selection of humane endpoints: If the relevant indications of clinical symptoms in the experiment had not yet appeared but the body weight of the mouse had decreased by >20% and the mouse was hunched, trembled, distanced from the group and unable to eat normally, the mouse was euthanized. Absence of heartbeat and breathing, and the disappearance of reflexes were used as the criteria for confirming death of the mice.
Detection of lactic acid (LA), ATP, triglyceride (TG), total cholesterol (T-CHO), HDL cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) content in tissues
The liver issues of WT healthy mice and WT tumor-forming mice were obtained. The levels of LA, ATP, TG, T-CHO, HDL-C and LDL-C were determined in tissues using kits purchased from Beijing Solarbio Science & Technology Co., Ltd. (cat. no. BC2235) and Nanjing Jiancheng Bioengineering Institute (cat. nos. A095-1-1, A110-1-1, A111-1-1, A112-1-1 and A113-1-1), according to the manufacturers' protocols.
H&E staining
Tissues were fixed in 10% formalin for 48 h at room temperature, embedded in paraffin and cut into 4-µM sections. Following deparaffinization and rehydration, the sections were stained using H&E and the morphology was observed under a microscope (Olympus Corporation).
Immunohistochemistry
The liver tissues of WT healthy mice and WT tumor-forming mice were embedded in paraffin and the sections were subsequently dissected into thin slices and deparaffinized in xylene. Tissue slides were incubated with SREBF1 antibody (cat. no. ab28481; 1:150 dilution; Abcam) overnight at 4°C. Following primary incubation, the slides were incubated with the secondary antibody (cat. no. GB23303; 1:200 dilution; Servicebio, Co., Ltd.) at 37°C for 50 min. The slides were subsequently incubated with DAB (1:50 dilution; Servicebio, Co., Ltd.) to visualize the staining. Samples were counterstained using hematoxylin solution (Servicebio, Co., Ltd.) for 90 sec and then differentiated using 1% hydrochloric acid alcohol for several seconds at room temperature. The slides were mounted using neutral gum (cat. no. G1403; Servicebio, Co., Ltd.) prior to being placed under a microscope (Olympus Corporation) to observe the expression of SREBF1 protein in each tissue. Image-Pro-Plus 6.0 software (Media Cybernetics, Inc.) was used for data analysis.
The Cancer Genome Atlas (TCGA) data screening
R software (version 3.6.3) [DESeq2 (version 1.26.0) 25516281] was used to enter the Level 3 HTSeq-Counts RNASeqV2 data in TCGA (https://portal.gdc.cancer.gov/). The hepatocellular carcinoma project and the target molecule ApoM (ENSG00000204444) were used for screening. The expression levels of SREBF1 and SREBF2 were calculated according to low and high expression levels of ApoM.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc.) and SPSS 26.0 (IBM Corporation) software. A total of three parallel experiments were set up in each group and these were performed as three repeats. Values are expressed as the mean ± standard deviation. The Kolmogorov-Smirnov test was applied to determine compliance with a normal distribution. Pairwise comparisons between groups in the presence of multiple groups were performed using one-way ANOVA followed by Tukey's post-hoc test. An unpaired Student's t-tests was used to determine significant differences between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
N-nitrosodiethylamine induces high expression of SREBF1 in liver cancer tissue derived from tumor-forming mice
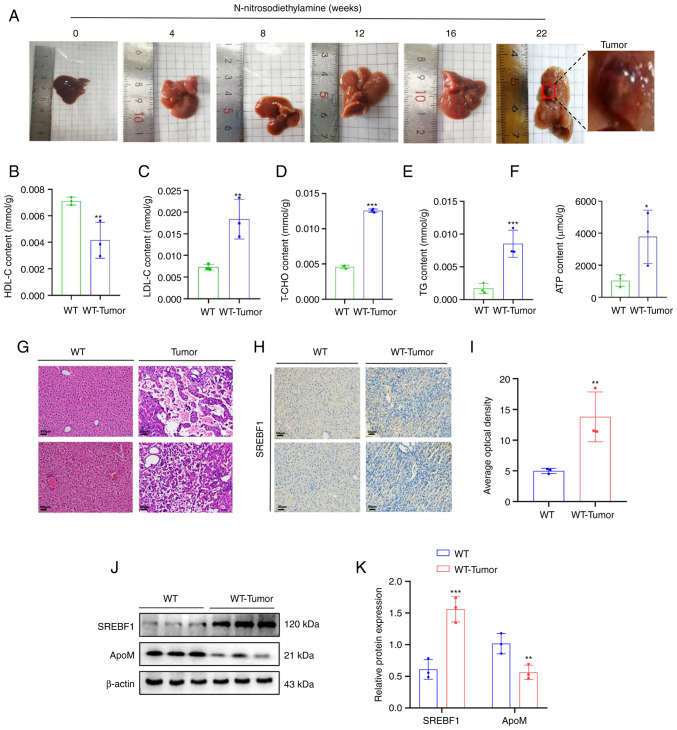
N-nitrosodiethylamine was used to induce liver cancer in mice. Mice induced by N-nitrosodiethylamine developed up to two tumors in the liver, but more frequently, one tumor was formed. The tumor diameter in mice with liver tumors was 0.3-0.6 cm (experimental period, 5–6 months) (Fig. 1A). In WT tumor-forming mice, it was observed that the levels of LDL-C, T-CHO, TG and ATP increased, while the levels of HDL-C decreased significantly (Fig. 1B-F), compared with the same indicators in liver tissue of healthy WT mice. Western blot analysis suggested that, compared with that in healthy WT mouse liver tissue, the expression of SREBF1 in liver cancer tissue was significantly increased and the expression of ApoM was significantly decreased (Fig. 1J). Furthermore, the results of the immunohistochemical analysis further verified that WT tumor-forming mice exhibited increased expression levels of SREBF1 compared with those of healthy WT mice (Fig. 1H).
Figure 1.
High expression of SREBF1 in N-nitrosodiethylamine-induced mouse hepatocellular carcinoma tissues. (A) Images of livers from mouse models with N-nitrosodiethylamine-induced liver cancer tumors. (B-F) A test kit was utilized to detect the content of (B) HDL-C, (C) LDL-C, (D) T-CHO, (E) TG and (F) ATP in liver cancer tissue of WT mice and liver tissue from healthy mice. (G) H&E staining revealed the morphology of liver tissue in mice prior to and after induction with N-nitrosodiethylamine (scale bar, 100 µm). (H) SREBF1 expression levels were determined using immunohistochemistry (scale bar, 50 µm). (J and K) Western blot analysis was performed to evaluate the expression levels of SREBF1 and ApoM in liver cancer tissue of WT mice and liver tissue from healthy mice. (J) Representative western blots and (K) quantified results. Analysis in each group was performed three times in parallel. *P<0.05, **P<0.03, ***P<0.01 vs. WT. HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; T-CHO, total cholesterol; TG, triglyceride; WT, wildtype; SREBF1, sterol regulatory element-binding protein 1; ApoM, apolipoprotein M.
ApoM knockout promotes SREBF1 to regulate the expression of PFKL
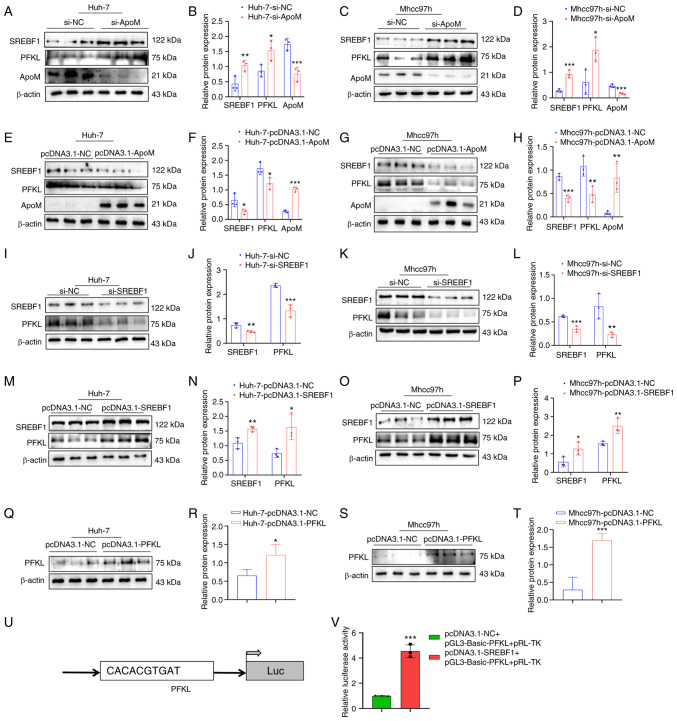
Results of a previous study by our group demonstrated that ApoM gene knockout in promoted the expression of SREBF1 in the liver (18). In addition, the levels of lactic acid in liver cancer tissue and serum of mice with liver cancer in which the ApoM gene was inhibited were significantly higher than those in WT mice with liver cancer (Fig. S1). These results suggested that the levels of glycolysis were increased. In order to further explore whether ApoM affects the expression levels of SREBF1 and PFKL in liver cancer cells, ApoM was silenced in liver cancer cells and the expression levels of SREBF1 and PFKL were markedly increased in Huh-7 and Mhcc97h cells (Fig. 2A-D). Following overexpression of the ApoM gene, the expression levels of SREBF1 and PFKL were decreased in Huh-7 and Mhcc97h cells (Fig. 2E-H). In addition, following SREBF1 gene knockout in liver cancer cells, the expression of PFKL in Huh-7 cells and Mhcc97h cells was significantly decreased (Fig. 2I-L). Following overexpression of the SREBF1 gene, the expression levels of PFKL were increased in Huh-7 and Mhcc97h cells (Fig. 2M-P). Western blot analysis validated the PFKL overexpression model in Huh-7 and Mhcc97h cells (Fig. 2Q-T). In order to explore whether the transcription factor SREBF1 has the ability to regulate PFKL at the transcriptional level, the JASPAR database was initially used to predict the binding sites. Of note, the results of the luciferase-based gene reporter assay demonstrated that SREBF1 enhanced PFKL promoter activity (Fig. 2U and V).
Figure 2.
SREBF1 is able to regulate the expression of PFKL. (A-H) Detection of the expression levels of SREBF1, PFKL and ApoM in Huh-7 and Mhcc97h cells using western blot analysis after knockdown or overexpression of ApoM. (A) Representative western blot image for Huh-7 cells with ApoM knockdown and (B) quantified expression levels. (C) Representative western blot image for Mhcc97h cells with ApoM knockdown and (D) quantified expression levels. (E) Representative western blot image for Huh-7 cells with ApoM overexpression and (F) quantified expression levels. (G) Representative western blot image for Mhcc97h cells with ApoM overexpression and (H) quantified expression levels. (I-P) Detection of the expression levels of SREBF1 and PFKL in Huh-7 and Mhcc97h cells using western blot analysis after knockdown or overexpression of SREBF1. (I) Representative western blot image for Huh-7 cells with SREBF1 knockdown and (J) quantified expression levels. (K) Representative western blot image for Mhcc97h cells with SREBF1 knockdown and (L) quantified expression levels. (M) Representative western blot image for Huh-7 cells with SREBF1 overexpression and (N) quantified expression levels. (O) Representative western blot image for Mhcc97h cells with SREBF1 overexpression and (P) quantified expression levels. (Q-T) Western blot analysis was used to validate the PFKL overexpression model in Huh-7 cells and Mhcc97h cells. (Q) Representative western blot image for Huh-7 cells with PFKL overexpression and (R) quantified expression levels. (S) Representative western blot image for Mhcc97h cells with PFKL overexpression and (T) quantified expression levels. (U) Prediction of the binding site of SREBF1 and PFKL. (V) Results of the luciferase-based gene reporter assay used to detect the promoter activity of PFKL through promoting SREBF1. Each group was set up three times in parallel. *P<0.05, **P<0.03, ***P<0.01 vs. NC group. ApoM, apolipoprotein M; SREBF1, sterol regulatory element-binding protein 1; PFKL, ATP-dependent 6-phosphofructokinase, liver type; NC, negative control; si-, small interfering RNA; Luc, luciferase.
ApoM regulates PFKL through the transcription factor SREBF1 to inhibit liver cancer cell proliferation
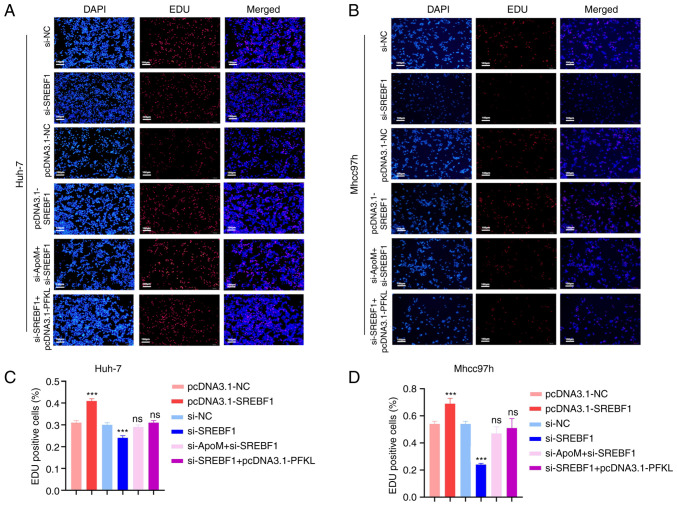
Results of a previous study by our group demonstrated that ApoM gene knockout significantly increased the proliferation of liver cancer cells (17). These results demonstrated that ApoM affects the expression of SREBF1 and thereby changes the promoter activity of PFKL. Using EDU staining, the role of ApoM in the proliferation of liver cancer cells through this pathway was verified (Fig. 3). In Huh-7 cells, the proliferation activity in the pcDNA3.1-SREBF1 group was significantly increased compared with that in the pcDNA3.1-NC group (Fig. 3A and C). Of note, the levels of Mhcc97h cell proliferation were also increased (Fig. 3B and D). In the si-SREBF1 group, the proliferation activity significantly decreased compared with that in the si-NC group (Fig. 3A and C). Consistent with these results, there was also a downward trend in Mhcc97h cells (Fig. 3B and D). The aforementioned results indicated that SREBF1 interference inhibited the proliferation activity of hepatoma cells; on the contrary, SREBF1 overexpression promoted the proliferation activity of liver cancer cells. However, there was no significant difference between the proliferation activity in the si-SREBF1+pcDNA3.1-PFKL group compared with that in the si-NC group. Thus, it was hypothesized that the transcription factor SREBF1 regulated PFKL to promote the proliferation of liver cancer cells (Fig. 3A and C). These results were consistent in both Mhcc97h and Huh-7 cells (Fig. 3B and D). In addition, there was no significant difference between the proliferation activity of the si-ApoM+si-SREBF1 group and the si-NC group (Fig. 3A and C). These results were also consistent between both Mhcc97h and Huh-7 cells (Fig. 3B and D), suggesting that ApoM may inhibit the proliferation of liver cancer cells through the transcription factor SREBF1.
Figure 3.
Knockdown of ApoM promotes SREBF1 to regulate PFKL to promote the proliferation of hepatoma cells. An EDU staining assay was used to detect the proliferation activity of (A) Huh-7 cells or (B) Mhcc97h cells in each group (pcDNA3.1-NC, pcDNA3.1-SREBF1, si-NC, si-SREBF1, si-SREBF1+pcDNA3.1-PFKL and si-ApoM+si-SREBF1; scale bar, 100 µm). Statistical analysis of the results for (C) Huh-7 cells or (D) Mhcc97h cells using ImageJ. Each group was set up three times in parallel. ns, no significance; ***P<0.01 vs. NC. NC, negative control; SREBF1, sterol regulatory element-binding protein 1; siRNA, small interfering RNA; PFKL, ATP-dependent 6-phosphofructokinase, liver type; ApoM, apolipoprotein M.
ApoM regulates PFKL through the transcription factor SREBF1 to inhibit the migration and invasion of liver cancer cells
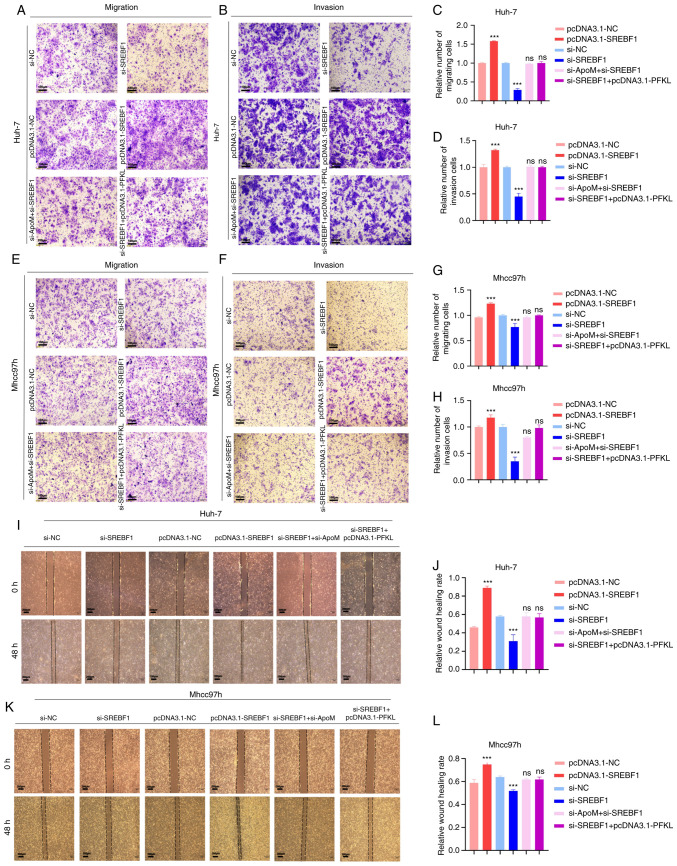
Metastasis is one of the most important causes of malignancy. Thus, Transwell assays were performed in the present study to detect whether ApoM regulates PFKL through the transcription factor SREBF1 and affects the migration and invasion of liver cancer cells. First, the role of SREBF1 in the development of liver cancer cells was identified (Fig. 4). The data suggested that inhibiting the expression of SREBF1 gene in Huh-7 cells resulted in a lower migration and invasion ability compared with the control group (siSREBF1 vs. si-NC; Fig. 4A-D, I and J), and the experimental results in Mhcc97h cells exhibited the same trend (siSREBF1 vs. si-NC; Fig. 4E-H, K and L). On the contrary, overexpression of SREBF1 significantly increased the migration and invasion ability in the two groups of cells (pcDNA3.1-SREBF1 vs. pcDNA3.1-NC; Fig. 4A-L). Therefore, transcription factor SREBF1 has a positive role in the development of hepatocellular carcinoma cells. A previous study by our group proved that inhibition of ApoM expression increased the expression of SREBF1 in the liver (18) and promoted the migration and invasion of liver cancer cells (17). In the present study, in order to verify whether the down-regulation of ApoM gene expression is able to drive the progression of hepatocellular carcinoma mediated by SREBF1, the expression of SREBF1 was inhibited by inhibiting ApoM gene expression and it was observed whether this affected the migration and invasion ability of liver cancer cells. Compared with the control group, there was no significant change in the migration or invasion ability of cells in both groups (si-ApoM + si-SREBF1 vs. si-NC; Fig. 4A-L). Therefore, combined with the above conclusions, it was indicated that the enhancement of the migration and invasion ability of liver cancer cells caused by the downregulation of the ApoM gene was achieved by promoting the expression of SREBF1. Finally, in order to explore whether SREBF1 is a key factor for PFKL in promoting the progression of liver cancer cells from another perspective, the migration and invasion abilities of the si-SREBF1+ pcDNA3.1-PFKL group and the siNC group were compared; the results indicated after the SREBF1 gene was inhibited, even overexpression of the key glycolysis enzyme PFKL did not enhance the ability of liver cancer cells to migrate and invade (Fig. 4A-L). Taken together, these results suggest that ApoM regulates PFKL by affecting the expression of SREBF1 and ultimately mediate the progression of HCC cells.
Figure 4.
Knockdown of ApoM promotes SREBF1 to regulate PFKL to promote the migration and invasion of hepatoma cells. Huh-7 cells or Mhcc97h cells were transfected with pcDNA3.1-NC, pcDNA3.1-SREBF1, si-NC, si-SREBF1, si-SREBF1 + pcDNA3.1-PFKL or si-ApoM + si-SREBF1. (A-H) Transwell assays. Representative images of Huh-7 cells transgressed through the membrane in the (A) migration and (B) invasion experiment and quantified results for (C) migration and (D) invasion. Representative images of Mhcc97h cells transgressed through the membrane in the (E) migration and (F) invasion experiment and quantified results for (G) migration and (H) invasion (scale bars, 100 µm). (I-L) Wound-healing assay. (I) Representative images of migration of Huh-7 cells and (J) quantified results. (K) Representative images of migration of Mhcc97h cells (scale bars, 200 µm) and (L) quantified results. Each group was set up three times in parallel. ns, no significance; ***P<0.01 vs. NC. NC, negative control; SREBF1, sterol regulatory element-binding protein 1; si-, small interfering RNA; PFKL, ATP-dependent 6-phosphofructokinase, liver type; ApoM, apolipoprotein M.
Discussion
Results of previous studies have demonstrated that ApoM is commonly associated with liver cancer. Previous reports indicated differential ApoM mRNA levels and ApoM protein mass in liver cancer tissue and adjacent tissues (14), and that loss of the ApoM gene increased the proliferation activity (16) and decreased the level of apoptosis (15). In summary, ApoM is considered to be a potential protective factor that may inhibit the occurrence and development of liver cancer.
Although ApoM has been confirmed to be involved in glucose and lipid metabolism (11,24), it has remained elusive whether ApoM affects liver cancer through metabolic pathways. To abnormally proliferate, tumor cells exhibit high levels of glycolysis, even in the presence of oxygen. This is a phenomenon named the Warburg effect (25). Based on a previous study by our group, the growth rate of tumors induced by diethylnitrosamine in ApoM (−/-) mice was significantly higher than that in WT mice (17). In a subsequent analysis, the lactic acid levels in ApoM(−/-) mice were indicated to be significantly higher than those in WT mice (Fig. S1). These data suggested that downregulation of the ApoM gene promotes glycolysis in tumor cells.
The glycolysis pathway contains three key rate-limiting enzymes: Hexokinase, phosphofructokinase (PFK) and pyruvate kinase. Results of previous studies have demonstrated that PFK may act as the most important regulator in the glycolysis pathway, including three PFK isoforms, platelet, liver and muscle isoform (4,26). In the present study, inhibiting ApoM expression upregulated the expression of PFKL in liver cancer cells. By contrast, overexpression of ApoM in liver cancer cells reduced PFKL expression. In accordance with the effects of ApoM on lactate production in tumor cells, results of the present study demonstrated that ApoM may attenuate the glycolytic pathway in tumor cells by suppressing PFKL expression.
The present study also aimed to explore the mechanism by which increased ApoM gene expression is negatively correlated with PFKL expression. Results of a previous study demonstrated that ApoM deficiency significantly suppressed the autophagy function in the mouse liver and caused lipid accumulation; furthermore, the expression levels of SREBF1 were significantly increased (18). As a transcription factor, SREBF1 has two different isoforms, SREBF1 and SREBF2 (also known as SREBP1 and SREBP2) (27). Using a bioinformatics analysis, the present study demonstrated that the upregulation of SREBF1 gene expression levels was more significant than that of SREBF2 in a low-ApoM gene expression group, compared with that in an ApoM high-expression group (Table SI). Previous studies have further highlighted that SREBF1 regulates lipid metabolism and it may provide energy for tumor cells through the lipogenesis pathway (28). In terms of glycolysis, it has been reported that SREBF1 and PKM2 are closely associated with tumor growth; however, the association between SREBF1 and PFKL has remained to be elucidated (29). To the best of our knowledge, the present study was the first to confirm that ApoM inhibits the expression of SREBF1 and PFKL. According to the results of the dual-luciferase reporter gene assay, SREBF1 has the ability to bind to PFKL and enhance its promoter activity. Furthermore, results of the EDU cell proliferation and Transwell assays indicated that ApoM may influence the growth and progression of liver cancer by regulating PFKL through SREBF1.
In conclusion, the present study further elucidated the potential mechanisms by which ApoM inhibits the development of liver cancer by regulating glycolysis; however, further investigations are required. Although metabolic enzymes are known to regulate metabolic processes, their non-metabolic activities require further investigation, which may include protein interactions and the crosstalk between different compartments of the signaling pathway. Similarly, this view is also particularly important for SREBF1. The present study confirmed that SREBF1 as a transcription factor may enhance the promoter activity of PFKL and its expression level was affected by ApoM. With regard to the question of how ApoM affects the expression of SREBF1, integration of existing research results provides noteworthy and highly relevant clues. First of all, SREBF1, as the central transcription factor regulating lipid metabolism, mainly regulates the expression of the factors required for fatty acid synthesis (30), which has recently been proved to be the reason why PI3K-AKT-mTOR signaling pathway endows cancer cells with the ability to resist iron death (31). Furthermore, ApoM is closely related to the AKT-mTOR signaling pathway and is associated with inflammation, apoptosis and insulin resistance (11,32,33). Future work will aim to prove that ApoM affects SREBF1 through certain pathways (at least the PI3K-AKT-mTOR signaling pathway). In addition, with the application of gene chip and sequencing technology, the downstream target genes of SREBF1 are gradually being identified, whose target roles may be roughly divided into direct regulation, indirect regulation and possible regulation. Therefore, the advancement of research on ApoM-SREBF1 - downstream effector will help to further expand and enrich the mechanism of ApoM participating in the regulation of liver cancer progression.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This study was supported by grants from the Natural Science Research Project of Anhui Universities (grant no. KJ2020A0612) and the Key Research and Development Program of Anhui, China (grant no. 1804h08020241).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and YB designed and supervised the study. WP performed the data analysis, statistical analysis and completed the manuscript writing. XZ, WZ and XL performed the experiments. XZ and WP performed plausibility checks and confirmed the authenticity of the raw data, which were further edited and later approved for publication by all authors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The use of the C57BL/6J mice required for the experiment was approved by the Experimental Animal Welfare and Ethics Committee of Wannan Medical College (Wuhu, China; approval no. LLSC-2020-001).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chao J, Zhao S, Sun H. Dedifferentiation of hepatocellular carcinoma: molecular mechanisms and therapeutic implications. Am J Transl Res. 2020;12:2099–2109. [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang H, Cao HJ, Ma N, Bao WD, Wang JJ, Chen TW, Zhang EB, Yuan YM, Ni QZ, Zhang FK, et al. Chromatin remodeling factor ARID2 suppresses hepatocellular carcinoma metastasis via DNMT1-snail axis. Proc Natl Acad Sci USA. 2020;117:4770–4780. doi: 10.1073/pnas.1914937117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Rossi JZ, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Li L, Li W, Chen T, Zou B, Zhao L, Wang H, Wang X, Xu L, Liu X, et al. TAp73-induced phosphofructokinase-1 transcription promotes the warburg effect and enhances cell proliferation. Nat Commun. 2018;9:4683. doi: 10.1038/s41467-018-07127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: Fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Wang H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016;379:191–197. doi: 10.1016/j.canlet.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 8.Center MM, Jemal A. International trends in liver cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2011;20:2362–2368. doi: 10.1158/1055-9965.EPI-11-0643. [DOI] [PubMed] [Google Scholar]
- 9.Marengo A, Rosso C, Bugianesi E. Liver Cancer: Connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67:103–117. doi: 10.1146/annurev-med-090514-013832. [DOI] [PubMed] [Google Scholar]
- 10.Xu N, Dahlback B. A novel human apolipoprotein (apoM) J Biol Chem. 1999;274:31286–31290. doi: 10.1074/jbc.274.44.31286. [DOI] [PubMed] [Google Scholar]
- 11.Kurano M, Tsukamoto K, Shimizu T, Kassai H, Nakao K, Aiba A, Hara M, Yatomi Y. Protection against insulin resistance by apolipoprotein M/sphingosine-1-phosphate. Diabetes. 2020;69:867–881. doi: 10.2337/db19-0811. [DOI] [PubMed] [Google Scholar]
- 12.Kurano M, Yatomi Y. Sphingosine 1-phosphate and atherosclerosis. J Atheroscler Thromb. 2018;25:16–26. doi: 10.5551/jat.RV17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y, Lam SM, Liu H, Luo G, Zhang J, Yao S, Li J, Zheng L, Xu N, Zhang X, Shui G. Comprehensive lipidomics in apoM(−/-) mice reveals an overall state of metabolic distress and attenuated hepatic lipid secretion into the circulation. J Genet Genomics. 2020;47:523–534. doi: 10.1016/j.jgg.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J, Wu C, Luo G, Zheng L, Chen L, Zhang X, Xu N. Expression of apolipoprotein M in human hepatocellular carcinoma tissues. Acta Histochem. 2011;113:53–57. doi: 10.1016/j.acthis.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Hu YW, Chen ZP, Hu XM, Zhao JY, Huang JL, Ma X, Li SF, Qiu YR, Wu XJ, Sha YH, et al. The miR-573/apoM/Bcl2A1-dependent signal transduction pathway is essential for hepatocyte apoptosis and hepatocarcinogenesis. Apoptosis. 2015;20:1321–1337. doi: 10.1007/s10495-015-1153-x. [DOI] [PubMed] [Google Scholar]
- 16.Yu M, Pan L, Sang C, Mu Q, Zheng L, Luo G, Xu N. Apolipoprotein M could inhibit growth and metastasis of SMMC7721 cells via vitamin D receptor signaling. Cancer Manag Res. 2019;11:3691–3701. doi: 10.2147/CMAR.S202799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai Y, Pei W, Zhang X, Zheng H, Hua C, Min J, Hu L, Du S, Gong Z, Gao J, Zhang Y. ApoM is an important potential protective factor in the pathogenesis of primary liver cancer. J Cancer. 2021;12:4661–4671. doi: 10.7150/jca.53115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Zhang P, Gao J, Huang Q. Autophagy dysregulation caused by ApoM deficiency plays an important role in liver lipid metabolic disorder. Biochem Biophys Res Commun. 2018;495:2643–2648. doi: 10.1016/j.bbrc.2017.12.148. [DOI] [PubMed] [Google Scholar]
- 19.Shimano H, Sato R. SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 20.Tang JJ, Li JG, Qi W, Qiu WW, Li PS, Li BL, Song BL. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 2011;13:44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond) 2018;38:27. doi: 10.1186/s40880-018-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin F, Feng F, Wang L, Wang X, Li Z, Cao Y. SREBP-1 inhibitor betulin enhances the antitumor effect of Sorafenib on hepatocellular carcinoma via restricting cellular glycolytic activity. Cell Death Dis. 2019;10:672. doi: 10.1038/s41419-019-1884-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An L, Zeng HM, Zheng RS, Zhang SW, Sun KX, Zou XN, Chen R, Wang SM, Gu XY, Wei WW, He J. Liver cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi. 2019;41:721–727. doi: 10.3760/cma.j.issn.0253-3766.2019.10.001. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 24.Borup A, Christensen PM, Nielsen LB, Christoffersen C. Apolipoprotein M in lipid metabolism and cardiometabolic diseases. Curr Opin Lipidol. 2015;26:48–55. doi: 10.1097/MOL.0000000000000142. [DOI] [PubMed] [Google Scholar]
- 25.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. doi: 10.1126/science.124.3215.269. [DOI] [PubMed] [Google Scholar]
- 26.Schoneberg T, Kloos M, Brüser A, Kirchberger J, Sträter N. Structure and allosteric regulation of eukaryotic 6-phosphofructokinases. Biol Chem. 2013;394:977–993. doi: 10.1515/hsz-2013-0130. [DOI] [PubMed] [Google Scholar]
- 27.Dorotea D, Koya D, Ha H. Recent insights Into SREBP as a direct mediator of kidney fibrosis via lipid-independent pathways. Front Pharmacol. 2020;11:265. doi: 10.3389/fphar.2020.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu C, Zhang L, Wang D, Jiang S, Cao D, Zhao Z, Huang M, Jin J. Lipidomics reveals that sustained SREBP-1-dependent lipogenesis is a key mediator of gefitinib-acquired resistance in EGFR-mutant lung cancer. Cell Death Discov. 2021;7:353. doi: 10.1038/s41420-021-00744-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao T, Su Q, Xu S, Deng J, Zhou S, Zhuang Y, Huang Y, He C, He S, Peng M, et al. Down-regulation of PKM2 decreases FASN expression in bladder cancer cells through AKT/mTOR/SREBP-1c axis. J Cell Physiol. 2019;234:3088–3104. doi: 10.1002/jcp.27129. [DOI] [PubMed] [Google Scholar]
- 30.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI0215593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi J, Zhu J, Wu J, Thompson CB, Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc Natl Acad Sci USA. 2020;117:31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Z, Zeng Y, Zhu X, Tan Y, Li Y, Li Q, Yi G. ApoM-S1P modulates Ox-LDL-induced inflammation through the PI3K/Akt signaling pathway in HUVECs. Inflammation. 2019;42:606–617. doi: 10.1007/s10753-018-0918-0. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Tang X, Li S. Propofol promotes migration, alleviates inflammation, and apoptosis of lipopolysaccharide-induced human pulmonary microvascular endothelial cells by activating PI3K/AKT signaling pathway via upregulating APOM expression. Drug Dev Res. 2021;83:397–406. doi: 10.1002/ddr.21869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.