Abstract
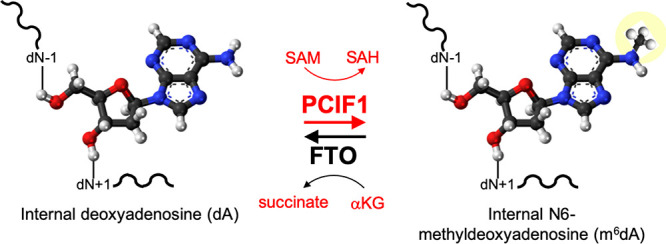
PCIF1 and FTO are a pair of human mRNA cap-specific modification enzymes that have opposing activities. PCIF1 adds a methyl group to the N6-position of 2′O-methyladenosine (Am), generating N6, 2′O-dimethyladenosine (m6Am), when Am is the cap-proximal nucleotide. FTO removes the N6-methyl group from m6Am. In addition, FTO has a demethylase activity on a broad spectrum of various RNA substrates, as well as on DNA N6-methyldeoxyadenosine (m6dA). While the existence of m6dA in mammalian DNA remains controversial, we show here that PCIF1 has significant methylation activity on single stranded DNA deoxyadenosine, double stranded RNA/DNA hybrids, and double stranded DNA, though with lower catalytic efficiency than that on its preferred RNA substrate. PCIF1 has activities in the order ssRNA > RNA/DNA hybrid > ssDNA > dsDNA. We discuss the implications of PCIF1 generation, and FTO removal, of DNA adenine methylation.
Introduction
While detected immunochemically as early as 1983,1 the existence of N6-methyl deoxyadenosine (m6dA) in mammalian DNA remains a controversial topic2−7 ever since it was reported again in 2016.8 One key issue is uncertainty regarding the methyltransferases (MTases) responsible for generating m6dA in mammalian DNA, suggesting the inclusion of N6AMT1/HemK2/KMT9 (henceforth HemK2), MettL4, and MettL3-MettL14 heterodimers, as well as regarding the enzymes removing the methyl group from m6dA (demethylases). We note that many nucleic acid-modifying enzymes modify both DNA and RNA (ref (9) and references therein), including members of the AlkB family (see below) involved in the direct reversal of alkylation damage to DNA and RNA10 and members of the Apobec family of cytidine deaminases.11 Tet2, one of the ten-eleven translocation proteins initially discovered as DNA 5-methylcytosine (5mC) dioxygenases,12 also mediates oxidation of 5mC in mRNA.13,14 These considerations prompted us to investigate whether known human RNA m6A MTases also possesses methyl transfer activity on DNA adenine.
Human HemK2 forms a heterodimer with Trm112 and was thought to be a DNA m6dA MTase (N6AMT1 activity) based on sequence comparisons15 but is actually a protein MTase active on glutamine (HemK2 activity) and lysine (KMT9 activity)16−19 and has no N6AMT1 activity. Murine MettL4 is responsible for DNA m6dA deposition in gene elements associated with transcriptional silencing.20 Intriguingly, recombinant human MettL4 expressed in HEK293T cells has in vitro enzymatic activity on mitochondrial DNA,21 whereas recombinant human MettL4 purified from Escherichia coli has RNA MTase activity on U2 snRNA that adds a methyl group to the N6-position of 2′O-methyloxyadenosine (Am), generating N6, 2′O-dimethyloxyadenosine (m6Am).22,23 [We note that, in searching of the Protein Data Bank (PDB), a homolog of Arabidopsis thaliana MettL4 has been crystallized in complex with the mononucleotide Am (which could occur in the context of RNA) (PDB 7CV6) and in complex with dsDNA (PDB 7DPE).] While MettL3-MettL14 had been characterized extensively as generating internal m6A in mRNA (ref (24) and references therein), human MettL3-MettL14 is also active in vitro on single-stranded DNA, as well as on double-stranded DNA containing short lesion-associated bubbles.25−27
From the opposing demethylation side, nine mammalian AlkB homologs exist (ALKBH1–8, and FTO), but only a subset functions as DNA/RNA (de)modification enzymes.10 The AlkB family members are Fe(II)- and α-ketoglutarate-dependent dioxygenases that direct reversal of DNA/RNA damage, removing alkyl adducts from nucleobases via oxidative dealkylation, including in some cases direct demethylation.28−31 Focusing on the m6dA substrates (Table 1), ALKBH1 is active on DNA containing unpaired lesions.32−34 In a study using HEK293T cells, DNA m6dA deposition and removal are catalyzed by the METTL4 MTase and ALKBH4 dioxygenase, respectively, corresponding with transcriptional silencing.20 An in vitro study found that ALKBH5 and FTO do not discriminate between RNA and DNA substrates.35
Table 1. Examples of Mammalian MTases and Demethylases Tested for DNA m6dA Activity.
| enzymes | adenine in DNA | references | |
|---|---|---|---|
| MTases | MettL3–14 | GGACT | Woodcock et al.25 |
| Yu et al.26 | |||
| Qi et al.27 | |||
| MettL4 | NNAGNN (nuclear) | Kweon et al.20 | |
| CTHATC (mitochondrial) | Hao et al.21 | ||
| PCIF1 | A | this study | |
| demethylases | FTO | m6dA | Jia et al.38 |
| Zou et al.35 | |||
| Zhang et al.41 | |||
| ALKBH1 | m6dA in dsDNA lesions | Li et al.32 | |
| Zhang et al.33 | |||
| Tian et al.34 | |||
| ALKBH4 | m6dA | Kweon et al.20 | |
| ALKBH5 | m6dA | Zou et al.35 | |
FTO, a fat mass and obesity-associated protein,36 was initially characterized as a repair enzyme active on 3-methyldeoxythymidine (m3dT) in single-stranded (ss) DNA,36 or on 3-methyloxyuracil (m3U) in ssRNA.37 Later, five additional FTO activities were identified: (a) a demethylase that removes the methyl group from internal m6A residues in mRNA,38 (b) a cap-dependent demethylase, which preferentially removes the N6 methyl group of m6Am,39 (c) a demethylase active on internal m6A and m6Am in small nuclear RNA (snRNA) and a demethylase of (d) 1-methyloxyadenosine (m1A) in tRNA40 and of (e) m6dA in ssDNA.35,38,41 Besides the important effects on the stability of mRNAs having m6Am at the 5′ end (in HEK293T cells39), FTO modulates mRNA splicing, is required for adipogenesis,42 and mediates stem-like properties in colorectal cancer cells.43 Among all the known FTO substrates, including multiple RNA species, structures have only been determined for human FTO bound to the mononucleotide m3dT44 or to ssDNA containing an internal m6dA.41 However, these structures do not explain the preference of FTO for either mRNA caps or for 2′O-methylation.
Although the demethylase activity of FTO on mRNA cap-specific m6Am modification was characterized first,39 the opposing activity of the MTase PCIF1 was discovered soon after. PCIF1 adds a methyl group to the N6-position of 2′O-methylated Am, generating dimethylated m6Am, when Am is the cap-proximal nucleotide.45−49 FTO removes the N6-methyl group from m6Am.39 Since FTO has a broad spectrum of substrates, including the demethylase activity on DNA m6dA,35,38,41 we asked whether PCIF1 is active on DNA substrates in vitro.
Materials and Methods
Reagents
The purified recombinant protein of human PCIF1 (NCBI: NP_071387.1; pXC2055) used in this study was recently characterized.50 The catalytic dead mutant of Asn553-to-Ala (N553A; pXC2303) was generated by PCR, confirmed by sequencing, and purified similar as the wild type. The oligonucleotides were synthesized by Integrated DNA Technologies (IDT). N6-methyladenosine (D9D9W) Rabbit mAb #56593 was purchased from Cell Signaling Technology. MettL3 inhibitors, two racemates of UZH1(R) and UZH1(S),51 and STM245752 were purchased from MCE (MedChemExpress).
Methylation Assays
Methylation assays of PCIF1 on different RNA and DNA oligonucleotides were conducted in the reaction buffer containing 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM DTT, 20 μM, or 100 μM S-adenosyl-l-methionine (SAM), with various PCIF1 and substrate concentrations, at 37 °C (for reaction time up to 4 h) or at room temperature (∼22 °C overnight). Methylation assays of MettL3-MettL14 (NCBI: NP_062826.2 and NP_066012.1) and HemK2-Trm112 (NCBI: NP_037372.4 and NP_057488.1) were conducted under the conditions as previously characterized in our laboratory.18,25,26,50
To calibrate SAH concentration and luminescence, the SAH standard solution within the MTase-Glo Methylation Assay Kit (Promega) was subjected to serial twofold dilutions, starting from 4 μM. Luminescence signals from a standard concentration curve of SAH were generated according to the manufacturer’s protocol. A 5 μL aliquot of sample was transferred into a low-volume 384-well plate, and the luminescence signal was detected using a Synergy 4 Multi-Mode microplate reader (BioTek). A linear regression of the SAH standard was plotted against luminescence.
Methylation Activity Detection by Antibody
Methylation reactions were visualized by dot blotting of reaction products onto an Amersham Hybond-N+ membrane (GE Healthcare) followed by immunoblotting with anti-methyladenosine antibody (Cell Signaling Technology, catalog D9D9W). Methylation reactions were carried out at room temperature (∼22 °C) for 4 h in a 20 μL mixture containing 10 μM substrates (RNA or DNA oligos or cap analogs), 20 μM SAM, 2 μM PCIF1 or HemK2-Trm112, or 0.1 μM Mettl3–14 in the reaction buffer. An aliquot of 5 μL of reaction products was spotted onto the membrane and air dried for 15 min. The membrane was blocked in 5% non-fat milk in TBST (a mixture of tris-buffered saline (TBS) and Tween 20) at room temperature for 1 h. The membrane was incubated overnight with the antibody (1:1000, 1% BSA in TBST) at 4 °C. After washing the membrane with TBST buffer three times for 10 min, it was incubated at room temperature for 1 h with HRP-linked anti-rabbit IgG antibody (Cell Signaling catalog 7074S, 1:5000, 1% BSA in TBST). The membrane was again washed three times with TBST buffer. Signals were detected with a Clarity Western ECL substrate (Bio-Rad Laboratories, #1705061) and imaged using the ChemiDoc imaging system (Bio-Rad Laboratories).
Results
Reanalysis of Existing Structures for FTO and PCIF1
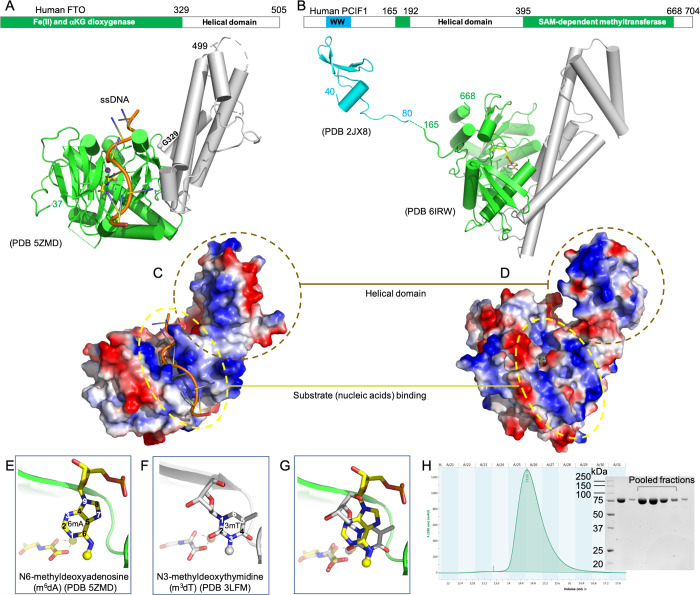
Human FTO and PCIF1 have both been structurally characterized.41,44,45 FTO contains an N-terminal double-strand β-helix fold, conserved among almost all Fe(II) and α-ketoglutarate (αKG) dependent dioxygenases,53 and a C-terminal helical domain44 (Figure 1A). PCIF1 contains a small N-terminal WW domain for binding the polymerase II C-terminal domain54 and a large C-terminal region consisting of a helical domain and MTase domain, the latter having a canonical Rossmann fold containing a conserved catalytic motif for binding of SAM45 (Figure 1B). Because the WW domain did not affect the in vitro MTase activity of PCIF1,45 the overall topological arrangement of FTO and PCIF1 are remarkably similar: a catalytic domain (with opposite activities) associated with a helical domain comprising six helices. Potential functions of the helical domains have been suggested, including protein stability and mediating interactions with the catalytic domain in FTO,44 or binding the RNA substrate in PCIF1.45 Both helical domains have additional basic- and acidic-rich surfaces (Figure 1C,D), which could mediate interactions with factors common to both proteins including chromatin remodeling and DNA repair55 (see Discussion).
Figure 1.
Structural comparison of FTO and PCIF1. (A) Schematic of human FTO with an N-terminal dioxygenase domain (green) coupled with a helical domain (gray cylinders). The bound ssDNA is illustrated with a ribbon model (orange), a metal ion as a small sphere, and αKG as a yellow stick. (B) Schematic of human PCIF1 with a smaller N-terminal WW domain (cyan) and a larger C-terminal MTase domain (green) coupled with a helical domain (gray cylinders). A bound SAH molecule is in stick (yellow). (C, D) Electrostatic surfaces of FTO (C) and PCIF1 (D) with blue for positive and red for negative charges. Two dashed circles indicate substrate (nucleic acids) binding sites and helical domains, respectively. (E–G) In the active site of FTO: m6dA of ssDNA (E), mononucleotide m3dT (F), and superimposition of m6dA and m3dT (G). (H) A profile of Superdex S200 10/300 chromatography and a 12% SDS-PAGE showing the pooled fractions of recombinant human PCIF1.
The active site of FTO could accommodate both m6dA of ssDNA and mononucleotide m3dT (which could occur in the context of DNA) (Figure 1E,F). The (removable) methyl groups of m6dA and m3dT occupy similar positions relative to the metal ion and αKG, implying that the same oxidative reaction would occur to both methyl groups upon the binding of dioxygen. However, the conformations of the deoxyribose differ markedly between the two deoxynucleotides (Figure 1G). Thus, the current structural information is insufficient to explain the preference of FTO for either mRNA caps or for RNA 2′O-methylation.
PCIF1 Activity on DNA Substrates
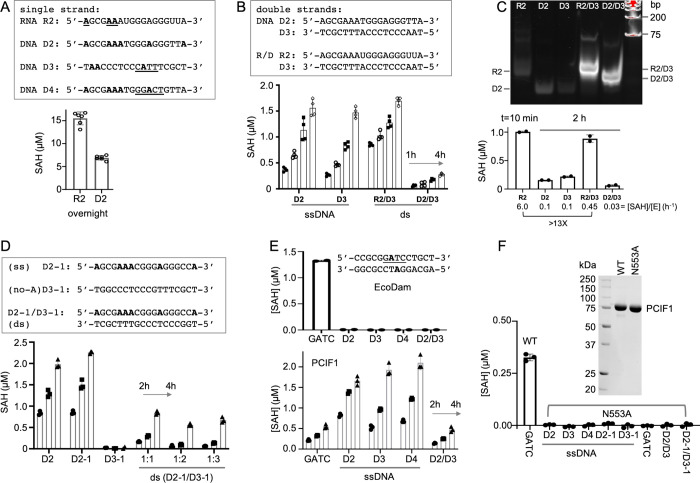
Inspired by the dual activity of FTO (on RNA and DNA), as well as by the overall similarity of FTO and PCIF1 architecture, we asked whether PCIF1 is also active on DNA. Using the recombinant human PCIF1 (Figure 1H), we first tested PCIF1 activity on an ssDNA sequence (oligo D2 in Figure 2A). The sequence of the D2 DNA oligo corresponds to that of the 18-nt uncapped RNA oligo R2 that contains a 5′-adenosine and additional internal Ade residues, which was used in our recent study of PCIF1 activity on uncapped RNAs.56 For the RNA R2 oligo, PCIF1 showed high activity, resulting in fully N6-methylated 5′ A, with a second methylation site at the internal adenosine, three or four residues away from the 5′ end (AGCGAA).56 In the case of the DNA D2 oligo, PCIF1 exhibited substantial activity, at about half the level, after a saturated overnight reaction, which was observed on the corresponding RNA (Figure 2A). The 5′ adenine is not required for PCIF1 activity on ssDNA, as the complimentary strand D3 yields similar activity to that of D2 (Figure 2B).
Figure 2.
PCIF1 activity on DNA. (A) Comparison of PCIF1 activity on oligo 2 of corresponding RNA and DNA sequences under the saturated overnight reactions ([E] = 5 μM, [S] = 10 μM and [SAM] = 100 μM) (N = 6). The underlined As in R2 oligo indicates the RNA methylation sites confirmed by mass spectrometry.56 The CATT site in D3 oligo is used in Figure 3, and the GGACT site in D4 oligo is used in Figure 4. (B) Comparison of PCIF1 activities on single-stranded oligos (D2 and D3) and double-stranded duplexes (R2/D3 and D2/D3) (N = 4). (C) A 15% native gel (top) showing substrates used in the assays (bottom) (N = 2). (D) PCIF1 activity on dsDNA (D2-1/D3-1) with an increased complementary strand (D3-1) that contains no As (N = 3). (E) EcoDam activity (top) and PCIF1 activity (bottom) on double-stranded GATC (N = 3). (F) The PCIF1 N553A mutant is inactive (N = 3). For panels (B–F), assay conditions were [E] = 1 μM, [S] = 10 μM, [SAM] = 40 μM, pH 8.0, T = 37 °C.
However, the double-stranded substrates, either dsDNA (D2/D3) or in a RNA/DNA hybrid (R2/D3), exhibit significantly reduced activity compared to the corresponding single stranded substrates (Figure 2B). We estimated that the rate of PCIF1 activity on R2/D3 is reduced by more than 13-fold comparing to that on R2 alone: the similar activities are reached by 2 h on R2/D3 and in merely 10 min on R2, under the same reaction conditions (Figure 2C). To ensure that the remaining activity is not due to unpaired R2 (in which PCIF1 has much higher activity), we added excess D3 to assure that R2 is fully annealed. Thus, the activity observed on R2/D3, which is in between R2 and D3 (R2 < R2/D3 < D3), is not from the single-stranded D3.
To further evaluate the weak activity of PCIF1 on dsDNA, we included three controls. First, we prepared dsDNA with the adenines contained only in one strand and paired it with no-A complementary strand excessively (Figure 2D). Second, because the E. coli strain used here expresses Dam MTase, which modifies GATC sites, we confirmed that there was no observable activity of Dam on non-GATC dsDNA (Figure 2E). Third, a catalytically dead mutant of PCIF1 (N553PPF to A553PPF) is inactive (Figure 2F).
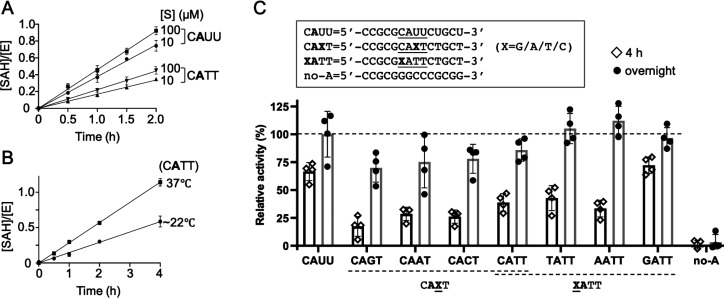
PCIF1 Is a Sequence-Independent MTase on Internal Adenine Nucleotides
Next, we designed a short 14-mer single-strand (ss)DNA oligo (CATT) containing a single internal A, and compared the PCIF1 enzymatic activity on this potential DNA substrate to that on the RNA equivalent (CAUU). First, we showed that PCIF1 activity is linear under the conditions used: like the RNA substrates (whether capped or uncapped),50,56 PCIF1 activity on DNA CATT is concentration- and temperature-dependent (Figure 3A,B). Second, we observed about half the PCIF1 activity on DNA that was observed on the corresponding RNA (up to 4 h), though eventually, the same level of methylation was reached in the saturated overnight reaction (Figure 3C). As a control, no activity was observed on a DNA oligo without any adenines (Figure 3C). Thus, PCIF1 has significant in vitro methylation activity on adenine in ssDNA.
Figure 3.
PCIF1 is a sequence-independent MTase on internal A. (A) PCIF1 activities on RNA (CAUU) and DNA (CATT) as a function of substrate concentration (N = 4). (B) PCIF1 activity on CATT as a function of temperature (N = 4). (C) PCIF1 activities on CAXT and XATT (X = G, A, T, C). ([E] = 1 μM, [S] = 10 μM and [SAM] = 40 μM) (N = 4).
Finally, we tested the effects of varying the nucleotides immediately preceding and following the target adenine. By varying the 5′ or 3′ nucleotides with all four possibilities (X = G, A, T, and C), we found that PCIF1 has roughly equal activity on all substitutions of CAXT or XATT) (Figure 3C). This observation agrees with FTO showing no specificity for the flanking sequence around the target methyladenine.41 We note that PCIF1 had slightly higher activity on GATT at an early time point (4 h), but methylation reached similar levels after saturated overnight reactions (Figure 3C).
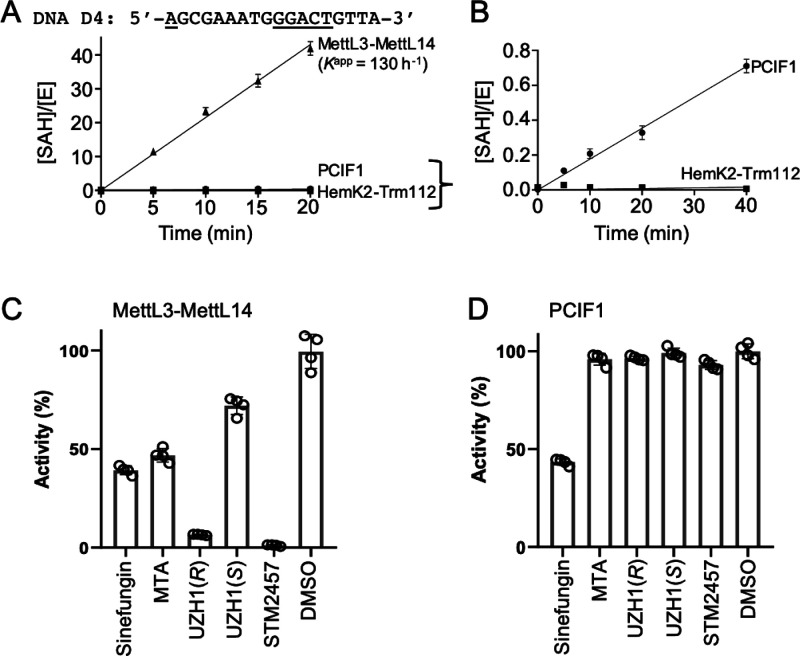
Comparison of PCIF1 and MettL3–14 Activities on DNA
To compare the PCIF1 DNA methylating activities with those of other MTases (HemK2-Trm112 and MettL3-MettL14 heterodimers), using the same DNA substrates, we prepared DNA oligo D4, containing GGACT, the consensus sequence for MettL3-MettL14. [At the time of this experiment, we did not have recombinant MettL4 for comparison.] First, we observed high activity of MettL3-MettL14 on the ssDNA (Kapp = 130 h–1; Figure 4A), which is the same as or even better than the corresponding RNA containing GGACU (Kapp = 120 h–1).56 Second, as expected, we observed no activity of HemK2-Trm112 on DNA (Figure 4B). Observations on both the positive control (MettL3-MettL14)25−27 and the negative control (HemK2-Trm1112)17−19 agree with previous publications. Third, we observed weak activity of PCIF1 on the DNA, which is marginal in comparison to MettL3-MettL14 on the same DNA molecule. In summary, MettL3-MettL14 has high DNA adenine methylation activity, at least in vitro. As a control, the DNA methylation activity of MettL3-MettL14 was inhibited by known selective MettL3–MettL14 inhibitors51,52 (Figure 4C), which had no effect on PCIF1 activity (Figure 4D).
Figure 4.
Comparison of PCIF1 and MettL3-MettL14 activities on the same DNA substrate. (A, B) Comparison of activities of three enzymes (MettL3-MettL14, PCIF1, and HemK2-Trm112). Panel (B) is an enlarged portion of panel A in order to visualize the low activity of PCIF1. (C) Inhibition of MettL3-MettL14. The compounds tested were known inhibitors of MettL3, and included sinefungin, 5′-deoxy-5′-methylthioadenosine (MTA), the two racemates UZH1(R) and UZH1(S),51 and STM2457.52 (D) The MettL3-selective inhibitors had no effect on PCIF1. Compounds were used at a concentration of [I] = 50 μM.
Methylation Activity Detection by Antibody
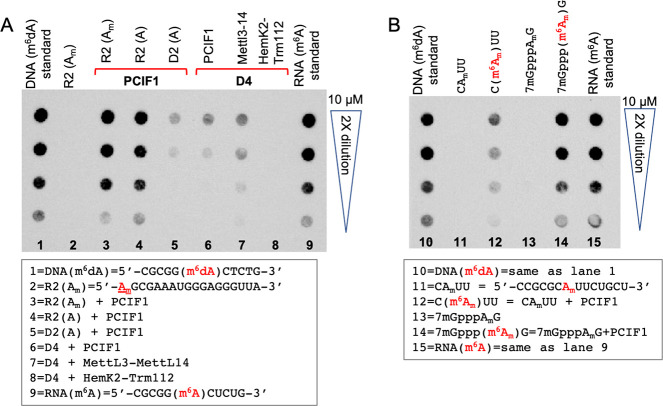
Anti-methyladenosine antibodies have been used to detect adenine modifications in mammalian DNA, and the results are controversial, partly because the antibodies cross-react with both m6dA and m6A.3 Here, we characterized one commercially available antibody (D9D9W from Cell Signaling Technology), using our in vitro enzyme reaction products (Figure 5A). First, the antibody detects but does not distinguish quantitatively an internal m6dA in synthesized DNA from m6A in RNA (lanes 1 and 9 of Figure 5A). However, it does not recognize 2′O-methylation in RNA, as 5′ Am (lane 2 of Figure 5A), at an internal position (lane 11 of Figure 5B), or in a capped structure (lane 13 of Figure 5B).
Figure 5.
Use of an anti-m6A antibody (D9D9W from Cell Signaling Technology) in dot blots. (A) The antibody recognizes both m6dA in DNA and m6A in RNA. (B) The antibody recognizes both m6A and m6Am in RNA modifications.
Second, PCIF1-catalyzed RNA modification is more intensive than its DNA modification (comparing lanes 3 and 4 to lane 5 in Figure 5A). Among the three enzymes tested for DNA methylation, MettL3-MettL14 has higher activity than PCIF1 (comparing lanes 6 and 7), while HemK2-Trm112 has no detectable activity (lane 8). Thus, the antibody-based dot blots agree with our enzymatic assays.
Third, we asked whether the antibody can distinguish monomethylated m6A and dimethylated m6Am, located either cap-proximally or internally. The m6Am can be cap-proximal (driven by PCIF1) or internal (in U2 snRNA added by MettL4). However, the transcriptome-wide distribution of m6Am and m6A sites do not overlap (initially observed in 2019 47 and reaffirmed in 202157), suggesting distinct functions between the two modifications. In any case, the tested antibody does not distinguish between these two RNA modifications. Shown in lanes 14 and 12 of Figure 5B, the antibody recognizes PCIF1-driven m6Am as the first nucleotide after the m7G cap as well as at an internal position.
Discussion
Here, we analyzed existing structures for human FTO and PCIF1, which illustrated a general arrangement of a well-characterized catalytic domain (methylase or dioxygenase) in association with a less-characterized helical domain. In sum, FTO and PCIF1 have opposite enzymatic activities on cap-specific m6Am,39,45 on internal adenosines (m6A or m6Am) in the context of RNA,38,40,56 and on DNA adenine (35,38,41 and this study). We note that some of the activities are observed in vitro.
In a recent comprehensive study of interactomes of FTO and PCIF1, in addition to their unique partners, both enzymes have protein-interaction networks that include overlapping DNA replication and DNA repair proteins.55 Among them are DNA helicase RECQ5 with roles in DNA repair and homologous recombination,58,59 a ssDNA-binding protein RADX that is recruited to sites of replication stress to promote genome stability,60,61 DNA double-strand break (DSB) repair enzyme XRCC4,62 the E3 ubiquitin-protein ligase RNF8 coordinating the repair of DNA lesions in specific chromatin topologies,63 the tyrosyl DNA phosphodiesterase 2 (TDP2) potentially involved in topoisomerase-mediated DNA damage,64 or PHRF1, a protein that contains a plant homeodomain (PHD) and a RING domain, possibly promoting genome integrity by modulating non-homologous end-joining.65 Covelo-Molares et al. noted that the identification of proteins involved in DNA replication and repair was the most striking result, with the most enriched and significant FTO interactor being the ssDNA-binding protein RADX.55
Indeed, we have recently proposed a role for DNA m6dA in DNA damage repair, which involves m6dA in ssDNA generated transiently, and in preventing or reducing incorporation of 8-oxo-2′-deoxyguanosine during the gap-filling DNA synthesis.66 We note that the initial observation in embryonic stem cells identified m6dA enrichment in H2A.X deposition regions,8 where H2A.X is a histone variant typically associated with DSBs. m6dA modification is also elevated in glioblastoma (a disease partially associated with the cumulative effects of high-dose exposure to ionizing radiation, or to chemical carcinogens), but not in normal adult tissues or mammalian cells.67 Recently, the same laboratory that made the initial observation has reported that m6dA increases during the development of mouse trophoblast stem cells (which eventually give rise to the placenta), specifically at regions of stress-induced DNA double helix destabilization (SIDD).32 ssDNA that persists in that state is one of the possible SIDD aftereffects.68
Although there is no direct evidence yet for PCIF1 involvement in DNA repair, there is growing evidence for a functional link between m6A-mediated activities of writers (MTases), readers (YTH domain proteins) and erasers (demethylases), and DNA damage repair after UV irradiation.69−72 The biochemically characterized candidates for the m6dA writer (MettL3-MettL14), reader (YTHDC1), and eraser (ALKBH1) all prefer locally unpaired DNA substrates in vitro.25,33,73 Of these three proteins, the activities of YTHDC1 and ALKBH1 are independent of sequence context aside from the methylated adenine itself. In contrast, MettL3-MettL14 is an adenine methylase complex, long studied for its activity on RNA, where it targets the sequence RRACH (R is purine and H is not G).74 Thus, while PCIF1 is the mRNA cap-proximal adenine methylase, this enzyme might also be a versatile methyltransferase for modifying internal adenine in RNA and DNA.
Acknowledgments
We thank Ms. Yu Cao for technical assistance, Dr. Clayton B. Woodcock for purified HemK2-Trm112, Dr. Taraneh Hajian and Dr. Masoud Vedadi for purified MettL3-MettL14, and Dr. John R. Horton for purified EcoDam.
Glossary
Abbreviations
- m6dA
N6-methyl deoxyadenosine in DNA
- m6A
N6-methyladenosine in RNA
- m6Am
N6, 2′O-dimethyladenosine in RNA
- Am
2′O-methyladenosine in RNA
Accession Codes
PCIF1 (NCBI: NP_071387.1; PDB: 2JX8 and 6IRW); MettL3 (NCBI: NP_062826.2); MettL14 (NCBI: NP_066012.1); HemK2 (NCBI: NP_037372.4); Trm112 (NCBI: NP_057488.1); FTO (PDB: 5ZMD and 3LFM).
Author Contributions
∥ D.Y. and J.Z. contribute equally to this work.
Author Contributions
D.Y. and J.Z. performed protein purification and enzymatic assays. D.Y. and Q.C. performed the antibody dot blot assay. T.W. participated in discussion and provided the antibody. R.M.B. participated in discussions and assisted in preparing the manuscript. X.Z and X.C organized and designed the scope of the study.
U.S. National Institutes of Health (NIH) grant [R35GM134744]; Cancer Prevention and Research Institute of Texas (CPRIT) [RR160029]. X.C. is a CPRIT Scholar in Cancer Research. Funding for open access charge: MD Anderson Cancer Center.
The authors declare no competing financial interest.
References
- Achwal C. W.; Iyer C. A.; Chandra H. S. Immunochemical evidence for the presence of 5mC, 6mA and 7mG in human, Drosophila and mealybug DNA. FEBS Lett. 1983, 158, 353–358. 10.1016/0014-5793(83)80612-7. [DOI] [PubMed] [Google Scholar]
- Schiffers S.; Ebert C.; Rahimoff R.; Kosmatchev O.; Steinbacher J.; Bohne A. V.; Spada F.; Michalakis S.; Nickelsen J.; Müller M.; Carell T. Quantitative LC-MS Provides No Evidence for m(6) dA or m(4) dC in the Genome of Mouse Embryonic Stem Cells and Tissues. Angew. Chem., Int. Ed. 2017, 56, 11268–11271. 10.1002/anie.201700424. [DOI] [PubMed] [Google Scholar]
- Douvlataniotis K.; Bensberg M.; Lentini A.; Gylemo B.; Nestor C. E. No evidence for DNA N (6)-methyladenine in mammals. Sci Adv 2020, 6, eaay3335 10.1126/sciadv.aay3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musheev M. U.; Baumgärtner A.; Krebs L.; Niehrs C. The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 2020, 16, 630–634. 10.1038/s41589-020-0504-2. [DOI] [PubMed] [Google Scholar]
- Bochtler M.; Fernandes H. DNA adenine methylation in eukaryotes: Enzymatic mark or a form of DNA damage?. BioEssays 2021, 43, e2000243 10.1002/bies.202000243. [DOI] [PubMed] [Google Scholar]
- Kong Y.; Cao L.; Deikus G.; Fan Y.; Mead E. A.; Lai W.; Zhang Y.; Yong R.; Sebra R.; Wang H.; Zhang X. S.; Fang G. Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution. Science 2022, 375, 515–522. 10.1126/science.abe7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K.; Greer E. L. The adenine methylation debate. Science 2022, 375, 494–495. 10.1126/science.abn6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. P.; Wang T.; Seetin M. G.; Lai Y.; Zhu S.; Lin K.; Liu Y.; Byrum S. D.; Mackintosh S. G.; Zhong M.; Tackett A.; Wang G.; Hon L. S.; Fang G.; Swenberg J. A.; Xiao A. Z. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333. 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P., Grosjean H. (2009) The Interplay between RNA and DNA modifications, InDNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution (edited byH., Grosjean)Landes Bioscience., 259–274. [Google Scholar]
- Fedeles B. I.; Singh V.; Delaney J. C.; Li D.; Essigmann J. M. The AlkB Family of Fe(II)/alpha-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J Biol Chem 2015, 290, 20734–20742. 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. D.; Li S.; Goedderz C.; Chen X. S. The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity. NAR Cancer 2020, 2, zcaa027. 10.1093/narcan/zcaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M.; An J.; Bandukwala H. S.; Chavez L.; Äijö T.; Pastor W. A.; Segal M. F.; Li H.; Koh K. P.; Lähdesmäki H.; Hogan P. G.; Aravind L.; Rao A. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 2013, 497, 122–126. 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D.; Bi X.; Pardavila J. A.; Huang X.; Saenz C.; Shi X.; Zhou H.; Faiola F.; Ding J.; Haruehanroengra P.; Yang F.; Li D.; Sanchez-Priego C.; Saunders A.; Pan F.; Valdes V. J.; Kelley K.; Blanco M. G.; Chen L.; Wang H.; Sheng J.; Xu M.; Fidalgo M.; Shen X.; Wang J. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018, 50, 443–451. 10.1038/s41588-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.; Zhang Q.; Shi Y.; Shi Q.; Jiang Y.; Gu Y.; Li Z.; Li X.; Zhao K.; Wang C.; Li N.; Cao X. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 2018, 554, 123–127. 10.1038/nature25434. [DOI] [PubMed] [Google Scholar]
- Xiao C. L.; Zhu S.; He M.; Chen; Zhang Q.; Chen Y.; Yu G.; Liu J.; Xie S. Q.; Luo F.; Liang Z.; Wang D. P.; Bo X. C.; Gu X. F.; Wang K.; Yan G. R. N(6)-Methyladenine DNA Modification in the Human Genome. Mol. Cell 2018, 71, 306. 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- Liu P.; Nie S.; Li B.; Yang Z. Q.; Xu Z. M.; Fei J.; Lin C.; Zeng R.; Xu G. L. Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality. Mol. Cell. Biol. 2010, 30, 4245–4253. 10.1128/MCB.00218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E.; Wang S.; Urban S.; Willmann D.; Schmidt A.; Offermann A.; Allen A.; Sum M.; Obier N.; Cottard F.; Ulferts S.; Preca B. T.; Hermann B.; Maurer J.; Greschik H.; Hornung V.; Einsle O.; Perner S.; Imhof A.; Jung M.; Schüle R. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat. Struct. Mol. Biol. 2019, 26, 361–371. 10.1038/s41594-019-0219-9. [DOI] [PubMed] [Google Scholar]
- Woodcock C. B.; Yu D.; Zhang X.; Cheng X. Human HemK2/KMT9/N6AMT1 is an active protein methyltransferase, but does not act on DNA in vitro, in the presence of Trm112. Cell Discovery 2019, 5, 50. 10.1038/s41421-019-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Shi Y.; Zhang T.; Ye J.; Ding J. Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase. Cell Discovery 2019, 5, 51. 10.1038/s41421-019-0121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon S. M.; Chen Y.; Moon E.; Kvederaviciutė K.; Klimasauskas S.; Feldman D. E. An Adversarial DNA N(6)-Methyladenine-Sensor Network Preserves Polycomb Silencing. Mol. Cell 2019, 74, 1138–1147.e6. 10.1016/j.molcel.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z.; Wu T.; Cui X.; Zhu P.; Tan C.; Dou X.; Hsu K. W.; Lin Y. T.; Peng P. H.; Zhang L. S.; Gao Y.; Hu L.; Sun H. L.; Zhu A.; Liu J.; Wu K. J.; He C. N(6)-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol. Cell 2020, 78, 382–395. 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Gu L.; Orellana E. A.; Wang Y.; Guo J.; Liu Q.; Wang L.; Shen Z.; Wu H.; Gregory R. I.; Xing Y.; Shi Y. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020, 30, 544–547. 10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh Y. T.; Koh C. W. Q.; Sim D. Y.; Roca X.; Goh W. S. S. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 2020, 48, 9250–9261. 10.1093/nar/gkaa684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Dou X.; Chen C.; Chen C.; Liu C.; Xu M. M.; Zhao S.; Shen B.; Gao Y.; Han D.; He C. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 2020, 367, 580–586. 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. B.; Yu D.; Hajian T.; Li J.; Huang Y.; Dai N.; Corrêa I. R. Jr.; Wu T.; Vedadi M.; Zhang X.; Cheng X. Human MettL3-MettL14 complex is a sequence-specific DNA adenine methyltransferase active on single-strand and unpaired DNA in vitro. Cell Discovery 2019, 5, 63. 10.1038/s41421-019-0136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.; Horton J. R.; Yang J.; Hajian T.; Vedadi M.; Sagum C. A.; Bedford M. T.; Blumenthal R. M.; Zhang X.; Cheng X. Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic Acids Res. 2021, 49, 11629–11642. 10.1093/nar/gkab460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S.; Mota J.; Chan S. H.; Villarreal J.; Dai N.; Arya S.; Hromas R. A.; Rao M. K.; Corrêa I. R. Jr.; Gupta Y. K. RNA binding to human METTL3-METTL14 restricts N(6)-deoxyadenosine methylation of DNA in vitro. eLife 2022, 11, e67150 10.7554/eLife.67150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan T.; Trewick S. C.; Koivisto P.; Bates P. A.; Lindahl T.; Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 16660–16665. 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes P. Ø.; Johansen R. F.; Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 2002, 419, 178–182. 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- Trewick S. C.; Henshaw T. F.; Hausinger R. P.; Lindahl T.; Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 2002, 419, 174–178. 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- Aas P. A.; Otterlei M.; Falnes P. Ø.; Vagbo C. B.; Skorpen F.; Akbari M.; Sundheim O.; Bjørås M.; Slupphaug G.; Seeberg E.; Krokan H. E. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 2003, 421, 859–863. 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- Li Z.; Zhao S.; Nelakanti R. V.; Lin K.; Wu T. P.; Alderman M. H.; Guo C.; Wang P.; Zhang M.; Min W.; Jiang Z.; Wang Y.; Li H.; Xiao A. Z. N6-methyladenine in DNA antagonizes SATB1 in early development. Nature 2020, 583, 625–630. 10.1038/s41586-020-2500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Yang S.; Nelakanti R.; Zhao W.; Liu G.; Li Z.; Liu X.; Wu T.; Xiao A.; Li H. Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res 2020, 30, 197–210. 10.1038/s41422-019-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L. F.; Liu Y. P.; Chen L.; Tang Q.; Wu W.; Sun W.; Chen Z.; Yan X. X. Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res 2020, 30, 272–275. 10.1038/s41422-019-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S.; Toh J. D.; Wong K. H.; Gao Y. G.; Hong W.; Woon E. C. N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci. Rep. 2016, 6, 25677. 10.1038/srep25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T.; Girard C. A.; Tung Y. C.; Webby C. J.; Saudek V.; Hewitson K. S.; Yeo G. S.; McDonough M. A.; Cunliffe S.; McNeill L. A.; Galvanovskis J.; Rorsman P.; Robins P.; Prieur X.; Coll A. P.; Ma M.; Jovanovic Z.; Farooqi I. S.; Sedgwick B.; Barroso I.; Lindahl T.; Ponting C. P.; Ashcroft F. M.; O’Rahilly S.; Schofield C. J. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007, 318, 1469–1472. 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Yang C. G.; Yang S.; Jian X.; Yi C.; Zhou Z.; He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008, 582, 3313–3319. 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Fu Y.; Zhao X.; Dai Q.; Zheng G.; Yang Y.; Yi C.; Lindahl T.; Pan T.; Yang Y. G.; He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J.; Luo X.; Blanjoie A.; Jiao X.; Grozhik A. V.; Patil D. P.; Linder B.; Pickering B. F.; Vasseur J. J.; Chen Q.; Gross S. S.; Elemento O.; Debart F.; Kiledjian M.; Jaffrey S. R. Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.; Liu F.; Lu Z.; Fei Q.; Ai Y.; He P. C.; Shi H.; Cui X.; Su R.; Klungland A.; Jia G.; Chen J.; He C. Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985. 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wei L. H.; Wang Y.; Xiao Y.; Liu J.; Zhang W.; Yan N.; Amu G.; Tang X.; Zhang L.; Jia G. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 2919–2924. 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Yang Y.; Sun B. F.; Shi Y.; Yang X.; Xiao W.; Hao Y. J.; Ping X. L.; Chen Y. S.; Wang W. J.; Jin K. X.; Wang X.; Huang C. M.; Fu Y.; Ge X. M.; Song S. H.; Jeong H. S.; Yanagisawa H.; Niu Y.; Jia G. F.; Wu W.; Tong W. M.; Okamoto A.; He C.; Rendtlew Danielsen J. M.; Wang X. J.; Yang Y. G. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relier S.; Ripoll J.; Guillorit H.; Amalric A.; Achour C.; Boissiere F.; Vialaret J.; Attina A.; Debart F.; Choquet A.; Macari F.; Marchand V.; Motorin Y.; Samalin E.; Vasseur J. J.; Pannequin J.; Aguilo F.; Lopez-Crapez E.; Hirtz C.; Rivals E.; Bastide A.; David A. FTO-mediated cytoplasmic m(6)Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat. Commun. 2021, 12, 1716. 10.1038/s41467-021-21758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.; Niu T.; Chang J.; Lei X.; Zhao M.; Wang Q.; Cheng W.; Wang J.; Feng Y.; Chai J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 2010, 464, 1205–1209. 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- Akichika S.; Hirano S.; Shichino Y.; Suzuki T.; Nishimasu H.; Ishitani R.; Sugita A.; Hirose Y.; Iwasaki S.; Nureki O.; Suzuki T.. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase, Science 363, eaav0080. 2019. [DOI] [PubMed]
- Boulias K.; Toczydlowska-Socha D.; Hawley B. R.; Liberman N.; Takashima K.; Zaccara S.; Guez T.; Vasseur J. J.; Debart F.; Aravind L.; Jaffrey S. R.; Greer E. L. Identification of the m(6)Am Methyltransferase PCIF1 Reveals the Location and Functions of m(6)Am in the Transcriptome. Mol. Cell 2019, 75, 631–643. 10.1016/j.molcel.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendinc E.; Valle-Garcia D.; Dhall A.; Chen H.; Henriques T.; Navarrete-Perea J.; Sheng W.; Gygi S. P.; Adelman K.; Shi Y. PCIF1 Catalyzes m6Am mRNA Methylation to Regulate Gene Expression. Mol. Cell 2019, 75, e629 10.1016/j.molcel.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R. R.; Delfino E.; Homolka D.; Roithova A.; Chen K. M.; Li L.; Franco G.; Vagbo C. B.; Taillebourg E.; Fauvarque M. O.; Pillai R. S. The Mammalian Cap-Specific m(6)Am RNA Methyltransferase PCIF1 Regulates Transcript Levels in Mouse Tissues. Cell Rep. 2020, 32, 108038 10.1016/j.celrep.2020.108038. [DOI] [PubMed] [Google Scholar]
- Sun H.; Zhang M.; Li K.; Bai D.; Yi C. Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res 2019, 29, 80–82. 10.1038/s41422-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.; Kaur G.; Blumenthal R. M.; Zhang X.; Cheng X. Enzymatic characterization of three human RNA adenosine methyltransferases reveals diverse substrate affinities and reaction optima. J. Biol. Chem. 2021, 296, 100270 10.1016/j.jbc.2021.100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz-Omori E. V.; Huang D.; Kumar Bedi R.; Cheriyamkunnel S. J.; Bochenkova E.; Dolbois A.; Rzeczkowski M. D.; Li Y.; Wiedmer L.; Caflisch A. METTL3 inhibitors for epitranscriptomic modulation of cellular processes. ChemMedChem 2021, 16, 3035–3043. 10.1002/cmdc.202100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankova E.; Blackaby W.; Albertella M.; Rak J.; De Braekeleer E.; Tsagkogeorga G.; Pilka E. S.; Aspris D.; Leggate D.; Hendrick A. G.; Webster N. A.; Andrews B.; Fosbeary R.; Guest P.; Irigoyen N.; Eleftheriou M.; Gozdecka M.; Dias J. M. L.; Bannister A. J.; Vick B.; Jeremias I.; Vassiliou G. S.; Rausch O.; Tzelepis K.; Kouzarides T. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. S.; Leissing T. M.; Chowdhury R.; Hopkinson R. J.; Schofield C. J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. 10.1146/annurev-biochem-061516-044724. [DOI] [PubMed] [Google Scholar]
- Fan H.; Sakuraba K.; Komuro A.; Kato S.; Harada F.; Hirose Y. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophys. Res. Commun. 2003, 301, 378–385. 10.1016/S0006-291X(02)03015-2. [DOI] [PubMed] [Google Scholar]
- Covelo-Molares H.; Obrdlik A.; Poštulková I.; Dohnálková M.; Gregorová P.; Ganji R.; Potěšil D.; Gawriyski L.; Varjosalo M.; Vaňáčová Š. The comprehensive interactomes of human adenosine RNA methyltransferases and demethylases reveal distinct functional and regulatory features. Nucleic Acids Res. 2021, 49, 10895–10910. 10.1093/nar/gkab900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.; Dai N.; Wolf E. J.; Correa I. R. Jr.; Zhou J.; Wu T.; Blumenthal R. M.; Zhang X.; Cheng X. Enzymatic characterization of mRNA cap adenosine-N6 methyltransferase PCIF1 activity on uncapped RNAs. J. Biol. Chem. 2022, 298, 101751 10.1016/j.jbc.2022.101751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.; Li K.; Zhang X.; Liu J.; Zhang M.; Meng H.; Yi C. m(6)Am-seq reveals the dynamic m(6)Am methylation in the human transcriptome. Nat Commun 2021, 12, 4778. 10.1038/s41467-021-25105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson H. C.; Davis L.; Kiianitsa K.; Khoo K. J.; Liu Y.; Knijnenburg T. A.; Maizels N. Increased levels of RECQ5 shift DNA repair from canonical to alternative pathways. Nucleic Acids Res. 2018, 46, 9496–9509. 10.1093/nar/gky727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbakry A.; Juhász S.; Chan K. C.; Löbrich M. ATRX and RECQ5 define distinct homologous recombination subpathways. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2010370118 10.1073/pnas.2010370118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert L.; Ho T.; Hoffmann S.; Haahr P.; Guérillon C.; Mailand N. RADX interacts with single-stranded DNA to promote replication fork stability. EMBO Rep. 2017, 18, 1991–2003. 10.15252/embr.201744877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H.; Bhat K. P.; Le Meur R.; Chazin W. J.; Ding X.; Sharan S. K.; Wessel S. R.; Sathe A. A.; Zhao R.; Cortez D. RADX Promotes Genome Stability and Modulates Chemosensitivity by Regulating RAD51 at Replication Forks. Mol. Cell 2017, 67, e375 10.1016/j.molcel.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Alt F. W. Identification of the XRCC4 gene: complementation of the DSBR and V(D)J recombination defects of XR-1 cells. Curr. Top Microbiol. Immunol. 1996, 217, 143–150. 10.1007/978-3-642-50140-1_10. [DOI] [PubMed] [Google Scholar]
- Luijsterburg M. S.; van Attikum H. Close encounters of the RNF8th kind: when chromatin meets DNA repair. Curr. Opin. Cell Biol. 2012, 24, 439–447. 10.1016/j.ceb.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Crewe M.; Madabhushi R. Topoisomerase-Mediated DNA Damage in Neurological Disorders. Front. Aging Neurosci. 2021, 13, 751742 10.3389/fnagi.2021.751742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. F.; Chu P. C.; Wu P. Y.; Yu M. Y.; Lee J. Y.; Tsai M. D.; Chang M. S. PHRF1 promotes genome integrity by modulating non-homologous end-joining. Cell Death Dis. 2015, 6, e1716 10.1038/cddis.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Blumenthal R. M.; Cheng X. A Role for N6-Methyladenine in DNA Damage Repair. Trends Biochem. Sci. 2021, 46, 175–183. 10.1016/j.tibs.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q.; Wu T. P.; Gimple R. C.; Li Z.; Prager B. C.; Wu Q.; Yu Y.; Wang P.; Wang Y.; Gorkin D. U.; Zhang C.; Dowiak A. V.; Lin K.; Zeng C.; Sui Y.; Kim L. J. Y.; Miller T. E.; Jiang L.; Lee C. H.; Huang Z.; Fang X.; Zhai K.; Mack S. C.; Sander M.; Bao S.; Kerstetter-Fogle A. E.; Sloan A. E.; Xiao A. Z.; Rich J. N. N(6)-methyladenine DNA Modification in Glioblastoma. Cell 2018, 175, 1228–1243. 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F.; Wojtowicz D.; Baranello L.; Yamane A.; Nelson S.; Resch W.; Kieffer-Kwon K. R.; Benham C. J.; Casellas R.; Przytycka T. M.; Levens D. Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome. Cell Syst. 2017, 4, 344–356. 10.1016/j.cels.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.; Laurent B.; Hsu C. H.; Nachtergaele S.; Lu Z.; Sheng W.; Xu C.; Chen H.; Ouyang J.; Wang S.; Ling D.; Hsu P. H.; Zou L.; Jambhekar A.; He C.; Shi Y. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Chen L.; Peng D.; Jiang A.; He Y.; Zeng Y.; Xie C.; Zhou H.; Luo X.; Liu H.; Chen L.; Ren J.; Wang W.; Zhao Y. METTL3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of DSBs by Modulating DNA-RNA Hybrid Accumulation. Mol. Cell 2020, 79, 425–442.e7. 10.1016/j.molcel.2020.06.017. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Yang S.; Cui Y. H.; Wei J.; Shah P.; Park G.; Cui X.; He C.; He Y. Y. METTL14 facilitates global genome repair and suppresses skin tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2025948118 10.1073/pnas.2025948118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abakir A.; Giles T. C.; Cristini A.; Foster J. M.; Dai N.; Starczak M.; Rubio-Roldan A.; Li M.; Eleftheriou M.; Crutchley J.; Flatt L.; Young L.; Gaffney D. J.; Denning C.; Dalhus B.; Emes R. D.; Gackowski D.; Correa I. R. Jr.; Garcia-Perez J. L.; Klungland A.; Gromak N.; Ruzov A. N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat. Genet. 2020, 52, 48–55. 10.1038/s41588-019-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. B.; Horton J. R.; Zhou J.; Bedford M. T.; Blumenthal R. M.; Zhang X.; Cheng X. Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA. Nucleic Acids Res. 2020, 48, 10329–10341. 10.1093/nar/gkaa604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U.; Kelley D. E.; Perry R. P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 1977, 115, 695–714. 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]