Abstract
The objective of this study was to determine whether chitosan (poly-β-1,4-glucosamine) and hydrolysates of chitosan can be used as novel preservatives in foods. Chitosan was hydrolyzed by using oxidative-reductive degradation, crude papaya latex, and lysozyme. Mild hydrolysis of chitosan resulted in improved microbial inactivation in saline and greater inhibition of growth of several spoilage yeasts in laboratory media, but highly degraded products of chitosan exhibited no antimicrobial activity. In pasteurized apple-elderflower juice stored at 7°C, addition of 0.3 g of chitosan per liter eliminated yeasts entirely for the duration of the experiment (13 days), while the total counts and the lactic acid bacterial counts increased at a slower rate than they increased in the control. Addition of 0.3 or 1.0 g of chitosan per kg had no effect on the microbial flora of houmous, a chickpea dip; in the presence of 5.0 g of chitosan per kg, bacterial growth but not yeast growth was substantially reduced compared with growth in control dip stored at 7°C for 6 days. Improved antimicrobial potency of chitosan hydrolysates like that observed in the saline and laboratory medium experiments was not observed in juice and dip experiments. We concluded that native chitosan has potential for use as a preservative in certain types of food but that the increase in antimicrobial activity that occurs following partial hydrolysis is too small to justify the extra processing involved.
Chitosan (poly-β-1,4-glucosamine) is prepared commercially by alkaline deacetylation of chitin obtained from the exoskeletons of marine crustaceans (6, 17). Chitosan has a pKa value of approximately 6.3 (18); at lower pH values, the molecule is cationic due to protonation of the amino groups. Previous reports have indicated that when chitosan is dissolved in saline, distilled water, or laboratory media, it exhibits antimicrobial activity against some strains of filamentous fungi (2, 9, 15), yeasts (15, 18), and bacteria (21). Considerable variation in the MICs and/or minimum bactericidal concentrations of chitosan, both between and within genera, has been described. Nevertheless, on the basis of the available evidence, bacteria appear to be generally less sensitive to the antimicrobial action of chitosan than fungi are. The antifungal activity of chitosan is greater at lower pH values (15).
Chitosan has a mean molecular mass of up to 1 MDa, which corresponds to a chain length of approximately 5,000 U, but there is considerable variation between commercial batches. There is limited in vitro evidence which suggests that partial depolymerization may enhance the antimicrobial activity of chitosan against phytopathogenic fungi and some bacteria of medical significance. For example, Uchida et al. (20) produced chitosan hydrolysates with total reducing sugar (TRS) contents of 50, 508, and 590 mg/g by using Bacillus chitosanase. The antifungal activities of the hydrolysates were tested on solid laboratory media at pH 6 by using three Fusarium spp. isolates. The MIC (around 0.4 g/liter) of the mildly hydrolyzed chitosan (TRS content, 50 mg/g) was approximately one-half the MIC of the native chitosan. The most highly degraded hydrolysate (TRS content, 590 mg/g) was not effective at a concentration of 10 g/liter, the highest concentration tested. The same chitosan preparations were also tested by using four bacteria in liquid laboratory media. The native chitosan and the mildly hydrolyzed chitosan (TRS content, 50 mg/g) were equally inhibitory for Escherichia coli, but the hydrolysate was more active against Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus than the native chitosan.
In another study, Kendra and Hadwiger (9) determined the extent to which the degree of polymerization of chitosan could be reduced before antifungal activity was adversely affected. Chitosan oligomers were prepared by digesting chitosan with 6 N HCl and were separated on a Fractogel size exclusion column. The oligomers were tested at a range of concentrations in liquid medium by using two strains of the plant pathogen Fusarium solani. The shortest oligomer which exhibited the maximum antifungal activity was the heptamer. Antimicrobial activity then decreased with chain length, and the dimer and trimer of N-acetylglucosamine were inactive. The results of Hirano and Nagao (7) also suggested that low-molecular-weight chitosan in an agar system inhibited a range of phytopathogenic fungi more effectively than high-molecular-weight chitosan inhibited the organisms, although low-molecular-weight chitosan was not defined.
Most reports on the antimicrobial effects of hydrolyzed chitosan have focused on plant-pathogenic fungi and a few bacteria that have medical significance. The effectiveness of chitosan hydrolysates against food-associated microorganisms in vitro or in foods has not been reported previously.
Several enzymic and chemical methods for producing chitosan oligomers have been described. The chemical methods include acid hydrolysis with either cold nitrous acid (16) or hot hydrochloric acid (9); both of these methods involve rather long and harsh treatments. Nordtveit et al. (11) used hydrogen peroxide, which, in the presence of Fe(III), generates hydroxyl radicals which cleave the molecule by nucleophilic attack. Enzymes from a variety of sources have also been used for chitosan degradation. Lysozyme from hens' eggs has been investigated and has been shown to be most efficacious when the chitosan is only partially deacetylated (11, 12). Chitosan-degrading enzymes have been isolated from diverse bacteria, including Streptomyces griseus, Bacillus circulans, and members of the actinomycetes (4, 13, 22). Chitosan-degrading enzymes can also be isolated from vegetable sources. Latex sap from Carica papaya contains lysozyme (10), as well as chitinase enzymes (3). Papaya latex itself has been reported to exhibit antifungal activity against Candida albicans (5).
We found that few studies of chitosan and chitosan hydrolysates have been concerned with inhibition of food-borne microorganisms in real food systems. The objective of this study was to investigate the antimicrobial activity of degraded and native chitosans against spoilage microorganisms in foods and beverages, as well as in laboratory systems, in order to assess their potential for use as novel food preservatives.
MATERIALS AND METHODS
Materials.
Chitosan hydrochloride was obtained from Pronova Biopolymer (Drammen, Norway). The chloride content was 15.6%, and the degree of acetylation was 0.11 (data provided by the manufacturer). All microbiological media and diluents (maximum recovery diluent) were obtained from Oxoid Ltd. (Basingstoke, United Kingdom), and chemicals were obtained from Sigma Chemical Company Ltd. unless otherwise indicated. Pasteurized apple-elderflower juice and houmous (a dip containing pureed chickpeas, vegetable oil [fat content, 25.9%], sesame seed pulp, lemon juice, water, salt, and garlic) were purchased from the chill cabinets of a local retailer.
Microorganisms and cultivation.
The microorganisms used in this study included Saccharomyces cerevisiae 3085 from spoiled fruit juice and Zygosaccharomyces bailii 906 from a spoiled carbonated beverage; both of these strains were gifts from Pepsico International (Valhalla, N.Y.). Saccharomycodes ludwigii, an isolate from spoiled cider, was a gift from Ron Board (University of Bath, Bath, United Kingdom). Pseudomonas fragi, an isolate from spoiled meat, was obtained from the Leatherhead Food Research Association, Leatherhead, United Kingdom. Cryptococcus albidus and Bacillus sp. were isolated in our laboratory from a tomato-based dip, Candida sp. was isolated from houmous, and Rhodotorula sp. was isolated from a mayonnaise-based dip; our own isolates were identified by using API biochemical profile strips (Biomerieux, Marcy l'Etoile, France). The bacteria were cultured on plate count agar (PCA) and in nutrient broth, while the yeasts were cultured on malt extract agar and in malt extract broth supplemented with 2.5 g of glucose per liter (MEBG).
Degradation of chitosan.
The following three methods were used to degrade chitosan: oxidative-reductive degradation with hydrogen peroxide and iron(III), treatment with crude papaya latex, and hydrolysis with lysozyme.
In the oxidative-reductive method (11), 5.0 g of chitosan hydrochloride per liter was dissolved in 1.0% (vol/vol) acetic acid. Aliquots (8.5 ml) of the chitosan solution were mixed with 1.0 ml of 10 mM aqueous FeCl3. Hydrogen peroxide (1 M) was added to final concentrations of 0.0, 0.05, 0.2, 1.0, 5.0, 10.0, 25.0, and 50.0 mM, and the volume of each reaction mixture was increased to 10.0 ml by using distilled water. After 18 h of incubation at the ambient temperature, the viscosity of each mixture was measured by using a U-tube viscometer at 25°C. Samples were drawn up above the upper etched line of the glass tube of the viscometer and were allowed to fall due to gravity. The interval between the time that the meniscus of a sample crossed the upper etched line and the time that the meniscus crossed the lower etched line was measured in seconds. Distilled water was used as a blank, and its viscosity was subtracted from all experimental measurements. Degraded chitosan solutions were stored at 4°C for no more than 24 h before antimicrobial activity was tested.
Freeze-dried crude papaya latex powder (catalog no. P3250; Sigma Chemical Company Ltd.) was dissolved in distilled water at a concentration of 2.0 g/liter and was filtered through Whatman no. 1 paper. A 5.0-g/liter solution of chitosan hydrochloride was prepared in 10 mM acetic acid–sodium acetate buffer (pH 4.5). The papaya latex and chitosan solutions were mixed at a 1:4 ratio and then allowed to stand at room temperature for 20 h. Our initial investigations showed that the reduction in viscosity followed first-order kinetics; rapid hydrolysis occurred within the first 10 min, and this was followed by a slower but steady reaction for up to 20 h. The degraded chitosan solution was autoclaved and stored at 4°C for no more than 24 h before antimicrobial activity was tested.
Lysozyme from the albumen of hens' eggs (catalog no. L6876; Sigma Chemical Company Ltd.), which had an activity of 50,000 U/mg of protein (1 U was defined as the amount of enzyme that produced a change in absorbance at 450 nm of 0.001 U per min in 2.6 ml of a suspension of Micrococcus luteus at pH 6.24 and 25°C in a cuvette with a 1-cm light path), was dissolved in 0.1 M potassium chloride and added to a solution containing 2.5 g of chitosan hydrochloride per liter in 10 mM acetic acid–sodium acetate buffer (pH 4.5) so that the final enzyme concentration was 0.025% (wt/vol). The reaction mixture was stirred at room temperature, and samples were taken periodically for up to 10 min in order to determine the viscosity with a U-tube viscometer as described above. Samples were boiled for 30 min to inactivate the enzyme and were stored at 4°C for no more than 24 h before antimicrobial activity was tested.
Evaluation of antimicrobial activity in vitro.
Experiments to determine inactivation in saline solutions (9.0 g of NaCl per liter in distilled water, pH 6.4) were carried out by using Candida sp., Rhodotorula sp., C. albidus, P. fragi, and Bacillus sp. Native chitosan and chitosan hydrolyzed by papaya latex were used for these inactivation experiments. Cultures were grown to the stationary phase (for up to 3 days, depending on the growth rate of the organism) in MEBG at 25°C (yeasts) or in nutrient broth at 30°C (bacteria). The cells were washed three times by using centrifugation and a sterile saline solution to remove the residual culture medium. Washed cells were added to sterile saline solutions at 25°C, and the viable counts were determined before the antimicrobial agent was added. Yeasts were enumerated on malt extract agar plates that were incubated at 25°C for 5 days, and bacteria were enumerated on PCA plates that were incubated at 30°C for 3 days. Native and degraded chitosan solutions that had been autoclaved at 121°C for 15 min were added to the cell suspensions at final concentrations of 0.5 g/liter for yeasts and 1.0 g/liter for bacteria. Samples were removed periodically for up to 1 h, and viable counts were determined as described above. When we calculated viable count values, the initial counts were adjusted to compensate for the volume changes resulting from addition of the chitosan solutions.
Inhibition of growth of microorganisms by chitosan and chitosan hydrolysates produced by using hydrogen peroxide or lysozyme was assessed by using MEBG (pH 5.5) and monitoring absorbance at 620 nm. Aliquots (250 μl) of sterile MEBG containing the required concentration of chitosan were dispensed into wells containing either 30 μl of an overnight culture of a test organism or 30 μl of sterile MEBG alone (blanks). Inoculated wells were prepared in quintuplicate, and blanks were prepared in triplicate. Inocula were first counted with a hemocytometer and then diluted with MEBG so that the final inoculum contained 105 cells/ml. The microtiter plates were incubated at 25°C for up to 5 days, and we measured the absorbance of the wells periodically at 620 nm after 1 min of shaking by using an automated plate reader (Dynatech, Chantilly, Va.). For each sample, the mean value obtained for the blanks was subtracted from the mean value obtained for the inoculated wells.
Evaluation of antimicrobial activities in beverages and foods.
Sterile native chitosan and degraded chitosan (hydrolyzed by papaya latex) were added to pasteurized apple-elderflower juice at a final concentration of 0.3 g/liter. The juice was stored at 7°C for up to 13 days and was tested periodically by spread plating to determine the total viable count (on PCA), the lactic acid bacterial count (on de Man-Rogosa-Sharpe agar), and the yeast count (on oxytetracycline-glucose-yeast extract agar). All microbiological media were obtained from Oxoid Ltd. The agar plates were incubated aerobically at 25°C for 5 days. The PCA and oxytetracycline-glucose-yeast extract agar plates were incubated aerobically, while the de Man-Rogosa-Sharpe agar plates were placed in candle jars in order to provide a low-O2–high-CO2 atmosphere.
We also investigated the antimicrobial effects of chitosan and chitosan hydrolysates (produced by using papaya latex) at concentrations of 0.3, 1.0, and 5.0 g/kg in houmous (see above). Duplicate houmous samples were stored at 7°C for 6 days and were tested periodically to determine the total viable count, the lactic acid bacterial count, and the yeast count as described above for the juice experiments.
RESULTS
Preparation of chitosan hydrolysates.
Oxidative-reductive depolymerization of chitosan with hydrogen peroxide in the presence of iron(III) produced several chitosan hydrolysates, and the viscosities obtained ranged from >800 s for native chitosan to <9 s for the highly degraded samples. Overnight treatment of chitosan with crude papaya latex also resulted in degraded forms of chitosan with viscosities as low as 87 s. Depolymerized chitosan with a viscosity of 140 s was also obtained by using lysozyme. However, boiling for up to 30 min failed to inactivate lysozyme, and thus it was difficult to obtain chitosan oligomers with intermediate viscosities (and hence intermediate sizes). Consequently, all samples were ultimately degraded to the same extent as the enzyme continued to depolymerize the chitosan during the heating procedure.
Inactivation of microorganisms in saline solutions.
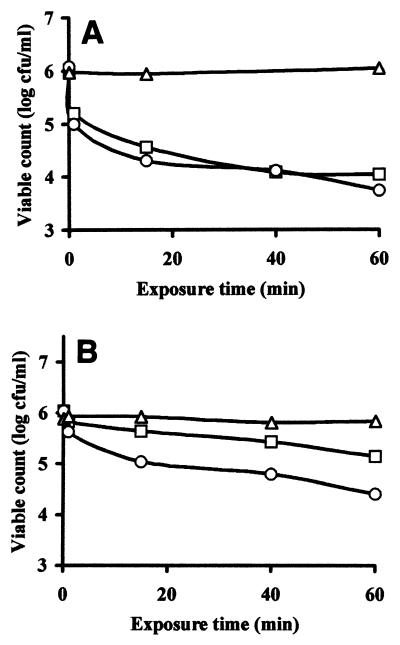
The antimicrobial activities of native chitosan and degraded chitosan in saline solutions (pH 6.4) were assessed by using the following five target organisms: Candida sp., Rhodotorula sp., P. fragi, Bacillus sp., and C. albidus. Neither native chitosan nor degraded chitosan at a concentration of 1 g/liter inactivated the latter three organisms. Approximately 2 log CFU of Candida sp. per ml was inactivated by 0.5 g of native chitosan per liter or 0.5 g of degraded chitosan per liter (Fig. 1A). In contrast, the level of inactivation of Rhodotorula sp. in the presence of 1 g of native chitosan per liter was low (0.5 log CFU/ml), but the level of inactivation in the presence of the same concentration of degraded chitosan was 1 log CFU/ml (Fig. 1B). Figure 1 shows that chitosan degraded with papaya latex exhibited somewhat greater antimicrobial activity than native chitosan, but the effect was not great.
FIG. 1.
Inactivation of Candida sp. (A) and Rhodotorula sp. (B) in saline solutions at 25°C (▵), in the presence of 0.5 g of native chitosan per liter (□), and in the presence of 0.5 g of a chitosan hydrolysate per liter prepared by using papaya latex (○).
Inhibition of yeast growth in laboratory media.
The inhibitory activities of native chitosan and chitosan hydrolysates prepared by oxidative-reductive degradation were assessed by using the following concentrations: 0.2 g/liter for S. ludwigii (Fig. 2) and 0.1, 0.2, and 0.3 g/liter for Z. bailii (Fig. 3) and S. cerevisiae (Fig. 4). These experiments were performed with MEBG at room temperature by using absorbance to measure growth. In addition to the results shown in Fig. 2 through 4, growth curves for controls containing only MEBG, only N-acetylglucosamine, only FeCl3, and N-acetylglucosamine plus hydrogen peroxide were obtained for all three spoilage yeasts; since the growth data for all of the control cultures were very similar, not all of the results are shown in Fig. 2 to 4 in order to keep the presentation clear.
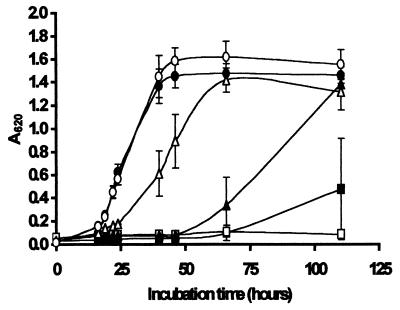
FIG. 2.
Growth of S. ludwigii in laboratory media at 25°C in the presence of no chitosan (control) (○), 0.2 g of native chitosan per liter (■), and chitosan hydrolysates with viscosities of 96 s (□), 24 s (▴), 17 s (▵), and 9 s (●). The hydrolysates were prepared by the oxidative-reductive degradation method. The results are means based on five replicate values for absorbance at 620 nm (A620). Bars indicate standard error.
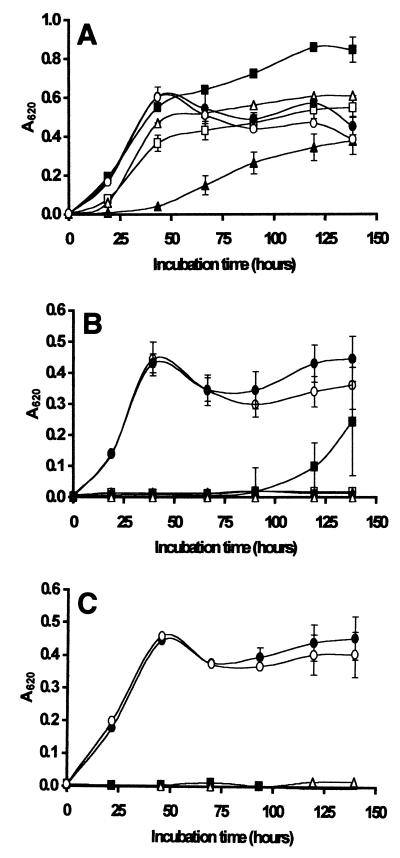
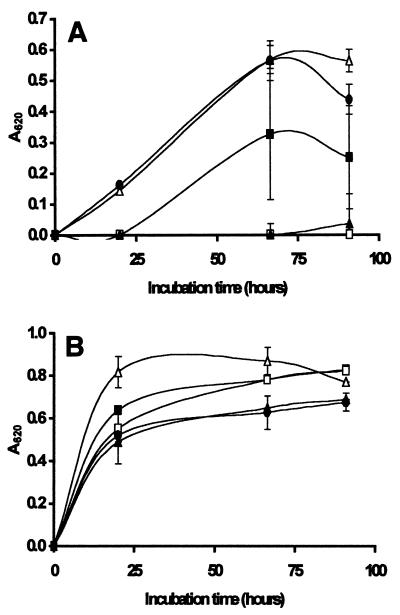
FIG. 3.
Growth of Z. bailii in laboratory media at 25°C in the presence of native chitosan (□) and hydrolyzed chitosan at concentrations of 0.1 g/liter (A), 0.2 g/liter (B), and 0.3 g/liter (C). The viscosities of the hydrolysates were 154 s (■), 96 s (▴), 54 s (▵), 11 s (●), and 0 s for the control containing N-acetylglucosamine plus hydrogen peroxide (○). The hydrolysates were prepared by the oxidative-reductive degradation method. The results are means based on five replicate values for absorbance at 620 nm (A620). Bars indicate standard error.
FIG. 4.
Growth of S. cerevisiae in laboratory media at 25°C in the presence of native chitosan (□) and hydrolyzed chitosan at concentrations of 0.1 g/liter (A), 0.2 g/liter (B), and 0.3 g/liter (C). The viscosities of the hydrolysates were 154 s (■), 96 s (▴), 54 s (▵), 11 s (●), and 0 s for the control containing N-acetylglucosamine plus hydrogen peroxide (○). The hydrolysates were prepared by the oxidative-reductive degradation method. The results are means based on five replicate values for absorbance at 620 nm (A620). Bars indicate standard error.
Overall, Figures 2 to 4 show that extensive degradation of chitosan to a viscosity of 11s or less (compared with 802s for the native polymer) resulted in a complete loss of inhibitory activity against all three spoilage yeasts. Hydrolysates with viscosities of 17 s and 24 s exhibited some antimicrobial activity but inhibited growth of S. ludwigii less than native chitosan inhibited growth of this organism (Fig. 2). Sensitivity to the antimicrobial action of the hydrolysate with a viscosity of 54 s depended on the target organism, and S. cerevisiae was less sensitive than Z. bailii at concentrations of 0.2 g/liter (Fig. 3B and 4B) and 0.3 g/liter (Fig. 3C and 4C). In general, the hydrolysates with viscosities of 96 s and 154 s were more effective in inhibiting the growth of all three spoilage organisms than native chitosan was. In most cases, the hydrolysates with viscosities of >96 s completely prevented growth of all three target organisms for the duration of the experiments (5 days). Notably, all of the hydrolysates except the very highly degraded 11 s hydrolysate inhibited growth of Z. bailii and S. cerevisiae at a concentration of 0.3 g/liter (Fig. 3C and 4C).
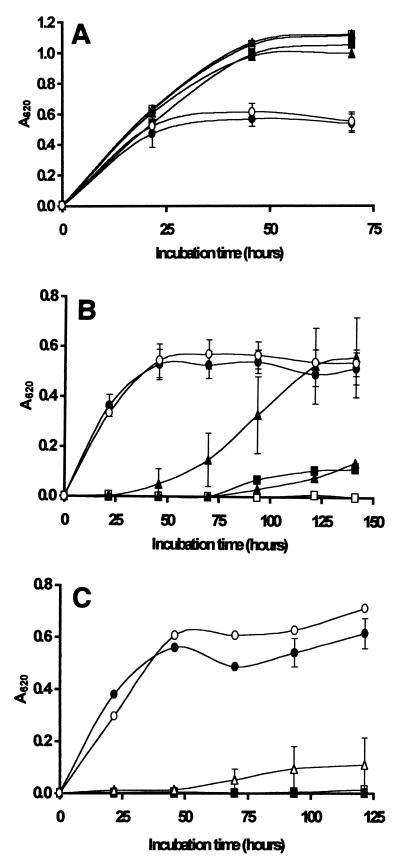
Figure 5A shows that growth of Z. bailii was completely inhibited in the presence of 0.1 g of lysozyme-degraded chitosan per liter for up to 4 days, while growth occurred in the presence of native chitosan, albeit at a slower rate than in the control preparation. Autoclaved lysozyme alone had no effect on the growth of the organism, but when native chitosan and autoclaved lysozyme were added together, they completely inhibited growth for the duration of the experiment (4 days). These results differed from the results shown in Fig. 3A, which shows that Z. bailii growth was largely unaffected by the presence of 0.1 g of native chitosan per liter or 0.1 g of hydrolysates per liter prepared by the oxidative-reductive depolymerization method. In contrast, S. cerevisiae growth was not inhibited in the presence of 0.1 g of chitosan per liter, whether the chitosan was in the native form or was degraded by either oxidative-reductive depolymerization or lysozyme (Fig. 4A and 5B).
FIG. 5.
Growth of Z. bailii (A) and S. cerevisiae (B) in laboratory media at 25°C in the presence of 0.1 g of native chitosan per liter (■) and lysozyme-degraded chitosan (□). The controls consisted of unsupplemented laboratory medium (●), autoclaved lysozyme (▵), and chitosan plus autoclaved lysozyme (▴). The results are means based on five replicate values for absorbance at 620 nm (A620). Bars indicate standard error.
Inhibition of spoilage in beverages and foods.
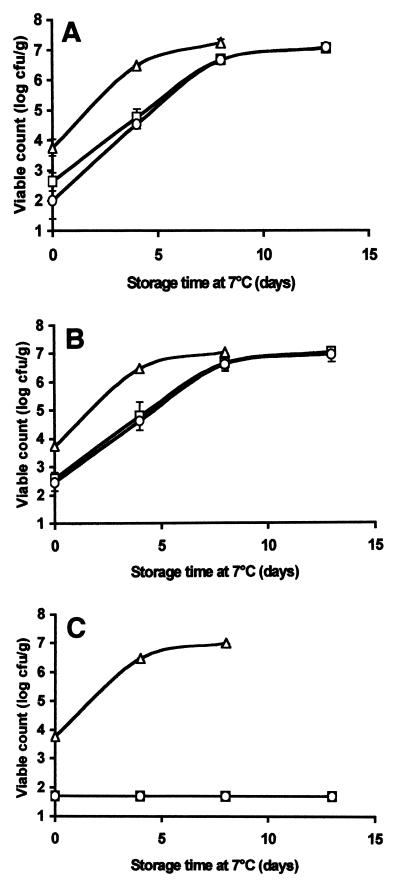
We assessed the antimicrobial activities of native chitosan and degraded chitosan against the natural microbial flora in a beverage (pasteurized apple-elderflower juice, pH 3.3) and in houmous (a popular chickpea-based dip, pH 4.2). Both products are sold refrigerated and have a relatively short shelf life once they are opened. These products were selected as model foods due to their low pH values and relative homogeneity (which allowed easy mixing with chitosan). As shown in Fig. 6, the total counts, lactic acid bacterial counts, and yeast counts in the apple-elderflower juice were reduced from the initial level (about 3 to 4 log CFU/ml) to a level below the sensitivity limit of the plating technique when either native or degraded chitosan was added at a concentration of 0.3 g/liter. After 4 days of storage at 7°C (this temperature was selected to represent domestic refrigeration conditions), the total counts and the lactic acid bacterial counts were approximately 2 log CFU/ml lower in the chitosan-supplemented juice samples than in the control samples; however, after 8 days, there were no differences in the total and lactic acid bacterial counts between the controls and the chitosan-supplemented samples (both reached a maximum population size of around 7 log CFU/ml). In contrast, yeast growth was completely suppressed; the levels of yeast were below the sensitivity limit of the plating technique for nearly 2 weeks in juice supplemented with chitosan (Fig. 6C). No substantial differences between the antimicrobial activities of native chitosan and degraded chitosan were noted.
FIG. 6.
Effects of native chitosan and degraded chitosan at a concentration of 0.3 g/liter on the natural flora of apple-elderflower juice stored at 7°C. The total mesophilic organisms (A), lactic acid bacteria (B), and yeasts (C) were enumerated on selective media. Symbols: ▵, juice alone; □, juice treated with native chitosan; ○, juice treated with chitosan degraded with papaya latex. The results are means based on duplicate viable count values.
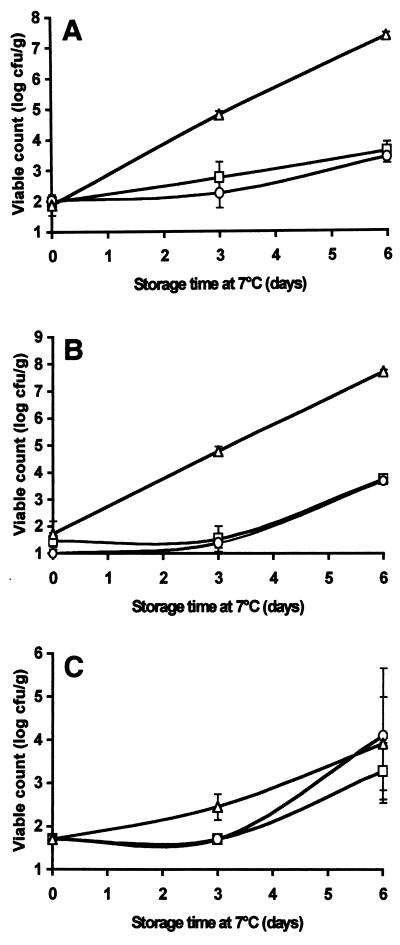
Native chitosan and degraded chitosan added at concentrations of 0.3 and 1.0 g/kg had no detectable effect on the development of the natural microflora of houmous (pH 4.2) stored at 7°C (results not shown). However, at a concentration of 5.0 g/kg, both native chitosan and degraded chitosan inhibited the growth of the total flora in general (Fig. 7A) and of lactic acid bacteria in particular (Fig. 7B). After 6 days of storage, the total viable counts and the lactic acid bacterial counts in the chitosan-containing samples were approximately 4 log CFU/g lower than the counts in the control (Fig. 7A and B). Notably, the yeast population was not affected by chitosan (either native or degraded), although the overall counts were generally low compared with the total counts and the lactic acid bacterial counts (Fig. 7C). We detected no differences in the counts for any of the microorganisms tested when we compared samples treated with native chitosan and samples treated with degraded chitosan.
FIG. 7.
Effects of native chitosan and degraded chitosan at a concentration of 5.0 g/liter on the natural flora of a houmous-water slurry stored at 7°C. Total mesophilic organisms (A), lactic acid bacteria (B), and yeasts (C) were enumerated on selective media. Symbols: ▵, houmous alone; □, houmous treated with native chitosan; ○, houmous treated with chitosan degraded with papaya latex. The results are means based on duplicate viable count values.
DISCUSSION
The inactivation experiments performed in simple saline solutions at pH 6.4 showed that Bacillus sp., P. fragi, and C. albidus were not sensitive to chitosan or chitosan degradation products at the limited number of concentrations tested in this study. Since some bacteria have been shown to exhibit sensitivity to the biocidal action of chitosan in relatively clean systems, such as buffers and distilled water (14, 21), it is possible that greater bacterial inactivation rates might have been observed in this study if higher concentrations of chitosan had been tested.
In contrast, the initial rapid decrease in the Candida sp. population (Fig. 1A) suggested that exposure to chitosan either rapidly killed the cells or rendered the cells nonculturable in a short time (i.e., less than 2 min). The surviving cells may have been members of a more resistant subpopulation or may have survived because much of the chitosan had been removed from solution by binding to the other cells or both. Interpretation of experimental results in terms of cell death is hampered by the lack of definitive information concerning the kinetics of chitosan-cell interactions and the mode of chitosan antimicrobial action.
Whereas the Candida sp. was affected equally by native chitosan and hydrolyzed chitosan, Rhodotorula sp. (Fig. 1B) was inactivated to a greater extent by degraded chitosan than by native chitosan. However, the antimicrobial activity observed with the degraded polymer was not extraordinarily greater. Additional simple saline-based inactivation experiments were not performed as we thought that results obtained with nutrient-deficient experimental systems were a poor indication of likely microbial responses to preservatives in the nutrient-rich environments found in most foods and beverages.
The results shown in Fig. 2 to 5 suggest that mild hydrolysis of chitosan enhanced the inhibitory activity of chitosan against some species of spoilage yeasts grown in complex laboratory media. Thus, we estimated that for Z. bailii, mild hydrolysis of chitosan decreased the MIC by a factor of about 2 (Fig. 3). However, highly degraded forms of chitosan exhibited no antimicrobial activity; this observation emphasized the importance of maintaining strict control over the process conditions used during chitosan degradation (Fig. 2 to 4). These results agree with the results of other authors obtained with nonfood organisms, such as the plant-pathogenic organisms Fusarium spp. (9, 20).
Degradation of chitosan by hydrogen peroxide differs fundamentally from enzymic depolymerization in three important respects. First, the chemical method is a random process, while enzymic attack is nonrandom and bond specific. Second, chitosan can be completely degraded to its constituent monomers by chemical methods provided that the reagents are present in excess amounts, as shown by the almost total loss of viscosity relative to water of the most highly hydrolyzed samples prepared in this study (viscosity, 11 s) (Fig. 2 to 4). In contrast, hexamers containing three or four acetylated residues must be present to initiate the lysozyme hydrolysis reaction (11, 12), and so lysozyme cannot be used to degrade chitosan completely, as shown by the generally higher viscosities obtained for the lysozyme-degraded chitosans in this study. Third, the hydroxyl radicals generated from hydrogen peroxide may react with parts of the chitosan molecule other than the β-1,4 linkage to form oligomers with nonnative oxidized groups. In this study, it was not possible to establish whether the improvement in the antimicrobial potency of chitosan degraded by hydrogen peroxide was due to variation in the degree of polymerization or to the presence of oxidized groups on the oligomers. It is possible that oxidation increased the biological activity of the polymer, while excessive hydrolysis of the polymeric backbone resulted in a loss of activity. The most active hydrolysates may represent the optimum balance between these two factors. In contrast, enzyme-degraded chitosan would not be expected to contain oxidized groups.
In more general terms, using hydrogen peroxide to degrade chitosan could be problematic in terms of potential toxicity and product licensing. The oligomers prepared by this method would probably need to be purified further to remove the excess iron, which could lead to off-flavors in foods. Additional processing of this nature would lead to extra costs for the ingredient manufacturer, the food processor, and ultimately the consumer. Enzymic methods of depolymerization may, in this context, provide a more “natural” and cheaper means of producing oligomers of chitosan. Lysozyme is readily available and is used as a food preservative in its own right to guard against clostridial spoilage of hard-cooked cheeses (8).
Although a degraded form of chitosan was successfully produced in this study by using lysozyme, oligomers having intermediate viscosities (and therefore intermediate molecular weights) could not be prepared because it was difficult to inactivate the enzyme by conventional heating methods. Nevertheless, increased antimicrobial activity of lysozyme-degraded chitosan against Z. bailii in laboratory media was observed (Fig. 5A), although we could not entirely rule out the possibility that the lysozyme in the oligomer preparation played a role in the inhibition of the organism.
Lysozymes are found in many plants, animals, and microorganisms (10, 19), and papaya latex is only one of many possible alternative sources of the enzyme. The enzyme structure differs slightly depending on source, and different structures can be expected to lead to different heat stabilities. Unlike the depolymerizing activity of lysozyme from hens' eggs, the depolymerizing activity of papaya latex can be arrested by heating by using conventional laboratory techniques (1). Papaya latex also contains other enzymes, such as papain, and has itself been reported to exhibit antifungal activity against C. albicans (5).
In this study, both native chitosan and papaya-degraded chitosan inactivated the natural yeast population of pasteurized apple-elderflower juice stored at 7°C to the extent that the organisms could not be detected for up to 2 weeks when conventional plating techniques were used (Fig. 6). These results are in agreement with the results of previous work that demonstrated that several spoilage yeasts were very sensitive to chitosan in clear ultrahigh-temperature-processed apple juice spiked with each organism individually and stored at room temperature (15).
Although the total counts and the lactic acid bacterial counts in the apple-elderflower juice were reduced by about 2 log CFU/ml immediately after exposure to chitosan, some of the remaining viable organisms appeared to recover and to resume growing at the same rate as the organisms in the control culture containing no added chitosan (Fig. 6A and B). It is possible that chitosan was irreversibly bound by some of the microbial cells, particulates, or negatively charged compounds present in the juice and thus rendered inactive against the remaining unbound microorganisms.
We noted that much higher concentrations of chitosan were required to inhibit microbial growth in houmous (5 g/kg) than in apple-elderflower juice (0.3 g/liter) in spite of the lower initial microbial load in houmous (<102 CFU/ml) than in juice (<104 CFU/ml). It is possible that the more particulate nature of the dip compared with the juice restricted mass transfer of the relatively large, polymeric chitosan molecules, which reduced the chance of contact with a microbial cell in the houmous. In addition, a number of other factors, such as pH (pH 4.2 in houmous and pH 3.3 in juice), fat content (25.6% fat in houmous and no fat in juice), and type of acid present (citric acid from the lemon juice in houmous and malic acid in apple juice), may have contributed to the differences in the antimicrobial activities of chitosan. In both juice and houmous, the activities of native chitosan and hydrolyzed chitosan did not differ.
Conclusions.
Chitosan has potential for use as a preservative in low-pH foods, either alone or in combination with other preservative systems. The constituents of the food matrix appear to have an important effect on the antimicrobial efficacy of chitosan, and this observation should be investigated further. Mild depolymerization of chitosan may slightly enhance its antimicrobial action, but the improvement is not considered sufficient to justify the extra processing steps involved.
ACKNOWLEDGMENTS
This work was carried out with the support of the European Union's FAIR Programme (contract CT96-1066) and with the sponsorship of Aplin and Barrett Ltd. (United Kingdom), CPC International (United Kingdom), Gist-brocades (The Netherlands), the Meat and Livestock Commission (United Kingdom), and Norsk Hydro (Norway).
We thank Kim Ilsley for technical assistance.
REFERENCES
- 1.Ahern T J, Klibanov A M. The mechanism of irreversible enzyme inactivation at 100°C. Science. 1985;228:1280–1284. doi: 10.1126/science.4001942. [DOI] [PubMed] [Google Scholar]
- 2.Allan C R, Hadwiger L A. The fungicidal effect of chitosan on fungi of varying cell wall composition. Exp Mycol. 1979;3:285–287. [Google Scholar]
- 3.Azarkan M, Amrani A, Nijs M, Vandermeers A, Zerhouni S, Smoulders N, Looze Y. Carica papaya latex is a rich source of class II chitinase. Phytochemistry. 1997;46:1319–1325. doi: 10.1016/s0031-9422(97)00469-x. [DOI] [PubMed] [Google Scholar]
- 4.Brzezinski R, Boucher I, Dupuy A, Plouffe B. Actinomycetes as model organisms for the study of chitosanases. In: Muzzarelli R A A, Peter M G, editors. Chitin handbook. Grottammare, Italy: European Chitin Society; 1997. pp. 291–296. [Google Scholar]
- 5.Giordani R, Cardenas M L, Moulin-Traffort J, Régli P. Fungicidal activity of latex sap from Carica papaya and antifungal effect of d(+)-glucosamine on Candida albicans growth. Mycoses. 1996;39:103–110. doi: 10.1111/j.1439-0507.1996.tb00110.x. [DOI] [PubMed] [Google Scholar]
- 6.Goosen M F A, editor. Applications of chitin and chitosan. Lancaster, Pa: Technomic Publishing Co., Inc.; 1997. [Google Scholar]
- 7.Hirano S, Nagao N. Effects of chitosan, pectic acid, lysozyme, and chitinase on the growth of several phytopathogens. Agric Biol Chem. 1989;53:3065–3066. [Google Scholar]
- 8.Johansen C, Gram L, Meyer A S. The combined inhibitory effect of lysozyme and low pH on growth of Listeria monocytogenes. J Food Prot. 1994;57:561–566. doi: 10.4315/0362-028X-57.7.561. [DOI] [PubMed] [Google Scholar]
- 9.Kendra D F, Hadwiger L A. Characterisation of the smallest chitosan oligomer that is maximally antifungal to Fusarium solani and elicits pisatin formation in Pisum sativum. Exp Mycol. 1984;8:276–281. [Google Scholar]
- 10.Lakhtin V M, Kostanova E A, Arbatski N P. Isolation and characterisation of multiple forms of papaya latex lysozyme. Appl Biochem Microbiol. 1995;31:214–220. [Google Scholar]
- 11.Nordtveit R J, Vårum K M, Smidsrød O. Degradation of fully water-soluble, partially N-acetylated chitosans with lysozyme. Carbohydr Polymers. 1994;23:253–260. [Google Scholar]
- 12.Nordtveit R J, Vårum K M, Smidsrød O. Degradation of partially N-acetylated chitosans with hen egg white and human lysozyme. Carbohydr Polymers. 1996;29:163–167. [Google Scholar]
- 13.Ohtakara A, Matsunaga H, Mitsutomi M. Action pattern of Streptomyces griseus chitinase on partially N-acetylated chitosan. Agric Biol Chem. 1990;54:3191–3199. [PubMed] [Google Scholar]
- 14.Papineau A M, Hoover D G, Knorr D, Farkas D F. Antimicrobial effect of water-soluble chitosans with high hydrostatic pressure. Food Biotechnol. 1991;5:45–57. [Google Scholar]
- 15.Roller S, Covill N. The antifungal properties of chitosan in laboratory media and apple juice. Int J Food Microbiol. 1999;47:67–77. doi: 10.1016/s0168-1605(99)00006-9. [DOI] [PubMed] [Google Scholar]
- 16.Sekiguchi S, Miura Y, Kaneko H, Nishimura S-I, Nishi N, Iwase M, Tokura S. Molecular weight dependency of antimicrobial activity by chitosan oligomers. In: Nishinari K, Doi E, editors. Food hydrocolloids: structures, properties, and functions. New York, N.Y: Plenum Press; 1994. pp. 71–76. [Google Scholar]
- 17.Skjak-Braek G, Anthonsen T, Sandford P. Chitin and chitosan. London, United Kingdom: Elsevier Applied Science; 1989. [Google Scholar]
- 18.Sudarshan N R, Hoover D G, Knorr D. Antibacterial action of chitosan. Food Biotechnol. 1992;6:257–272. [Google Scholar]
- 19.Tranter H S. Lysozyme, ovotransferrin and avidin. In: Dillon V M, Board R G, editors. Natural antimicrobial systems and food preservation. Wallingford, United Kingdom: CAB International; 1994. pp. 65–97. [Google Scholar]
- 20.Uchida Y, Izume M, Ohtakara A. Preparation of chitosan oligomers with purified chitosanase and its application. In: Skjåk-Brœk G, Anthonsen T, Sandford P, editors. Chitin and chitosan: sources, chemistry, biochemistry, physical properties and applications. London, United Kingdom: Elsevier Applied Science; 1989. pp. 373–382. [Google Scholar]
- 21.Wang G-H. Inhibition and inactivation of five species of foodborne pathogens by chitosan. J Food Prot. 1992;55:916–919. doi: 10.4315/0362-028X-55.11.916. [DOI] [PubMed] [Google Scholar]
- 22.Yabuki M. Characterisation of chitosanase produced by Bacillus circulans MH-K1. In: Skjåk-Brœk G, Anthonsen T, Sandford P, editors. Chitin and chitosan: sources, chemistry, biochemistry, physical properties and applications. London, United Kingdom: Elsevier Applied Science; 1989. pp. 197–206. [Google Scholar]