Abstract

A characteristic feature of cytochromes P450* is that the complex formed between the ferrous heme iron and carbon monoxide generates an intense absorption band at 450 nm. This unique feature of P450s is due to the proximal thiolate Cys ligand coordinated to the heme iron. Various harsh treatments shift this band to 420 nm, thereby giving P420 which is most often associated with an inactive form of the enzyme. Various explanations have been put forward to explain the P450-to-P420 change ranging from protonation of the Cys heme ligand, displacement of the Cys ligand, or replacement of the Cys ligand with His. There are two crystal structures of the well-studied cytochrome P450cam that have a high fraction of P420. In one, P450cam is cross-linked to its redox partner, putidaredoxin (Pdx), and the second is P450cam crystallized in the absence of substrate. In both of these structures, a significant part of the substrate pocket is disordered and the poor quality of the electron density for the substrate indicates substantial disorder. However, in both structures there is no detectable change in the Cys-iron ligation or surrounding structure. These results indicate that the P450-to-P420 switch is due primarily to an opening and disordering around the substrate binding pocket and not ligand displacement or ligand swapping. Since it remains a possibility that ligand swapping could be responsible for P420 in some cases, we mutated to Gln the 3 His residues (352, 355, and 361) close enough to the proximal side of the heme that could possibly serve as heme ligands. The triple variant forms P420 which indicates that swapping Cys for His is not a requirement for the P450-to-P420 switch.
Introduction
One of the main characteristics of cytochromes P450 is the hyperporphyrin spectra with an intense 450 nm absorption band with a smaller 360 nm feature upon reduction and coordination of carbon monoxide (CO) to the ferrous heme iron.1 The name P450 derives from this intense 450 nm band.2 This 450 nm band is unique among heme proteins and has been attributed to the Cys thiolate ligand coordinated to the heme iron, while other heme proteins use a neutral ligand, such as histidine. Various harsh treatments of a P450 often result in a reduced-CO band at 420 nm which is usually associated with an inactive “damaged” P450, called P420.3,4 Conversion of P420 back to P450 often is not possible. The difference between P420 and P450 has been attributed to protonation of the Cys thiolate ligand,5 complete dissociation of the Cys ligand,6 or replacement of the Cys ligand by His.6,7 Thus, changes could be large or subtle given that as little as a 0.2 Å increase in S–Fe bond length8 can lead to P420. In the case of P450cam, His355 is the closest to the heme (Figure 1) and is the most likely candidate as a P420 ligand.7
Figure 1.
Crystal structure of the P450cam (cyan)-Pdx (green) complex1 (4JWS). The 2.15 Å 2Fo-Fc map is contoured at 1.0σ. His355 in P450cam cross-links to Cys73 in Pdx by the homo bifunctional maleimide (yellow). Very little movement of His355 is required to form the cross-link. The covalently cross-linked complex1 was prepared and purified as previously described.9
A crystal structure of a P420 would help to settle the question, and fortunately, such structures have been solved (Figure 1). Sarvind et al.9 solved the crystal structure of P450cam complexed to its ferredoxin Fe2S2 redox partner, Pdx. In order to trap the complex, a cross-linking approach was used where engineered Cys residues were cross-linked with a homobifunctional maleimide. Where to place the Cys residues to be cross-linked required some knowledge on the orientation of Pdx relative to P450cam. A model of the P450cam-Pdx complex derived from mutagenesis and NMR studies was used as a guide.10 The resulting structure (called complex1, 4JWS) unexpectedly showed that His355 in P450cam cross-linked with Cys73 in Pdx (Figure 1). The reason His355 was cross-linked rather than the engineered Cys residue is because Pdx in the crystal structure was oriented quite differently than the model used to engineer the cross-linking sites.
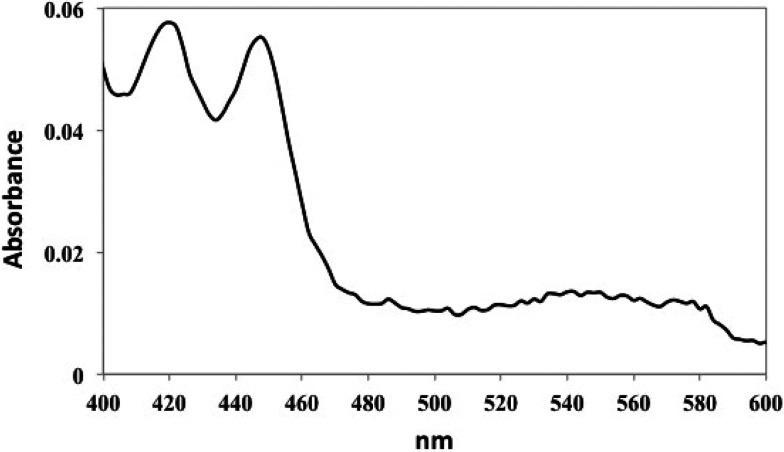
The UV/vis spectrum of the reduced CO-bound of complex1 clearly shows a large fraction of the cross-linked complex is P420 (Figure 2). At the time, there was concern that owing to the large amount of P420, the complex1 P450cam-Pdx structure does not reflect the active form of the enzyme. Therefore, Cys residues were engineered that would place the cross-link far from the heme and avoid cross-linking His355. The resulting 2.09 Å crystal structure (4JWU), called complex2, shows that the engineered Cys residues cross-linked as expected9 and the reduced CO spectrum of complex2 is fully P450 (ref (5), Figure 2B). Although complex1 and complex2 have different cross-linking sites, the Pdx binds the same, but there are major differences in the P450. In complex2, the entire polypeptide is visible and well-ordered, and product, 5-exo-hydroxycamphor, is bound in the active site, presumably because X-ray-generated reducing equivalents drives O2 activation and substrate hydroxylation in crystallo. However, in complex1, which gives a split P450 and P420 spectrum, the B′ helix (residues 91–101) and part of the F/G loop (residues 187–188) are disordered, and other helical segments such as the F, G, and I helices are displaced such that the active site is more open (Figure 3). In addition, there is an ill-defined density in the heme pocket that could be a disordered substrate/product. Similarly, the structure of the P450cam crystallized in the absence of substrate has an active site that is partially disordered and open. This open form of P450cam also generates a large fraction of P420.11
Figure 2.
Sample of the P450cam-Pdx complex1 was reduced with sodium dithionite followed by treatment with carbon monoxide. Spectra were recorded on an Agilent Cary 100/300 UV/vis spectrophotometer. The spectrum clearly indicates a large fraction of P420.
Figure 3.
(A) Comparison of complex1 (P420, cyan) and complex2 (P450, magenta). The B′ helix is disordered in complex1, and the F and G helices are displaced further from the main body of the protein which provides a wider opening to the active site. Part of the I helix which runs over the surface of the heme and provide key residues for substrate binding and O2 activation also is displaced. (B) Plot of α-carbon RMSD between complex1 and complex2 vs residue number. Regions of large changes are highlighted.
However, the heme and thiolate ligand environments are, within the limits of ∼2 Å resolution, identical in complex1, complex2, and the open form of P450cam crystallized in the absence of substrate.11 These results show that P420 can be generated without ligand swapping or displacement of the Cys thiolate ligand. In addition, treatment of P450cam with the well-known His modifying reagent, diethylpyrocabonate, actually promotes P420 formation which suggests that His ligation is not a requirement for P420 formation.4 Such chemical modification results, however, can be difficult to interpret owing to the possibility of incomplete His modification and/or modification of other residues. It thus remains a possibility that in some cases the switch to His ligand may occur in the P450-to-P420 switch. We therefore converted the three top candidates as a heme ligand, His352, 355, and 361, to Gln to determine if the variant would still form P420.
Methods and Materials
The P450cam wild-type and triple mutant genes were ordered in pET28a vectors from Genscript. The triple variant (His352, 355, and 361 to Gln) gene was used as a template for generation of the double variant gene by restoring Gln355 to His355 using the Takara PrimeSTAR Max DNA Polymerase kit for PCR. All three proteins followed the same expression and purification steps. The protein was overexpressed in E. coli C41 cells in TB media. Protein production was induced with 1 mM IPTG at an OD600 = 0.8–1.0, and the cells were grown at 25 °C and 80 rpm for 48 h. The cells were harvested and resuspended in lysis buffer (50 mM KPi pH 7.4, 1 mM camphor, 250 mM NaCl, and 2 mM β-mercaptoethanol (BME)) and lysed through a microfluidizer. The protein was loaded onto a HisPur Ni-NTA Resin (Thermo Scientific) column and washed with 3–5 column volumes of lysis buffer, which was followed by elution with 250 mM imidazole in lysis buffer. The protein was then dialyzed against 50 mM KPi pH 7.4, 8 mM camphor (saturated), 250 mM NaCl, and 2 mM BME. The N-terminal 6-His-tag was cleaved with thrombin. The proteins were loaded back onto a Ni-NTA column to collect the Histag-cleaved sample in the flow-through. The protein was dialyzed against 50 mM KPi pH 7.4, 8 mM camphor (saturated), and 5 mM BME. The protein was loaded onto a Q Sepharose (Cytiva) column and eluted with a gradient of 0–500 mM NaCl in 50 mM KPi pH 7.4, 1 mM camphor, and 5 mM BME. Finally, the protein was run on a size exclusion (S-200) chromatography (Cytiva) column in 50 mM KPi pH 7.4, 1 mM camphor, 150 mM NaCl, and 5 mM BME. The protein was concentrated, flash-frozen, and stored at −80 °C. The P450-to-P420 conversion was induced by the addition of 30% v/v acetone.12
All UV–vis spectroscopy was performed with an Agilent Cary 3 spectrophotometer at room temperature. The following protocol was used for all 3 P450cam proteins. The buffer was 50 mM KPi pH 7.4 and 1 mM camphor, and CO was bubbled in the buffer for 5–10 s. The protein was added to a final concentration of ∼6 μM. A few granules of dithionite powder were added to the cuvette, and the solution was inverted for mixing. The P450/P420 peaks appeared immediately, and reached a maximum after ∼10 min.
Results and Discussion
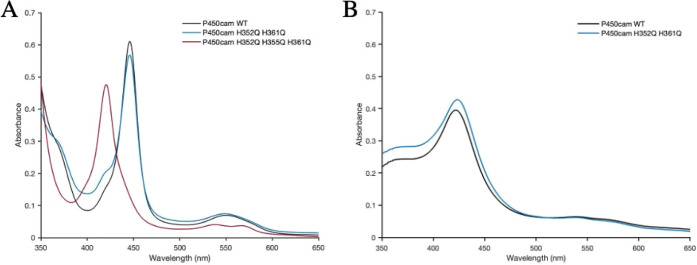
Figure 4 shows the His residues on the proximal side of the heme that are the most likely candidates for a heme ligand. We generated the H352Q/H361Q double variant and the H352Q/H355Q/H361Q triple variant. As shown in Figure 5A, formation of the reduced-CO complex of either the WT or the H352Q/H361Q double variant gives the expected 450 nm absorption band, while the triple variant is fully P420. Figure 5B shows that the double variant converts to P420 upon treatment with acetone exactly as wild-type. These spectral data indicate that mutating His352 and His361 have no effect on either the thiolate-Fe bond or the active site. This is not surprising given that both His residues are oriented toward the surface and play no direct role in stabilizing the active site. In sharp contrast, His355 plays a significant role as might be expected, since His355 H-bonds with a heme propionate and thus plays a direct role in stabilization of the heme pocket. The single variant of H355Q was not investigated in this work, but presumably it would display the same P420 results as the triple variant, as it is the only difference between the triple and double variants.
Figure 4.
Location of all the His residues in P450cam, colored cyan or green. The three most likely candidates to switch with the Cys ligand are His352, His355, or His 361, colored green. These three His residues are all on the same segment of polypeptide that is not associated with any regular secondary structure and could conceivably rearrange to enable His coordination without too much disruption of helices or sheets. The heme is shown in pink, and the substrate, camphor, is shown in yellow.
Figure 5.
UV–vis spectrum of P450cam WT and variants in the reduced-CO form: (A) wild-type, double, and triple variants; (B) acetone induced conversion to P420.
This is consistent with the complex1 crystal structure where the His355–heme interaction is disrupted since His355 is cross-linked to Pdx and complex1 generates a large fraction of P420. These results also clearly show that a switch from Cys to His ligation is not a requirement for the P450-to-P420 conversion. Instead, the disruption of the substrate binding pocket as evidenced in the complex1 crystal structure is the more likely reason for P420. That P420 is associated with a disordered and more open active site structure is evidenced by the fact that substrate binding results in no spin shift and that CO can more readily escape the P420cam active site.13 The substrate-free P450cam crystal structure11 and of the cyanide complex formed in the P450camPdx complex2 crystals (6NBL)14 in addition to the complex1 structure all show a rather dramatic restructuring around the distal substrate binding pocket with very little change around the Cys ligand on the proximal side. Even though our results indicate that ligand swapping is an unlikely reason for the P450-to-P420 switch, our results do not disprove this possibility.6,7 Such a switch, however, would require an energetically expensive major restructuring of the proximal pocket secondary structure and breakage of an S–Fe bond. A majority of structural and biochemical data thus favors changes in the substrate binding pocket and not major changes in Cys ligation that are detectable in ∼2 Å crystal structures as the source of the P450-to-P420 conversion.
Glossary
Abbreviations
- P450
cytochrome P450
- P420
an inactive form of P450
- Pdx
putidaredoxin
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- BME
β-mercaptoethanol; NTA, nitrilotriacetic acid
Author Present Address
# Department of Chemistry & Biochemistry, University of California, Santa Cruz, Santa Cruz, California 95064
Author Contributions
All authors have given approval to the final version of this manuscript
The authors acknowledge the National Institutes of Health Grant GM131920 for support of this work.
Associated Content: P450cam UniPro P00183; Pdx UniPro BAA00414
The authors declare no competing financial interest.
References
- Omura T.; Sato R. A new cytochrome in liver microsomes. J. Biol. Chem. 1962, 237, 1375–1376. 10.1016/S0021-9258(18)60338-2. [DOI] [PubMed] [Google Scholar]
- Hanson L. K.; Eaton W. A.; Sligar S. G.; Gunsalus I. C.; Gouterman M.; Connell C. R. Origin of the Anomalous Soret Spectra of Carboxy-Cytochrome P-450. J. Am. Chem. Soc. 1976, 98, 2672–2674. 10.1021/ja00425a050. [DOI] [PubMed] [Google Scholar]
- Martinis S. A.; Blanke S. R.; Hager L. P.; Sligar S. G.; Hui Bon Hoa G.; Rux J. J.; Dawson J. H. Probing the heme iron coordination structure of pressure-induced cytochrome P420cam. Biochemistry 1996, 35, 14530–14536. 10.1021/bi961511u. [DOI] [PubMed] [Google Scholar]
- Lipscomb J. D. Electron paramagnetic resonance detectable states of cytochrome P450cam. Biochemistry 1980, 19, 3590–3599. 10.1021/bi00556a027. [DOI] [PubMed] [Google Scholar]
- Perera R.; Sono M.; Sigman J. A.; Pfister T. D.; Lu Y.; Dawson J. H. Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 3641–3646. 10.1073/pnas.0737142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A. V.; Li P.; Champion P. M.; Martinis S. A.; Sligar S. G. Resonance Raman investigations of Escherichia coli-expressed Pseudomonas putida cytochrome P450 and P420. Biochemistry 1992, 31, 4384–4393. 10.1021/bi00133a002. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Zeng W.; Benabbas A.; Ye X.; Denisov I.; Sligar S. G.; Du J.; Dawson J. H.; Champion P. M. Investigations of heme ligation and ligand switching in cytochromes p450 and p420. Biochemistry 2013, 52, 5941–5951. 10.1021/bi400541v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C.; Friedrich J.; Ristau O. Quantum chemical interpretation of the spectral properties of the CO and O2 complexes of hemoglobin and cytochrome P-450. Acta Biol. Med. Ger. 1979, 38, 363–377. [PubMed] [Google Scholar]
- Tripathi S.; Li H.; Poulos T. L. Structural basis for effector control and redox partner recognition in cytochrome P450. Science 2013, 340, 1227–1230. 10.1126/science.1235797. [DOI] [PubMed] [Google Scholar]
- Pochapsky T. C.; Lyons T. A.; Kazanis S.; Arakaki T.; Ratnaswamy G. A structure-based model for cytochrome P450cam-putidaredoxin interactions. Biochimie 1996, 78, 723–733. 10.1016/S0300-9084(97)82530-8. [DOI] [PubMed] [Google Scholar]
- Lee Y. T.; Wilson R. F.; Rupniewski I.; Goodin D. B. P450cam visits an open conformation in the absence of substrate. Biochemistry 2010, 49, 3412–3419. 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.; Gunsalus I. C. Cytochrome P-450cam. II. Interconversion with P-420. J. Biol. Chem. 1974, 249, 102–106. 10.1016/S0021-9258(19)43096-2. [DOI] [PubMed] [Google Scholar]
- Tian W. D.; Wells A. V.; Champion P. M.; Di Primo C.; Gerber N.; Sligar S. G. Measurements of CO geminate recombination in cytochromes P450 and P420. J. Biol. Chem. 1995, 270, 8673. 10.1074/jbc.270.15.8673. [DOI] [PubMed] [Google Scholar]
- Follmer A. H.; Tripathi S.; Poulos T. L. Ligand and Redox Partner Binding Generates a New Conformational State in Cytochrome P450cam (CYP101A1). J. Am. Chem. Soc. 2019, 141, 2678–2683. 10.1021/jacs.8b13079. [DOI] [PMC free article] [PubMed] [Google Scholar]