Abstract
Background
The R2R3-MYB transcription factor is one of the largest gene families in plants and involved in the regulation of plant development, hormone signal transduction, biotic and abiotic stresses. Tobacco is one of the most important model plants. Therefore, it will be of great significance to investigate the R2R3-MYB gene family and their expression patterns under abiotic stress and senescence in tobacco.
Results
A total of 174 R2R3-MYB genes were identified from tobacco (Nicotiana tabacum L.) genome and were divided into 24 subgroups based on phylogenetic analysis. Gene structure (exon/intron) and protein motifs were especially conserved among the NtR2R3-MYB genes, especially members within the same subgroup. The NtR2R3-MYB genes were distributed on 24 tobacco chromosomes. Analysis of gene duplication events obtained 3 pairs of tandem duplication genes and 62 pairs of segmental duplication genes, suggesting that segmental duplications is the major pattern for R2R3-MYB gene family expansion in tobacco. Cis-regulatory elements of the NtR2R3-MYB promoters were involved in cellular development, phytohormones, environmental stress and photoresponsive. Expression profile analysis showed that NtR2R3-MYB genes were widely expressed in different maturity tobacco leaves, and however, the expression patterns of different members appeared to be diverse. The qRT-PCR analysis of 15 NtR2R3-MYBs confirmed their differential expression under different abiotic stresses (cold, salt and drought), and notably, NtMYB46 was significantly up-regulated under three treatments.
Conclusions
In summary, a genome-wide identification, evolutionary and expression analysis of R2R3-MYB gene family in tobacco were conducted. Our results provided a solid foundation for further biological functional study of NtR2R3-MYB genes in tobacco.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08658-7.
Keywords: Nicotiana tobacum L., R2R3-MYB transcription factors, Phylogenetic analysis, Stress response, Gene expression
Background
MYB transcription factor is one of the largest members of the plant transcription factor family [1]. MYB proteins share a highly conserved DNA-binding domain (MYB), mostly located at the N-terminus of the protein. The MYB domains are characterized by one to four incomplete tandem repeat (R) structures (termed R1, R2, R3, R4). Each repeat is composed of 50 ~ 53 conserved amino acid residues and forms three α-helices. The second and third helices can be folded to form a helix-turn-helix (HTH) structure, which is involved in DNA binding [2]. Three regularly spaced tryptophan residues (or other hydrophobic residues) form a hydrophobic core in the three-dimensional structure center of HTH, which is important for maintaining the configuration of HTH [3]. Sometimes, the tryptophan residues are replaced by other amino acids, such as aromatic amino acids and hydrophobic amino acids, especially in R3 domains. The C-terminal of MYB transcription factor usually contains a transcriptional activation region rich in acidic amino acids, which is responsible for multiple protein regulatory activities. Based on the number and types of MYB repeats, the MYB family is subdivided into four major groups, namely 4R-MYB, 3R-MYB/R1R2R3-MYB, 1R-MYB/MYB-related, 2R-MYB/R2R3-MYB [4].
R2R3-MYB transcription factor is predominantly present in plants, which contains R2 and R3 domains. It was reported that the R2R3-MYB genes probably evolved from the loss of the R1 repeat in the R1R2R3-MYB gene or from the duplication of the R1 repeat in 1R-MYB gene [5, 6]. R2R3-MYB gene family is widely involved in the regulation of various biological processes, including plant growth and development, hormone signaling, primary and secondary metabolism [7–9]. For example, AtMYB106, AtMYB16 and AtMYB17 are involved in the regulation of trichome branching, petal epidermal cell morphogenesis and early inflorescence development, respectively [10–12]. The biosynthesis of flavonoids is directly or indirectly controlled by three genes (P, C1, Pl) encoding R2R3-MYB domain in maize [1, 13, 14]. MYB7 gene in Actinidia deliciosa positively regulated the biosynthesis of carotenoids and chlorophyll [15]. Knockdown the expression of MYB305 of the ornamental tobacco which contains a conserved R2R3-MYB DNA binding domain resulted in the decrease expression of related genes in nectarins and flavonoid biosynthetic [16]. In addition, most R2R3-MYB genes have also been suggested to regulate plant responses to biotic and abiotic stress conditions. For instance, overexpression of AtMYB75 in Arabidopsis can increase secondary metabolites (anthocyanins and flavonols), which can protect against pests [17]. OsMYB6 gene of rice as a stress-responsive factor which plays the role as a positive regulator in response to drought and salt stress resistance [18]. Increasing the expression of NtMYB4a may promote anthocyanin accumulation, and thereby increase antioxidant capability and tolerance to low temperature of tobacco plant [19]. Tobacco NtMYB12 overexpression adapts to low Pi stress environment via regulating the contents of flavonol and phosphorus [20].
Tobacco is one of the most important model plants [21]. Like other plants, tobacco is often subjected to stress such as low temperature, salt, strong light, drought and other stresses, which leads to the reduction of production. Leaf senescence is an important production process with a positive and orderly process accompanied by the changes of leaf color, cell structure, biochemical metabolism and gene expression level, along with a series of degradations, which is related to secondary metabolites, such as flavonoids, carotenoids, chlorophyll, etc. [22, 23]. The regulatory role of R2R3-MYB genes in abiotic stress response, plant growth and development has been assessed in several studies [12, 18, 24]. The investigation of the R2R3-MYB gene family and their expression patterns under various stresses including abiotic stress and senescence in tobacco is of great significance for the study of plant physiology and development.
In this study, a comprehensive investigation of R2R3-MYB gene family, including gene structures, chromosomal localization, phylogenetic relationship, motif composition, duplication events and cis-element compositions was performed using the current tobacco genome sequence data. Moreover, the gene expression profile in different senescence stages of tobacco leaves and expression pattern under various abiotic stresses such as cold, salt, and drought treatments were analyzed. The objectives of this study were to systematically analyze the sequence structures of tobacco R2R3-MYB gene family and explore the evolutionary relationship of R2R3-MYB gene family in plant, and thereby, reveal the expression regulation of the R2R3-MYB gene family members under various stresses or adversity condition of senescence. The information derived from this study lay a foundation for further functional investigation on the R2R3-MYB gene family in tobacco.
Results
Characterization and distribution of R2R3-MYB genes in tobacco genome
In this study, a total of 174 NtR2R3-MYB genes were identified in tobacco and were renamed from NtMYB1 to NtMYB174 (Table 1). The information of these NtR2R3-MYB genes and their corresponding proteins are showed in Table 1 and Additional file 1: Table S1, namely, gene ID, location, number of exons, protein length (aa), molecular weight (MW), theoretical isoelectric point (pI) and subcellular location. The protein lengths varied greatly from 192aa (NtMYB37, NtMYB39) to 505aa (NtMYB96). The molecular weights ranged from 22,003.55 Da (NtMYB37) to 55,710.67 Da (NtMYB96), and the theoretical isoelectric point (pI) ranged from 4.80 (NtMYB56) to 9.77 (NtMYB3). Subcellular localization prediction indicated that the majority NtR2R3-MYB members were located in the nucleus. In addition, the chromosome positions of some NtR2R3-MYB genes could not yet be defined due to the incomplete sequencing of tobacco genome.
Table 1.
The R2R3-MYB family genes in Nicotiana tabacum L
| Gene Name | Gene ID | Location | Exon | Protein length (aa) | MW (Da) | pI | Subcellular location |
|---|---|---|---|---|---|---|---|
| NtMYB1 | Nitab4.5_0003163g0090.1 | Nitab4.5_0003163 | 2 | 279 | 31,833.62 | 6.10 | Nuclear |
| NtMYB2 | Nitab4.5_0001730g0050.1 | Nitab4.5_0001730 | 2 | 280 | 31,950.76 | 6.08 | Nuclear |
| NtMYB3 | Nitab4.5_0000673g0080.1 | Nt15 | 3 | 231 | 26,853.27 | 9.77 | Nuclear |
| NtMYB4 | Nitab4.5_0000109g0340.1 | Nt04 | 2 | 297 | 34,408.15 | 9.10 | Nuclear |
| NtMYB5 | Nitab4.5_0004437g0020.1 | Nitab4.5_0004437 | 3 | 270 | 30,067.05 | 9.05 | Nuclear |
| NtMYB6 | Nitab4.5_0002728g0020.1 | Nt23 | 2 | 299 | 34,523.17 | 8.98 | Nuclear |
| NtMYB7 | Nitab4.5_0006880g0040.1 | Nitab4.5_0006880 | 2 | 298 | 34,220.92 | 9.10 | Nuclear |
| NtMYB8 | Nitab4.5_0017504g0010.1 | Nitab4.5_0017504 | 3 | 215 | 24,922.58 | 8.99 | Nuclear |
| NtMYB9 | Nitab4.5_0000189g0200.1 | Nitab4.5_0000189 | 3 | 387 | 43,320.24 | 6.41 | Nuclear |
| NtMYB10 | Nitab4.5_0000303g0330.1 | Nt22 | 3 | 331 | 36,890.00 | 9.04 | Nuclear |
| NtMYB11 | Nitab4.5_0012287g0020.1 | Nitab4.5_0012287 | 3 | 215 | 24,927.65 | 8.83 | Nuclear |
| NtMYB12 | Nitab4.5_0002222g0050.1 | Nt23 | 4 | 279 | 31,844.87 | 9.46 | Nuclear |
| NtMYB13 | Nitab4.5_0004379g0020.1 | Nitab4.5_0004379 | 3 | 308 | 35,692.25 | 6.48 | Nuclear |
| NtMYB14 | Nitab4.5_0001007g0040.1 | Nt01 | 3 | 330 | 37,561.96 | 7.10 | Nuclear |
| NtMYB15 | Nitab4.5_0016704g0010.1 | Nitab4.5_0016704 | 3 | 277 | 32,076.03 | 5.68 | Nuclear |
| NtMYB16 | Nitab4.5_0000044g0310.1 | Nt14 | 3 | 281 | 31,984.67 | 5.08 | Nuclear |
| NtMYB17 | Nitab4.5_0000247g0080.1 | Nt04 | 2 | 264 | 29,833.73 | 8.96 | Nuclear |
| NtMYB18 | Nitab4.5_0004662g0010.1 | Nitab4.5_0004662 | 3 | 281 | 32,028.68 | 5.08 | Nuclear |
| NtMYB19 | Nitab4.5_0000436g0250.1 | Nt14 | 3 | 256 | 29,567.10 | 5.44 | Nuclear |
| NtMYB20 | Nitab4.5_0007938g0010.1 | Nitab4.5_0007938 | 3 | 320 | 36,739.11 | 5.75 | Nuclear |
| NtMYB21 | Nitab4.5_0008924g0010.1 | Nitab4.5_0008924 | 3 | 376 | 42,817.57 | 5.77 | Nuclear |
| NtMYB22 | Nitab4.5_0002250g0030.1 | Nt23 | 2 | 273 | 31,206.27 | 5.48 | Nuclear |
| NtMYB23 | Nitab4.5_0003631g0010.1 | Nt16 | 3 | 348 | 39,211.35 | 5.90 | Nuclear |
| NtMYB24 | Nitab4.5_0002714g0010.1 | Nt21 | 3 | 320 | 36,558.99 | 5.75 | Nuclear |
| NtMYB25 | Nitab4.5_0005304g0030.1 | Nitab4.5_0005304 | 4 | 270 | 30,750.81 | 8.95 | Nuclear |
| NtMYB26 | Nitab4.5_0000232g0160.1 | Nt12 | 3 | 376 | 42,584.38 | 7.78 | Nuclear |
| NtMYB27 | Nitab4.5_0000021g0690.1 | Nt04 | 2 | 358 | 40,501.32 | 6.37 | Nuclear |
| NtMYB28 | Nitab4.5_0000302g0200.1 | Nt22 | 3 | 278 | 32,005.06 | 5.53 | Nuclear |
| NtMYB29 | Nitab4.5_0010133g0010.1 | Nitab4.5_0010133 | 2 | 347 | 38,694.05 | 6.02 | Nuclear |
| NtMYB30 | Nitab4.5_0000769g0140.1 | Nitab4.5_0000769 | 3 | 335 | 38,067.87 | 5.74 | Nuclear |
| NtMYB31 | Nitab4.5_0005512g0020.1 | Nitab4.5_0005512 | 3 | 412 | 45,423.31 | 5.61 | Nuclear |
| NtMYB32 | Nitab4.5_0005819g0010.1 | Nitab4.5_0005819 | 2 | 361 | 40,247.79 | 6.11 | Nuclear |
| NtMYB33 | Nitab4.5_0000568g0090.1 | Nt17 | 3 | 406 | 44,589.22 | 5.41 | Nuclear |
| NtMYB34 | Nitab4.5_0000274g0200.1 | Nt17 | 4 | 324 | 36,112.10 | 5.45 | Nuclear |
| NtMYB35 | Nitab4.5_0000649g0090.1 | Nt17 | 3 | 377 | 43,062.99 | 5.74 | Nuclear |
| NtMYB36 | Nitab4.5_0002030g0030.1 | Nitab4.5_0005819 | 4 | 323 | 35,970.08 | 5.34 | Nuclear |
| NtMYB37 | Nitab4.5_0002529g0070.1 | Nt17 | 3 | 192 | 22,003.55 | 6.60 | Nuclear |
| NtMYB38 | Nitab4.5_0000121g0260.1 | Nt24 | 3 | 346 | 38,617.93 | 6.25 | Nuclear |
| NtMYB39 | Nitab4.5_0002217g0020.1 | Nt03 | 3 | 192 | 22,011.62 | 6.60 | Nuclear |
| NtMYB40 | Nitab4.5_0000634g0210.1 | Nt22 | 2 | 350 | 39,115.28 | 5.94 | Nuclear |
| NtMYB41 | Nitab4.5_0004704g0020.1 | Nitab4.5_0004704 | 3 | 339 | 37,732.30 | 5.83 | Nuclear |
| NtMYB42 | Nitab4.5_0000842g0120.1 | Nt04 | 3 | 355 | 39,487.19 | 5.85 | Nuclear |
| NtMYB43 | Nitab4.5_0000141g0070.1 | Nt09 | 3 | 358 | 40,625.05 | 8.66 | Nuclear |
| NtMYB44 | Nitab4.5_0000116g0020.1 | Nt19 | 4 | 384 | 43,672.85 | 5.03 | Nuclear |
| NtMYB45 | Nitab4.5_0000906g0090.1 | Nitab4.5_0000906 | 4 | 415 | 46,855.05 | 4.86 | Nuclear |
| NtMYB46 | Nitab4.5_0000889g0030.1 | Nt20 | 3 | 348 | 38,953.29 | 6.19 | Nuclear |
| NtMYB47 | Nitab4.5_0005586g0010.1 | Nt15 | 3 | 291 | 33,336.13 | 5.99 | Nuclear |
| NtMYB48 | Nitab4.5_0010251g0010.1 | Nitab4.5_0010251 | 5 | 294 | 33,247.57 | 8.10 | Nuclear |
| NtMYB49 | Nitab4.5_0001186g0080.1 | Nt22 | 2 | 349 | 38,905.01 | 6.00 | Nuclear |
| NtMYB50 | Nitab4.5_0000057g0160.1 | Nt12 | 3 | 297 | 34,210.97 | 6.27 | Nuclear |
| NtMYB51 | Nitab4.5_0001019g0020.1 | Nitab4.5_0001019 | 3 | 335 | 38,102.89 | 5.84 | Nuclear |
| NtMYB52 | Nitab4.5_0000083g0200.1 | Nt13 | 3 | 332 | 37,137.29 | 6.92 | Nuclear |
| NtMYB53 | Nitab4.5_0005842g0010.1 | Nitab4.5_0005842 | 3 | 322 | 37,158.58 | 7.62 | Nuclear |
| NtMYB54 | Nitab4.5_0001295g0260.1 | Nt04 | 4 | 338 | 38,240.08 | 6.55 | Nuclear |
| NtMYB55 | Nitab4.5_0001041g0130.1 | Nt14 | 3 | 261 | 30,652.74 | 9.44 | Nuclear |
| NtMYB56 | Nitab4.5_0018794g0010.1 | Nitab4.5_0018794 | 3 | 275 | 31,595.75 | 4.80 | Nuclear |
| NtMYB57 | Nitab4.5_0005224g0050.1 | Nitab4.5_0005224 | 3 | 294 | 33,426.74 | 7.80 | Nuclear |
| NtMYB58 | Nitab4.5_0002137g0010.1 | Nitab4.5_0002137 | 3 | 464 | 51,655.76 | 5.77 | Nuclear |
| NtMYB59 | Nitab4.5_0008050g0020.1 | Nitab4.5_0008050 | 3 | 298 | 33,438.64 | 6.20 | Nuclear |
| NtMYB60 | Nitab4.5_0002906g0080.1 | Nt04 | 3 | 293 | 33,161.35 | 7.01 | Nuclear |
| NtMYB61 | Nitab4.5_0001622g0080.1 | Nt01 | 3 | 322 | 36,081.65 | 7.61 | Nuclear |
| NtMYB62 | Nitab4.5_0002300g0110.1 | Nt22 | 3 | 449 | 48,659.87 | 5.24 | Nuclear |
| NtMYB63 | Nitab4.5_0000387g0040.1 | Nt05 | 3 | 290 | 32,869.09 | 6.79 | Nuclear |
| NtMYB64 | Nitab4.5_0006658g0010.1 | Nt08 | 3 | 274 | 31,241.42 | 4.84 | Nuclear |
| NtMYB65 | Nitab4.5_0000844g0110.1 | Nt17 | 2 | 360 | 40,537.50 | 6.60 | Nuclear |
| NtMYB66 | Nitab4.5_0000428g0150.1 | Nt19 | 3 | 450 | 48,565.84 | 5.26 | Nuclear |
| NtMYB67 | Nitab4.5_0000564g0080.1 | Nt17 | 3 | 304 | 34,305.37 | 5.93 | Nuclear |
| NtMYB68 | Nitab4.5_0009259g0010.1 | Nitab4.5_0009259 | 3 | 485 | 53,842.35 | 5.44 | Nuclear |
| NtMYB69 | Nitab4.5_0000365g0050.1 | Nt17 | 3 | 257 | 28,763.40 | 7.54 | Nuclear |
| NtMYB70 | Nitab4.5_0000614g0110.1 | Nt03 | 2 | 358 | 40,397.36 | 6.11 | Nuclear |
| NtMYB71 | Nitab4.5_0000081g0200.1 | Nt09 | 3 | 424 | 47,599.55 | 6.47 | Nuclear |
| NtMYB72 | Nitab4.5_0000103g0310.1 | Nt04 | 3 | 276 | 32,220.78 | 5.58 | Nuclear |
| NtMYB73 | Nitab4.5_0006318g0060.1 | Nt03 | 3 | 305 | 34,332.33 | 6.04 | Nuclear |
| NtMYB74 | Nitab4.5_0003511g0010.1 | Nt19 | 3 | 421 | 47,609.76 | 8.01 | Nuclear |
| NtMYB75 | Nitab4.5_0001863g0170.1 | Nt03 | 3 | 267 | 30,867.70 | 5.94 | Nuclear |
| NtMYB76 | Nitab4.5_0003683g0020.1 | Nitab4.5_0003683 | 4 | 323 | 35,789.44 | 4.97 | Nuclear |
| NtMYB77 | Nitab4.5_0001081g0060.1 | Nitab4.5_0001081 | 3 | 421 | 47,340.48 | 6.71 | Nuclear |
| NtMYB78 | Nitab4.5_0002714g0060.1 | Nt21 | 3 | 247 | 28,371.92 | 8.91 | Nuclear |
| NtMYB79 | Nitab4.5_0001048g0070.1 | Nt22 | 3 | 364 | 40,464.90 | 6.27 | Nuclear |
| NtMYB80 | Nitab4.5_0002721g0050.1 | Nitab4.5_0002721 | 3 | 283 | 32,031.79 | 7.63 | Nuclear |
| NtMYB81 | Nitab4.5_0008604g0010.1 | Nitab4.5_0008604 | 3 | 255 | 29,530.09 | 9.02 | Nuclear |
| NtMYB82 | Nitab4.5_0002352g0080.1 | Nt08 | 3 | 362 | 40,166.57 | 6.27 | Nuclear |
| NtMYB83 | Nitab4.5_0000578g0120.1 | Nt02 | 3 | 314 | 35,069.22 | 7.52 | Nuclear |
| NtMYB84 | Nitab4.5_0000541g0100.1 | Nt07 | 3 | 422 | 47,691.75 | 8.01 | Nuclear |
| NtMYB85 | Nitab4.5_0004051g0020.1 | Nitab4.5_0004051 | 3 | 335 | 38,513.93 | 5.58 | Nuclear |
| NtMYB86 | Nitab4.5_0001213g0100.1 | Nt21 | 5 | 481 | 53,651.94 | 5.66 | Nuclear |
| NtMYB87 | Nitab4.5_0000769g0210.1 | Nitab4.5_0000769 | 3 | 327 | 37,985.21 | 5.88 | Nuclear |
| NtMYB88 | Nitab4.5_0004482g0030.1 | Nt23 | 2 | 281 | 31,759.71 | 5.18 | Nuclear |
| NtMYB89 | Nitab4.5_0000041g0400.1 | Nt15 | 3 | 215 | 25,017.47 | 5.89 | Nuclear |
| NtMYB90 | Nitab4.5_0007679g0010.1 | Nitab4.5_0007679 | 3 | 317 | 35,491.55 | 6.45 | Nuclear |
| NtMYB91 | Nitab4.5_0001318g0060.1 | Nt05 | 3 | 241 | 27,878.46 | 5.54 | Nuclear |
| NtMYB92 | Nitab4.5_0004993g0020.1 | Nt09 | 3 | 336 | 38,085.43 | 8.57 | Nuclear |
| NtMYB93 | Nitab4.5_0003061g0020.1 | Nt08 | 3 | 245 | 28,208.46 | 8.06 | Nuclear |
| NtMYB94 | Nitab4.5_0007042g0010.1 | Nt01 | 2 | 284 | 31,955.02 | 5.17 | Nuclear |
| NtMYB95 | Nitab4.5_0004551g0030.1 | Nitab4.5_0004551 | 2 | 342 | 39,174.80 | 6.01 | Nuclear |
| NtMYB96 | Nitab4.5_0000271g0230.1 | Nitab4.5_0000271 | 3 | 505 | 55,710.67 | 5.31 | Nuclear |
| NtMYB97 | Nitab4.5_0004193g0070.1 | Nt06 | 3 | 273 | 31,079.67 | 6.31 | Nuclear |
| NtMYB98 | Nitab4.5_0001334g0020.1 | Nt22 | 3 | 282 | 32,969.78 | 8.54 | Nuclear |
| NtMYB99 | Nitab4.5_0011760g0030.1 | Nitab4.5_0011760 | 3 | 323 | 36,003.45 | 6.38 | Nuclear |
| NtMYB100 | Nitab4.5_0003242g0050.1 | Nt18 | 3 | 285 | 32,075.69 | 6.83 | Nuclear |
| NtMYB101 | Nitab4.5_0002160g0080.1 | Nitab4.5_0002160 | 3 | 307 | 34,335.39 | 7.11 | Nuclear |
| NtMYB102 | Nitab4.5_0000201g0150.1 | Nitab4.5_0000201 | 3 | 302 | 34,343.53 | 5.02 | Nuclear |
| NtMYB103 | Nitab4.5_0002560g0010.1 | Nitab4.5_0002560 | 3 | 235 | 27,290.34 | 6.08 | Nuclear |
| NtMYB104 | Nitab4.5_0000363g0320.1 | Nt17 | 3 | 351 | 39,800.29 | 4.96 | Nuclear |
| NtMYB105 | Nitab4.5_0000225g0010.1 | Nt14 | 3 | 295 | 34,323.28 | 7.23 | Nuclear |
| NtMYB106 | Nitab4.5_0001442g0010.1 | Nitab4.5_0001442 | 3 | 337 | 38,358.58 | 8.46 | Nuclear |
| NtMYB107 | Nitab4.5_0001074g0040.1 | Nitab4.5_0001074 | 3 | 371 | 42,214.45 | 6.16 | Nuclear |
| NtMYB108 | Nitab4.5_0006893g0010.1 | Nt13 | 3 | 345 | 39,161.20 | 5.04 | Nuclear |
| NtMYB109 | Nitab4.5_0000747g0030.1 | Nt09 | 3 | 283 | 32,046.96 | 5.95 | Nuclear |
| NtMYB110 | Nitab4.5_0006144g0030.1 | Nitab4.5_0006144 | 3 | 355 | 40,694.11 | 4.82 | Nuclear |
| NtMYB111 | Nitab4.5_0000205g0020.1 | Nt18 | 2 | 435 | 47,856.34 | 6.60 | Nuclear |
| NtMYB112 | Nitab4.5_0000103g0300.1 | Nt04 | 3 | 267 | 31,067.60 | 5.47 | Nuclear |
| NtMYB113 | Nitab4.5_0001050g0010.1 | Nt08 | 3 | 314 | 35,688.67 | 7.48 | Nuclear |
| NtMYB114 | Nitab4.5_0003781g0060.1 | Nitab4.5_0003781 | 3 | 270 | 30,619.96 | 5.06 | Nuclear |
| NtMYB115 | Nitab4.5_0006804g0010.1 | Nitab4.5_0006804 | 3 | 329 | 37,315.85 | 6.53 | Nuclear |
| NtMYB116 | Nitab4.5_0008289g0010.1 | Nitab4.5_0008289 | 2 | 446 | 49,114.75 | 6.32 | Nuclear |
| NtMYB117 | Nitab4.5_0003478g0050.1 | Nitab4.5_0003478 | 3 | 328 | 37,275.75 | 7.02 | Nuclear |
| NtMYB118 | Nitab4.5_0005004g0050.1 | Nitab4.5_0005004 | 3 | 355 | 40,635.36 | 6.47 | Nuclear |
| NtMYB119 | Nitab4.5_0012173g0010.1 | Nitab4.5_0012173 | 3 | 323 | 36,591.55 | 8.11 | Nuclear |
| NtMYB120 | Nitab4.5_0000610g0290.1 | Nt09 | 3 | 197 | 23,319.31 | 8.98 | Nuclear |
| NtMYB121 | Nitab4.5_0006516g0020.1 | Nitab4.5_0006516 | 3 | 258 | 29,584.34 | 8.74 | Nuclear |
| NtMYB122 | Nitab4.5_0000958g0010.1 | Nitab4.5_0000958 | 3 | 240 | 27,565.39 | 9.54 | Nuclear |
| NtMYB123 | Nitab4.5_0000063g0150.1 | Nt24 | 3 | 299 | 33,432.60 | 8.86 | Nuclear |
| NtMYB124 | Nitab4.5_0003212g0030.1 | Nt08 | 3 | 238 | 27,567.72 | 5.79 | Nuclear |
| NtMYB125 | Nitab4.5_0003324g0040.1 | Nitab4.5_0003324 | 3 | 280 | 32,332.22 | 5.69 | Nuclear |
| NtMYB126 | Nitab4.5_0003179g0030.1 | Nitab4.5_0003179 | 3 | 221 | 26,376.80 | 8.26 | Nuclear |
| NtMYB127 | Nitab4.5_0000958g0020.1 | Nitab4.5_0000958 | 3 | 271 | 31,650.18 | 6.34 | Nuclear |
| NtMYB128 | Nitab4.5_0002293g0010.1 | Nt06 | 3 | 268 | 31,228.87 | 5.93 | Nuclear |
| NtMYB129 | Nitab4.5_0005019g0040.1 | Nitab4.5_0005019 | 5 | 293 | 32,724.01 | 6.10 | Plasma Membrane |
| NtMYB130 | Nitab4.5_0000244g0260.1 | Nt18 | 3 | 376 | 42,425.45 | 5.93 | Nuclear |
| NtMYB131 | Nitab4.5_0001814g0020.1 | Nt11 | 3 | 311 | 35,910.95 | 6.60 | Nuclear |
| NtMYB132 | Nitab4.5_0000070g0110.1 | Nt17 | 3 | 252 | 29,568.27 | 8.52 | Nuclear |
| NtMYB133 | Nitab4.5_0001163g0150.1 | Nt22 | 3 | 309 | 35,134.18 | 7.42 | Nuclear |
| NtMYB134 | Nitab4.5_0002626g0050.1 | Nt05 | 6 | 276 | 31,084.17 | 6.77 | Nuclear |
| NtMYB135 | Nitab4.5_0005769g0040.1 | Nitab4.5_0005769 | 3 | 294 | 34,548.53 | 5.94 | Nuclear |
| NtMYB136 | Nitab4.5_0000080g0070.1 | Nt13 | 3 | 256 | 29,565.26 | 9.11 | Nuclear |
| NtMYB137 | Nitab4.5_0000895g0130.1 | Nt15 | 3 | 263 | 30,829.51 | 6.13 | Nuclear |
| NtMYB138 | Nitab4.5_0000103g0280.1 | Nt04 | 4 | 224 | 25,692.46 | 9.53 | Nuclear |
| NtMYB139 | Nitab4.5_0000642g0100.1 | Nitab4.5_0000642 | 3 | 354 | 39,785.35 | 6.10 | Nuclear |
| NtMYB140 | Nitab4.5_0000278g0070.1 | Nt04 | 2 | 400 | 43,099.14 | 5.62 | Nuclear |
| NtMYB141 | Nitab4.5_0005708g0020.1 | Nitab4.5_0005708 | 2 | 401 | 43,138.26 | 5.65 | Nuclear |
| NtMYB142 | Nitab4.5_0001373g0030.1 | Nt17 | 2 | 301 | 33,224.05 | 7.11 | Nuclear |
| NtMYB143 | Nitab4.5_0000569g0180.1 | Nt15 | 2 | 236 | 25,652.79 | 7.74 | Nuclear |
| NtMYB144 | Nitab4.5_0000210g0060.1 | Nt23 | 3 | 315 | 36,121.22 | 6.12 | Nuclear |
| NtMYB145 | Nitab4.5_0006450g0010.1 | Nitab4.5_0006450 | 3 | 315 | 36,204.08 | 5.43 | Nuclear |
| NtMYB146 | Nitab4.5_0002578g0010.1 | Nt03 | 1 | 357 | 39,264.88 | 7.12 | Nuclear |
| NtMYB147 | Nitab4.5_0008039g0010.1 | Nitab4.5_0008039 | 1 | 258 | 28,738.10 | 9.12 | Nuclear |
| NtMYB148 | Nitab4.5_0011004g0010.1 | Nitab4.5_0011004 | 3 | 243 | 28,553.78 | 7.25 | Nuclear |
| NtMYB149 | Nitab4.5_0000008g0380.1 | Nt02 | 2 | 364 | 39,638.73 | 5.33 | Nuclear |
| NtMYB150 | Nitab4.5_0005060g0020.1 | Nitab4.5_0005060 | 1 | 332 | 36,533.55 | 9.17 | Nuclear |
| NtMYB151 | Nitab4.5_0001818g0030.1 | Nt07 | 3 | 311 | 35,476.48 | 6.05 | Nuclear |
| NtMYB152 | Nitab4.5_0001180g0160.1 | Nt04 | 4 | 257 | 29,799.39 | 9.30 | Nuclear |
| NtMYB153 | Nitab4.5_0001984g0040.1 | Nt12 | 1 | 329 | 36,163.18 | 9.07 | Nuclear |
| NtMYB154 | Nitab4.5_0000068g0400.1 | Nt12 | 1 | 258 | 28,770.10 | 8.86 | Nuclear |
| NtMYB155 | Nitab4.5_0001176g0010.1 | Nt02 | 3 | 311 | 35,523.52 | 8.19 | Nuclear |
| NtMYB156 | Nitab4.5_0000683g0060.1 | Nt06 | 1 | 255 | 27,976.61 | 6.76 | Nuclear |
| NtMYB157 | Nitab4.5_0000797g0020.1 | Nt04 | 1 | 258 | 28,448.05 | 7.68 | Nuclear |
| NtMYB158 | Nitab4.5_0002152g0050.1 | Nt10 | 2 | 364 | 39,622.70 | 5.39 | Nuclear |
| NtMYB159 | Nitab4.5_0001966g0010.1 | Nitab4.5_0001966 | 1 | 290 | 31,772.73 | 6.33 | Nuclear |
| NtMYB160 | Nitab4.5_0005973g0020.1 | Nt06 | 5 | 275 | 32,382.32 | 9.70 | Nuclear |
| NtMYB161 | Nitab4.5_0000514g0130.1 | Nt04 | 3 | 377 | 43,759.81 | 5.91 | Nuclear |
| NtMYB162 | Nitab4.5_0002403g0060.1 | Nt06 | 3 | 373 | 43,268.20 | 6.39 | Nuclear |
| NtMYB163 | Nitab4.5_0001087g0070.1 | Nt16 | 2 | 316 | 36,177.08 | 9.40 | Nuclear |
| NtMYB164 | Nitab4.5_0006037g0010.1 | Nt05 | 3 | 338 | 38,823.57 | 6.98 | Nuclear |
| NtMYB165 | Nitab4.5_0000174g0270.1 | Nt12 | 2 | 320 | 36,827.87 | 9.54 | Nuclear |
| NtMYB166 | Nitab4.5_0001612g0030.1 | Nt02 | 4 | 423 | 47,479.16 | 5.92 | Nuclear |
| NtMYB167 | Nitab4.5_0000325g0020.1 | Nt02 | 3 | 435 | 49,747.63 | 6.15 | Nuclear |
| NtMYB168 | Nitab4.5_0000918g0020.1 | Nt10 | 3 | 445 | 50,496.34 | 5.57 | Nuclear |
| NtMYB169 | Nitab4.5_0007287g0010.1 | Nitab4.5_0007287 | 1 | 377 | 43,129.36 | 9.36 | Nuclear |
| NtMYB170 | Nitab4.5_0000010g0220.1 | Nt13 | 3 | 442 | 49,473.52 | 6.24 | Nuclear |
| NtMYB171 | Nitab4.5_0003500g0020.1 | Nitab4.5_0003500 | 3 | 436 | 49,151.29 | 6.90 | Nuclear |
| NtMYB172 | Nitab4.5_0007139g0020.1 | Nitab4.5_0007139 | 1 | 361 | 41,265.04 | 9.24 | Nuclear |
| NtMYB173 | Nitab4.5_0007777g0020.1 | Nt20 | 12 | 471 | 52,750.84 | 5.87 | Nuclear |
| NtMYB174 | Nitab4.5_0003062g0070.1 | Nt24 | 12 | 459 | 51,678.72 | 6.34 | Nuclear |
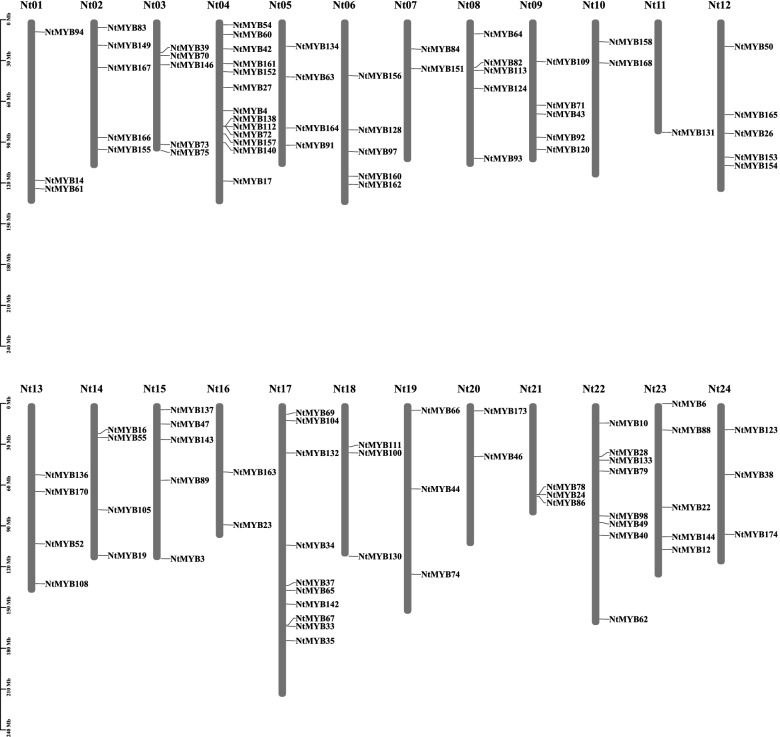
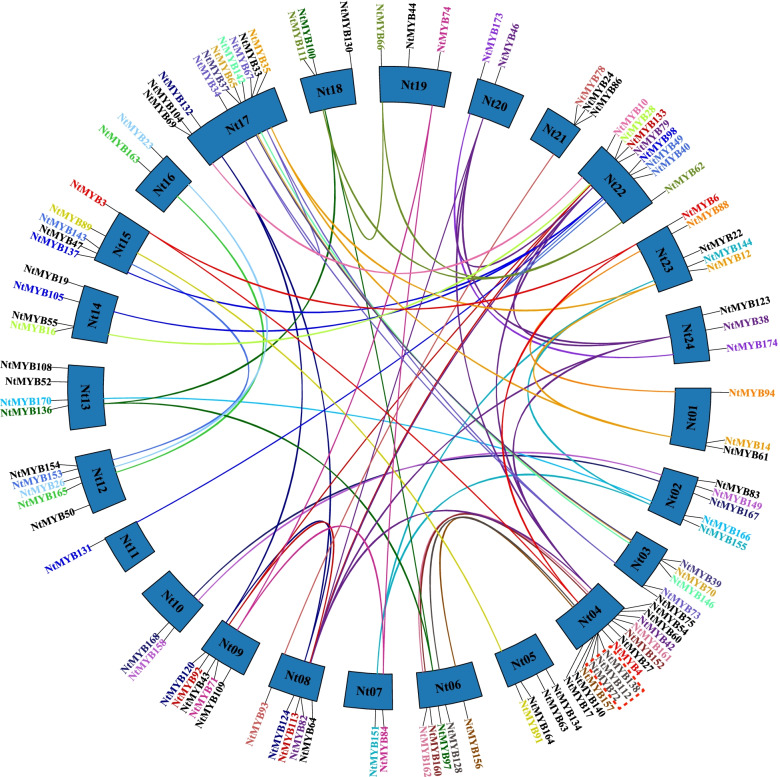
A total of 107 NtR2R3-MYB genes were unevenly distributed on 24 chromosomes of tobacco, while 67 genes were mapped to unattributed scaffolds (Fig. 1). Chromosome 4 contained the biggest number of NtR2R3-MYBs (13 genes), while chromosome 17 contained 10 NtR2R3-MYBs, and chromosome 22 had 8 NtR2R3-MYBs. In contrast, chromosome 11 only contained 1 NtR2R3-MYB gene. In our study, 3 pairs of tandem duplication genes on chromosome 4 (NtMYB72/112, NtMYB72/138, NtMYB112/138) and 62 pairs of segmental duplication genes were identified in tobacco R2R3-MYB gene family (Fig. 2, Additional file 2: Table S2).
Fig. 1.
The distribution of NtR2R3-MYB genes on 24 chromosomes in tobacco. The scale bar of the left displays the length of tobacco chromosomes
Fig. 2.
Gene duplication of NtR2R3-MYB genes on 24 chromosomes of tobacco genome. The segmental duplicated gene pairs are linked by different colored lines, and the tandem duplicated gene pairs of Nt04 are marked with red dashed lines. The corresponding relationships of duplicated gene are listed in Additional file 2: Table S2
Phylogenetics and gene structure of the NtR2R3-MYBs
To investigate the evolutionary relationships among NtR2R3-MYB genes, a phylogenetic tree was constructed based on the amino acid sequences of 174 NtR2R3-MYB genes by using the MEGA-X software (Fig. 3A). The NtR2R3-MYBs were classified into 24 subgroups (A to X) with at least 50% bootstrap supported on phylogenetic trees (Fig. 3A). However, 5 NtR2R3-MYBs (NtMYB139, NtMYB9, NtMYB61, NtMYB103 and NtMYB27) could not be assigned to any of the 24 subgroups due to the low bootstrap values (< 50%). Among these subgroups, subgroup A (27 members) and X (21 members) were the two largest groups and these two subgroups represented more than 27% of the total NtR2R3-MYB members. In contrast, subfamilies B, C, F, J, K, N and T only contained two members.
Fig. 3.
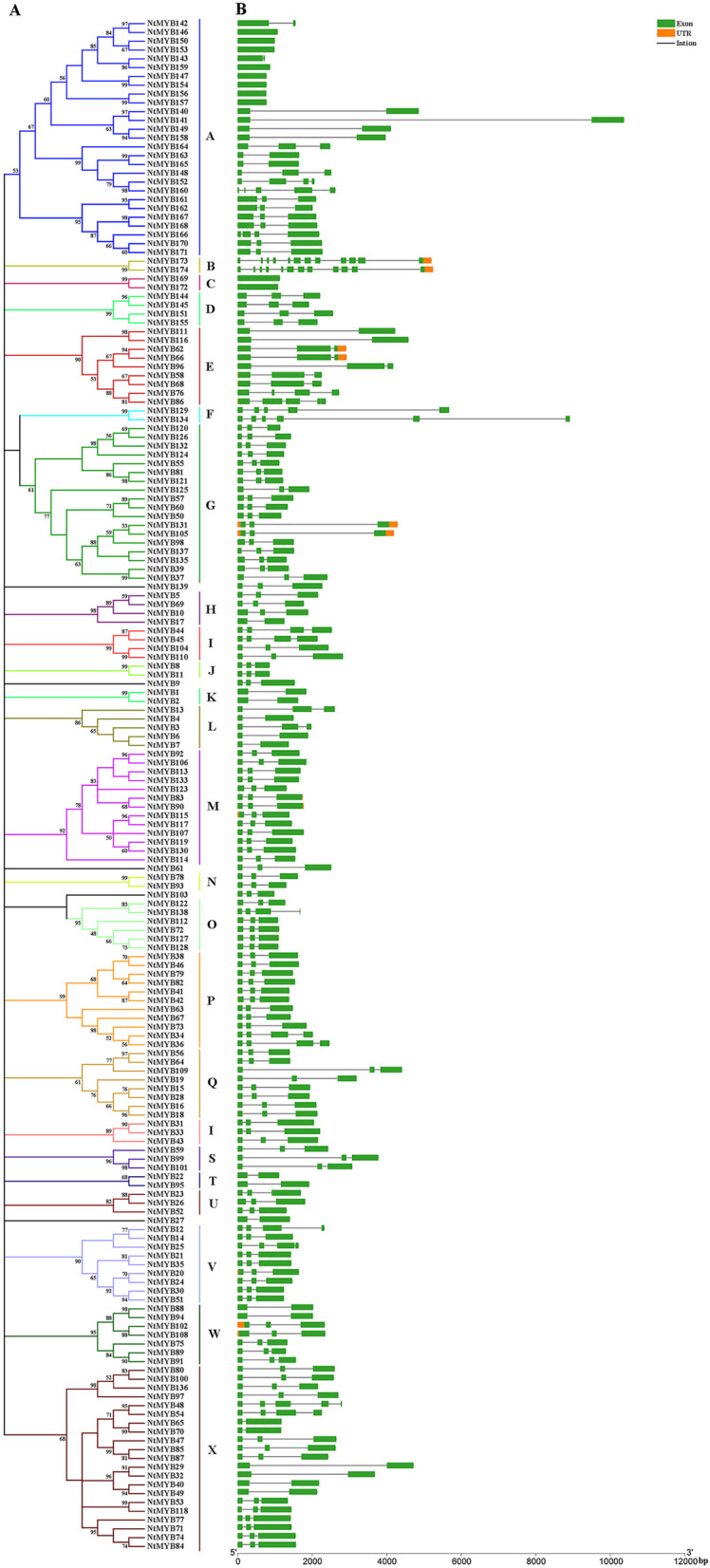
Gene structure and evolution of R2R3-MYB family in Nicotiana tabacum. A Phylogenetic relationships of NtR2R3-MYBs. Different subgroups were marked with different colors. B Intron-exon structure of NtR2R3-MYBs. Green boxes: exons; Yellow boxes: UTR; spaces between the boxes: introns. The scale bar of bottom demonstrates the length of exons and introns
Gene structure (Fig. 3B) analysis of NtR2R3-MYBs showed that the number of introns was varied from 0 to 11. The highest number (11) of introns was possessed by NtMYB173 and NtMYB174. Notably, a great number of R2R3-MYB genes (119 members, 68.39%) had a conserved gene structure with two introns and three exons, while 10 NtR2R3-MYBs completely lacked the introns. Similar exon-intron structural patterns were found among members within the same subgroup, especially the exon number and exon length were relatively conservative (Fig. 3).
Domain and motif analysis of the NtR2R3-MYBs
A total of 20 conserved motifs were predicted for 174 NtR2R3-MYB proteins using the online MEME program (Fig. 4). The lengths and conserved sequence of each motif is listed in Additional file 3: Table S3. The motif composition and distribution were found relatively conservative among members within the same subgroup (Fig. 4). The motif1, motif2 and motif3 located in the N-terminal of the majority NtR2R3-MYB protein sequences. There were conserved tryptophan residues in these three motifs, which were related to R2 and R3 domains. The type and number of motifs were similar in same subgroup, which suggested that motif pattern might be related to the function of MYB protein. Different subgroups usually possessed specific motifs, most of which located in the C-terminal. For example: motif7 and motif9 were specific to P; motif10 and motif13 only appear in A; motif8, motif14, and motif18 was presented in G, L, and Q alone, respectively.
Fig. 4.
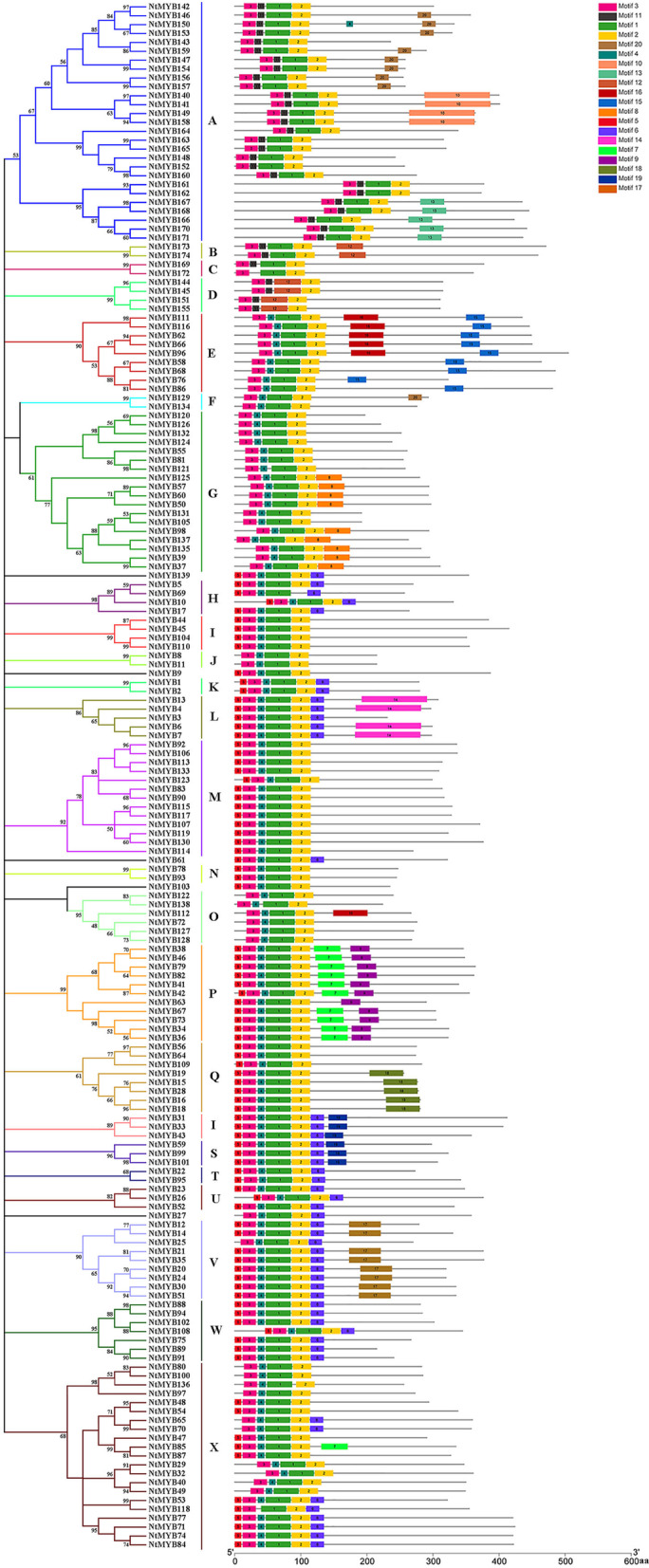
Conserved motifs for NtR2R3-MYB proteins in Nicotiana tabacum. Different motifs are showed with different colored boxes and numbers (1–20). The gray lines represent the non-conserved sequences. The lengths of motifs can be estimated using the scale at the bottom
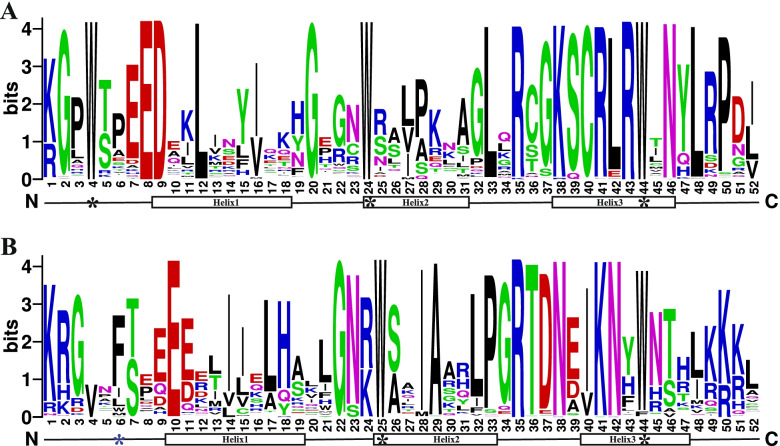
To further explore the conservative domain of the NtR2R3-MYB proteins, the multiple alignment of the 174 NtR2R3-MYB protein sequences was performed based on DNAMAN software, and the R2 and R3 sequence logos of MYB were generated by WebLogo (Fig. 5). As a result, NtR2R3-MYB members possess the typical characteristics of MYB conserved domains, and the R2 and R3 repeat of NtR2R3-MYB contain about 52 amino acid residues. The R2 repeat contains three highly conserved tryptophan residues (W), forming a hydrophobic core zin HTH structure. The first tryptophan residues (W) of R3 repeat often was replaced by a phenylalanine (F), isoleucine (I) or leucine (L) residues, whereas the second and third tryptophan residues were highly conserved, and this result was consistent with A. thaliana [4]. We also observed that some amino acid residues showed highly conservative, such as G-2, E-8, D-9, L-12, G-20, L-33, R-35, K-38, S-39, C-40, R-41, L-42, R-43, N-46, L-48 and P-50 in the R2 repeat and E-10, G-22, N-23, I-28, A-29, P-33, G-34, R-35, T-36, D-37, N-38, K-41 and N-42 in the R3 repeat. These highly conserved amino acid residues may be associated with conserved tryptophan residues to maintain the helix-turn-helix (HTH) structure of MYB transcription factor.
Fig. 5.
Sequence logos of the conserved R2 and R3 repeats of the NtR2R3-MYB domain. A Sequence logo of R2 in NtR2R3-MYBs. B Sequence logo of R3 in NtR2R3-MYBs. Asterisks indicate the conserved tryptophan residues (Trp) in the NtR2R3-MYB domain, and helix denote the helical structure among NtR2R3-MYBs
Promoter cis-elements analysis of NtR2R3-MYBs
Promoter cis-elements play critical roles in the initiation of gene expression. A total of 37 cis-regulatory were identified in the promoter region of NtR2R3-MYB genes, which could be classified into four categories elements, including cellular development, phytohormones, environmental stress and photoresponsive elements (Fig. 6, Additional file 4: Table S4). There were seven cis-acting elements related to cell development, such as CAT-box, MSA-like, GCN4_motif, CCAAT-box, MBSI, HD-Zip 1, and RY-element. Twelve phytohormone-responsive elements were identified, namely, CGTCA-motif, TGACG-motif, ABRE, P-box, TGA-element, TCA-element, AuxRR-core, TATC-box, GARE-motif, AuxRE, A-box and O2-site. These cis-elements are involved in JA/MeJA, abscisic acid, gibberellin, auxin, salicylic acid responsiveness and zein metabolism regulation. Meanwhile, the ABRE responsiveness elements were the most common in the NtR2R3-MYB gene promoters. In addition, ten light responsive elements were calculated, including GT1-motif, G-Box, Box 4, MRE, ATC-motif, Sp1, ATCT-motif, ACE, 3-AF1 binding site, and AAAC-motif. Almost all NtR2R3-MYB genes contained at least one phytohormone-responsive element in their promoter regions. There were 8 cis-regulatory elements that associated with response to external or environmental stresses were also present. This category includes low-temperature responsive element (LTR), anaerobic induction elements (ARE, GC-motif), drought-inducibility element (MBS), defense and stress responsive element (TC-rich repeats), circadian control element (circadian), wound-responsive element (WUN-motif), as well as AT-rich element. G-Box, Box 4, and ARE elements appear in most promoters of NtR2R3-MYB genes. The expression of these genes might be regulated by phytohormones, diverse light-responsiveness cis-elements, defense signaling transduction, and abiotic stresses during tobacco growth.
Fig. 6.
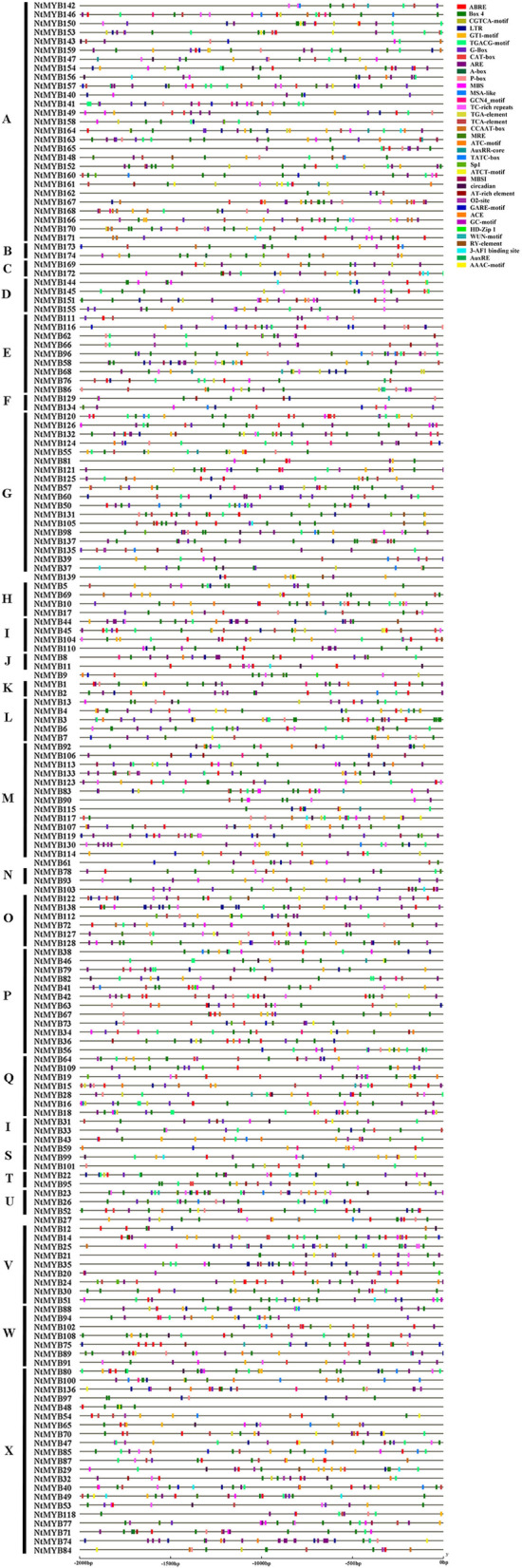
Predicted cis-elements in NtR2R3-MYBs promoters. Different shapes and colors represent the different types of cis-elements. Annotations of cis-elements were listed in Additional file 4: Table S4
Phylogenetic analysis of the NtR2R3-MYB gene family
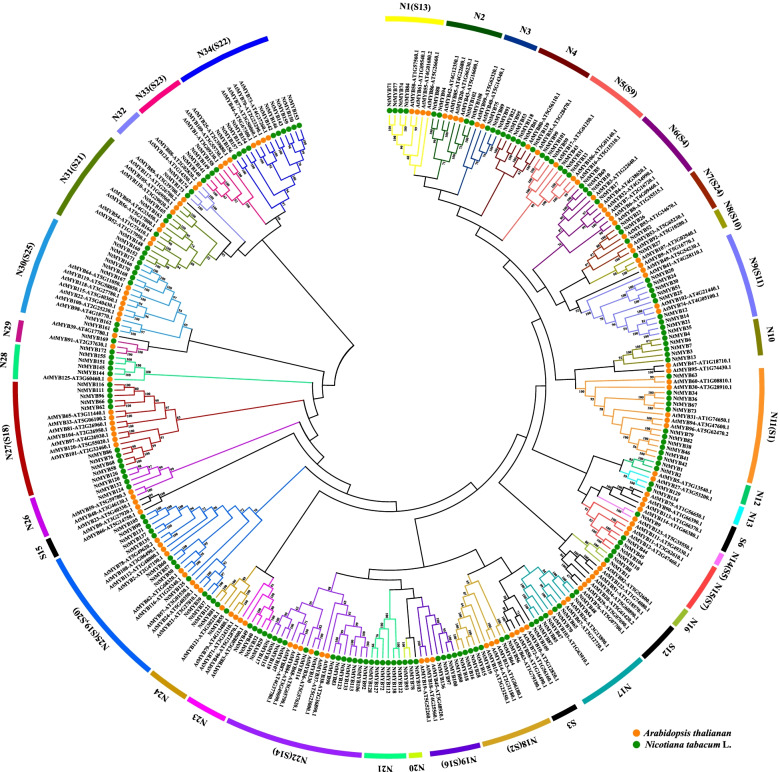
To investigate the phylogenetic relationships of the R2R3-MYB gene family, phylogenetic tree was generated based on the 174 tobacco R2R3-MYB protein sequences and 126 Arabidopsis R2R3-MYB protein sequences by using MEGA-X with maximum likelihood (ML) method (Fig. 7). According to the bootstrap values (> 50%) of the phylogenetic tree, the R2R3-MYB family were clustered into 38 subfamily. Among them, the R2R3-MYB members of tobacco were distributed in 34 subgroups (named N1-N34). There was no NtR2R3-MYB member distributed on the subfamily of S3, S6, S12 and S15, while three subfamilies (N10, N20, N21) only contain the R2R3-MYB members from tobacco. In addition, the NtR2R3-MYB members were mainly distributed in N9(9), N11(11), N22(13), N25(11), N27(9) and N34(10), and the number of R2R3-MYB members of tobacco were about twice to triple than that in Arabidopsis in these subfamilies. These results indicated that there were some common ancestors of R2R3-MYB genes between tobacco and Arabidopsis, and specific expansion and divergence also occurred after their separation during the evolution process.
Fig. 7.
Phylogenetic tree of Nicotiana tabacum and Arabidopsis thaliana R2R3-MYB genes. The phylogenetic relationships were constructed using MEGA-X by the maximum likelihood (ML) method (1000 bootstrap replicates). The amino acid sequences of 174 NtR2R3-MYB and 126 AtR2R3-MYB proteins were used. Arabidopsis and tobacco are marked with yellow circles and green circles respectively
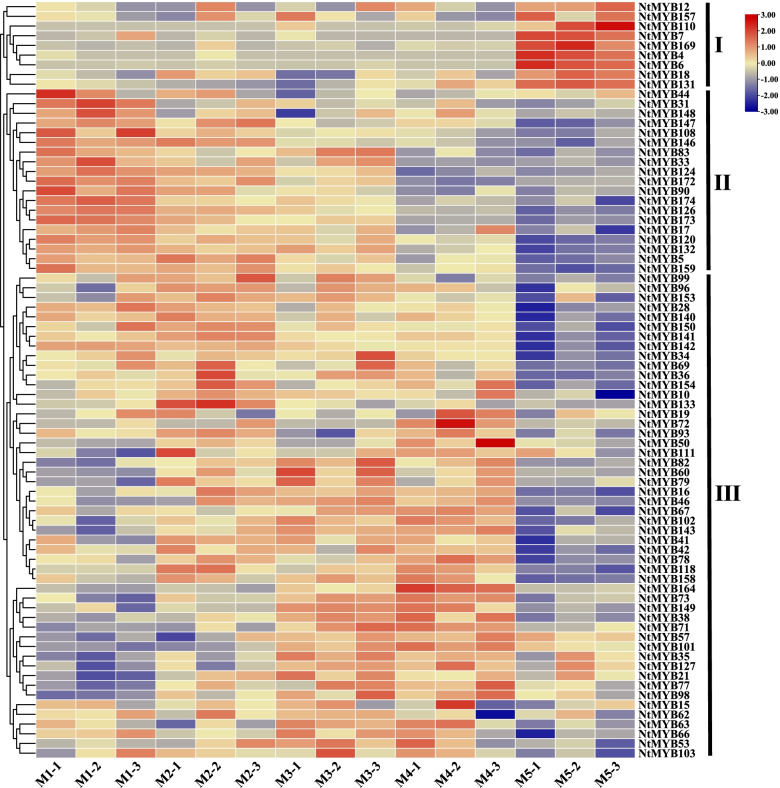
Expression changes of NtR2R3-MYB genes in five senescence stages of tobacco leaves
To analyze the expression pattern of NtR2R3-MYBs in tobacco leaves, the FPKM values of NtR2R3-MYB genes at five senescence stages of tobacco leaves were obtained from the transcriptome data (Additional file 5: Table S5), and NtR2R3-MYB genes with no expression or low expression level (FPKM < 0.5) were excluded. Finally, the expression profiles of NtR2R3-MYB genes of 78 NtR2R3-MYB genes were generated (Fig. 8). The results showed that the members of NtR2R3-MYB genes exhibited differential expression in tobacco leaves at different senescence stages (Fig. 8) and these 78 NtR2R3-MYB genes were classified into three groups (Fig. 8 I to III). A total of 9 NtR2R3-MYB members were including in group I, and these genes had high expression level at the M5 stage. In contrast, these genes showed relative low expression level at other four senescence stages (M1-M4). In group II, most of NtR2R3-MYB genes showed high expression level at the M1 stage and decreased regularly with the increase of maturity. In terms of group III, the expression level of NtR2R3-MYB genes showed increase first and then decreased with the increasing of the senescence degrees. The results indicated that there may be functional diversity among NtR2R3-MYB members during tobacco growth and development.
Fig. 8.
The expression of 78 NtR2R3-MYBs in tobacco leaves at five senescence stages. FPKM values for NtR2R3-MYB genes were transformed by log10. Red or blue colors represent the difference in expression levels in each sample (M1-M5), respectively
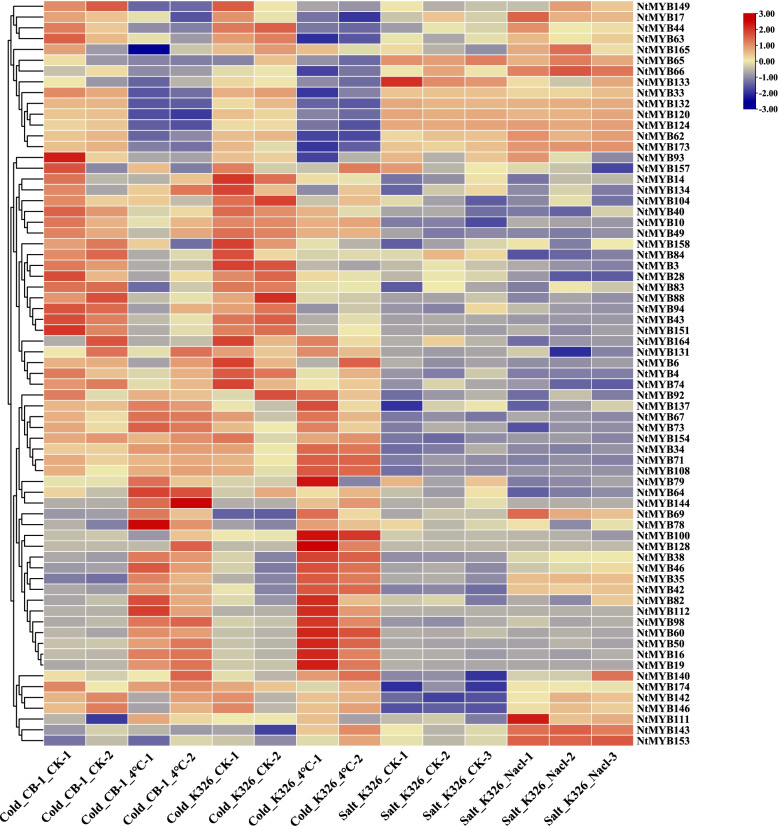
Expression of NtR2R3-MYB genes in response to abiotic stress
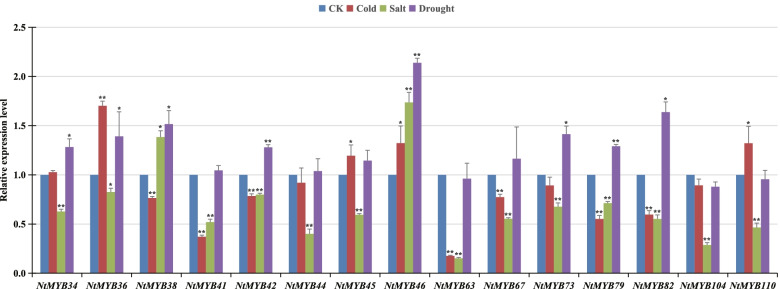
To analyze the expression pattern of NtR2R3-MYBs in response to abiotic stress, gene expression was investigated by using the Genevestigator tools based on transcriptome data. NtR2R3-MYB genes with no expression or low expression level (FPKM < 0.5) were excluded (Additional file 6: Table S6). Finally, the expression profiles of 69 NtR2R3-MYB genes were generated. The result showed that many genes showed significant up-regulated or down-regulated compared with the control group under cold and salt stress conditions (Fig. 9), and these genes including NtMYB34, NtMYB38, NtMYB42, NtMYB44, NtMYB46, NtMYB63, NtMYB67, NtMYB73, NtMYB79, NtMYB82 and NtMYB104 were clustered together with S1 and S7 subfamilies of Arabidopsis. In addition, it has been reported that the members of Arabidopsis R2R3-MYB gene family in S1 and S7 subgroups were related to various stress responses [25]. It is known that homologous genes in the same subgroup may have similar biological functions. To further explore the possible function of the tobacco R2R3-MYB genes, 15 tobacco R2R3-MYB genes that clustered with Arabidopsis R2R3-MYB gene members in S1 and S7 subgroups of the phylogenetic tree (Fig. 7) were selected for qRT-PCR analysis under cold, drought and salt stresses (Fig. 10). Compared with the control, the expression of four NtR2R3-MYB genes (NtMYB36, NtMYB45, NtMYB46 and NtMYB110) showed significantly up-regulated and seven NtR2R3-MYB genes (NtMYB38/41/42/63/67/79/82) showed significantly down-regulated under cold stress. As to the salt stress, the expression levels of 13 NtR2R3-MYB genes showed down-regulated, except MYB38 and MYB46 genes. In terms of drought stress, eight NtR2R3-MYB genes (NtMYB34/36/38/42/46/73/79/82) showed significantly up-regulated. Interestingly, the expression of NtMYB46 showed significantly up-regulated in response to all the stresses. In addition, the expression patterns of 15 genes in Fig. 9 and Fig. 10 were not completely consistent, and this phenomenon may be due to trial or sampling differences. The result implied the functional dissimilation among the tobacco R2R3-MYB genes.
Fig. 9.
Expression of 69 NtR2R3-MYB genes in response to cold and slat treatments. RNA-seq data was obtained by using Genevestigator software. Expression values from RNA-seq data were log10-transformed
Fig. 10.
Relative expression level of 15 NtR2R3-MYBs in response to cold, salt, and drought treatments. Error bars are standard deviations of three biological replicates. Asterisks were used to indicate the significant degree of the expression level compared to the value of the control (*P < 0.05, **P < 0.01)
Discussion
R2R3-MYB gene family members are widely distributed in eukaryotes [26]. With the development of genome sequencing, the whole-genome analysis of R2R3-MYB gene family has been identified in numerous species, including 126 in Arabidopsis thaliana, 89 in watermelon, 110 in rice, 157 in maize, 244 in soybean, and so on [7, 14, 27–29]. In this study, 174 R2R3-MYB genes of tobacco were identified, the number was greater than those identified in Arabidopsis, maize, rice, watermelon, but less than that in soybean. As an allotetraploid, the genome size of Nicotiana tabacum is 4.5Gb, while that of Arabidopsis, rice, maize, and soybean is 125 Mb, 430 Mb, 2300 Mb, and 1.025Gb, respectively [30–34]. In this case, it seems that there is no direct correlation between the number of R2R3-MYB genes and genome size in these plants. It was reported that polyploidization and gene region-specific duplication (tandem duplication and segmental duplications) were important mechanisms for the generation and expansion of gene families in plant [35]. In our study, phylogenetic analysis found that the majority subfamilies contained the R2R3-MYB members both from tobacco and Arabidopsis, and however, a few subfamilies possessed only the R2R3-MYB members either from tobacco or Arabidopsis, suggesting that they might be derived from a common ancestor, and moreover, the R2R3-MYB gene family could also be undergone species specific differentiation after their separation. A total of 3 pairs of tandem duplication genes and 62 pairs of segmental duplication genes were identified in tobacco R2R3-MYB gene family, implying that the segmental duplication events were the main source for the expansion of R2R3-MYB gene family in tobacco, and this result was possible due to the allotetraploid of tobacco.
The evolution of gene family largely depends on the organization of gene structure. The varied length of nucleotide sequence among 174 NtR2R3-MYBs indicated the complexity in the Nicotiana tabacum L. genome. The molecular weight and isoelectric point values of NtR2R3-MYB proteins were also different among family members, suggesting their functional divergence. In addition, NtR2R3-MYB proteins contained 20 conserved motifs with different compositions, and their members were clustered in the same subfamily containing similar type and number of motifs, demonstrating the conservation and diversification of R2R3-MYB gene family of tobacco. It has been reported that the ancestor MYB gene has no intron, but the intron insertion event occurred in the MYB domain region under very occasional condition, and this intron pattern was kept conserved during the long evolution process [5, 36]. In our study, the majority NtR2R3-MYB genes possess typically splicing pattern of three exons and two introns, which exists in the conserved R2 and R3 repeats, and this result was in consistent with the previous reports in other plants [14, 37]. In addition, the NtR2R3-MYB proteins are comprised of the highly conserved MYB domain (R2 repeat and R3 repeat), R2 repeat contained a conserved LRPD motif at the C-terminal and three highly conserved tryptophan residues (W), whereas R3 repeat exist diversity at the first tryptophan residues (W), which could be substituted by other residues such as phenylalanine (F), isoleucine (I) and leucine (L). Meanwhile, substitution of amino acid occurred frequently in some sites of the MYB domain, and these regions may play important roles for the evolution and functional differentiation of tobacco R2R3-MYB protein. Similar results have been reported in other species, including Arabidopsis, Zea mays (Fig. 5) [14, 25].
The cis-elements of promoter play a key role in initiating gene expression. Genes with different cis-regulatory elements in the promoter sequences of genes may result in different expression patterns in pepper [38]. In our study, a total of 37 cis-elements related sequences were identified in the promoter region of NtR2R3-MYB genes, and among them, 7 were related to cell development, 12 engaged in phytohormone-responsive, 10 for light responsive elements, and 8 involved in stresses cis-regulatory elements, suggesting the different function of regulatory elements of NtR2R3-MYB genes. These highly diverse cis-regulatory elements in the promoter region of NtR2R3-MYB genes may also reflect the functional divergence at the transcriptional level.
R2R3-MYB genes have been proved to be related to biotic and abiotic stress [8]. For example, TaMYB344-overexpressing of wheat enhanced the tolerance of transgenic tobacco to drought, heat and high salt stress [39]. OsMYB2 was induced by cold, salt, and dehydration stress in rice [40]. In present study, the profiling data of gene expression was used to dissect the functional roles of NtR2R3-MYB genes, and different expression patterns were identified among NtR2R3-MYB genes in response to various abiotic stresses (Fig. 9). This result indicated the functional differentiation of NtR2R3-MYB genes. In addition, the expression of 15 NtR2R3-MYB genes were analyzed by qRT-PCR in response to three abiotic stress conditions, including cold, salinity, and drought. A total of 13 NtR2R3-MYB genes could be significantly regulated by at least two treatments except NtMYB44 and NtMYB104, implied that NtR2R3-MYBs may be involved in the cross-talk of different signaling pathways under stress. Generally, genes with similar structure will be clustered in the same subfamily and these genes may have similar biological functions. It was reported that two R2R3-MYB genes of Arabidopsis (AtMYB60 and AtMYB94) clustered in subfamily S1 (Fig. 7) were involved in the physiological regulation under salt and drought treatments [41, 42], and therefore, the orthologous clustered in S1 may have similar function. In this study, two NtR2R3-MYB genes (NtMYB38 and NtMYB46) which clustered with these two R2R3-MYB genes of Arabidopsis (AtMYB60 and AtMYB94) in S1 subfamily showed similar response patterns under salt and drought stresses, suggesting that NtMYB38 and NtMYB46 may be involved in the response under salt and drought stresses, and further experiments showed be conducted to validation these functions.
Tobacco leaf senescence is often regarded as a kind of adversity, and R2R3-MYB genes were proved to be involved in the senescence accompany with secondary metabolites, including flavonoids, carotenoids, chlorophyll, and so on [23, 43]. In this study, diverse expression patterns were found among the NtR2R3-MYB genes inferring that the functional differentiation of family gene members should also be coexisting (Fig. 8). For example, NtMYB3/4 and NtMYB3/6 appeared to belong to segmental duplication genes pairs (Fig. 2, Additional file 2: Table S2), and our result showed that NtMYB4 and NtMYB6 expressed in all the five senescence stages, with especial high expression level in M5 stage, whereas NtMYB3 had no expression in all investigated senescence stages (M1-M5) (Fig. 8). It has been reported that there are three outcomes in the evolution of duplicate genes theoretically, including nonfunctionalization, neofunctionalization and subfunctionalization [44]. Therefore, it was inferred that NtMYB3 degenerated and lost its original function during the long evolution process. Notably, the expression level of majority members in group II including such as NtMYB146, NtMYB108, NtMYB90, NtMYB159, NtMYB120, NtMYB17 (Fig. 8) were decreased precisely corresponding consistent to the increasing of the senescence degrees, and these gene may closely relate to leaf senescence and can be further developed as the measure of leaf senescence or maturity. Whether or not the differential expression of NtR2R3-MYB genes leads to the changes of secondary metabolites such as flavonoids, carotenoids or chlorophyll still needs to be investigated. In short, our results provide using information for their further functional exploration.
Conclusions
In this study, a total of 174 R2R3-MYB genes were identified in tobacco (Nicotiana tabacum L.) genome and these genes were divided into 24 subfamilies. The NtR2R3-MYB genes were distributed randomly on 24 tobacco chromosomes. A total of 3 pairs of NtR2R3-MYB genes were founded to be originated from tandem duplication and 62 pairs NtR2R3-MYB genes were originated from segmental duplication. Cis-regulatory elements of the NtR2R3-MYB promoters were involved in cellular development, phytohormones, environmental stress and photoresponsive. The members of NtR2R3-MYB genes showed differential expression pattern in different maturity tobacco leaves, and differential response were also found under different abiotic stresses (cold, salt and drought) for 15 NtR2R3-MYBs. Our results provided valuable information for further functional study of NtR2R3-MYB genes in tobacco.
Methods
Identification of R2R3-MYB genes in tobacco
A total of 126 known Arabidopsis R2R3-MYB protein sequences were downloaded from the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/) database [25]. These sequences were used as queries using the online tool of BLASTP (E ≤ 1e− 5) for the identification of R2R3-MYB family members in the tobacco genome sequences of Sol Genomics Network database (https://solgenomics.net/organism/Nicotiana_attenuata/genome) [45, 46]. The redundant protein sequences of tobacco were removed manually, and then the candidate protein sequence which contained complete MYB domains (PF000249) were confirmed as the final R2R3-MYB protein sequence based on the Conserved Domain Database (CDD) of NCBI (https://www.ncbi.nlm.nih.gov/cdd/) [47]. These final tobacco R2R3-MYB genes were renamed (NtMYBs). The features and the subcellular localization information of the NtR2R3-MYB protein sequences were analyzed by the online ExPASY tool (http://Web.ExPASY.Org/protparam/) [48] and the Softberry service platform-ProtComp 9.0 (http://linux1.softberry.com/berry.phtml), respectively.
Gene structure and conserved motif analysis
The GFF format file of tobacco gene structure was downloaded from Solanaceae genome database (https://sol-genomics.net/) [46], and the NtR2R3-MYB gene structure (exon-intron) was defined using the online software Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/) [49]. The motifs of NtR2R3-MYB protein were obtained from the online MEME program (http://meme-suite.org/, v5.1.1) [50] and following parameters were used: the minimum width, maximum width and maximum number of motifs were set to 6 bp, 100 bp and 20, respectively. The conserved NtR2R3-MYB domain was visualized using the WebLogo platform (http://weblogo.berkeley.edu/) [51]. The cis-regulatory elements in the promoter region (2000 bp upstream of the starting codon) of the NtR2R3-MYBs were searched by the online program of PlantCARE (http://bioinformatics.psb.ugent.be/webtools /plantcare/html/) [52].
Chromosome localization and gene duplication
The physical position and chromosomal distribution information of NtR2R3-MYB genes were obtained by using the MapInspect software (http://mapinspect.software.informer.com/) [53]. The possible segmental duplication and tandem duplication events were defined based on the method reported by Wang et al. (2010) [54]. Both chromosomal localization and duplication events of the NtR2R3-MYB genes were graphic displayed using the TBtools software [55].
Multi-sequence alignment and phylogenetic classification
To explore the evolutionary relationship of R2R3-MYB gene family, the full protein sequences of R2R3-MYB from Arabidopsis and tobacco were used for the phylogenetic tree construction. Multiple sequence alignment was performed using ClustalW program in MEGA-X software [56], and the phylogenetic tree was constructed using the maximum likelihood (ML) with 1000 bootstraps.
NtR2R3-MYB genes expression analysis
The tobacco variety of Cuibi 1 (CB-1) was used in this study. To analyze the NtR2R3-MYB genes expression profile at different senescence stages, five senescence stages (M1, M2, M3, M4 and M5) of middle leaves (8th to 10th) judged by the appearance characteristics were collected for tests [57]. The FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) value of the NtR2R3-MYB genes at five senescence stages of tobacco leaves were extracted from our recent RNA-Seq data [58]. The expression profile of NtR2R3-MYB genes at different senescence stages were measured by their FPKM value, and the heat map was generated using the Heatmap function of R gplots package [59]. Additionally, expression analysis of NtR2R3-MYBs under salt (SRP193166) [60] and cold stress (SRP097876) [61] of tobacco were performed using Genevestigator software. Genes with low expression level (FPKM < 0.5) were filtered.
Plant treatments and quantitative real-time PCR analysis
To further decipher the expression pattern of NtR2R3-MYB genes in response to various abiotic, tobacco seeds were sown in sterilized mixed soil (vermiculite: humus = 1:1) under the condition of 22 °C and 16 h light/8 h dark photoperiod for 60 days [62]. The plantlets of 60 days were transplanted into a tray with a nutrient solution for 3 days in growth chamber, and then were exposed to the abiotic treatments, including cold (4 °C), drought (10% polyethylene glycol) and salt (200 mM NaCl), respectively. Untreated plantlets were used as control (CK). The samples for gene expression analysis were collected 6 h after treatment, and three biological replicates per treatment and 3 leaves for each sample from different plantlet were gathered and these samples were immediately stored at − 80 °C prior to RNA extraction.
Total RNA was extracted using the Hipure Plant RNA Mini Kit (Magen Biotech, Shanghai, China) and the cDNA was synthesized using the SMART kit (Takara) according to the manufacturer’s protocol. The qRT-PCR primers of NtR2R3-MYB genes were designed by online software primer3 (https://bioinfo.ut.ee/primer3-0.4.0/) [63] and were shown in Additional file 7: Table S7. Real-time quantitative RT-PCR (qRT-PCR) was performed with SYBR Green qPCR Premix (Low ROX). A total of 20 μl volume of reaction mixture for each PCR run was prepared, containing 1.5 μl cDNA, 1 × Taq SYBR Green qPCR Premix (Monad, China) and a primer pair with a concentration of 0.2 μM. The two-step thermal cycling profile used was 95 °C for 5 min, 40 cycles at 95 °C for 30 s, followed by 60 °C for 60 s. Three technical replicates were performed for each sample. The relative expression level was calculated by the 2-ΔΔCt method [64].
Supplementary Information
Acknowledgments
We appreciate the reviewers and editors for the patience to the work.
Abbreviations
- HTH
Helix-turn-helix structure
- MW
Molecular weight
- pI
Isoelectric points
- ML
Maximum likelihood
- NtR2R3-MYB
R2R3-MYB gene of Nicotiana tabacum
- FPKM
Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced
- GSDS
Gene Structure Display Server
- qRT-PCR
Quantitative real-time PCR
Authors’ contributions
Jiahan Yang designed and conducted the experiments, analyzed the data, wrote the manuscript, Binghui Zhang and Gang Gu contributed plant materials, performed the experiments and analyzed the data, Jiazheng Yuan helped to draft the manuscript, Shaojun Shen, Liao Jin, Zhiqiang Lin and Jianfeng Lin contributed analysis tools and analyzed the data, Xiaofang Xie the corresponding author, conceived and designed the experiments, wrote the manuscript. All authors have approved the manuscript.
Funding
This research was financially supported by Fujian Tobacco Company (2019350000240137) and Nanping Tobacco Company (NYK2021-10-03). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the the NCBI Sequence Read Archive repository, https://www.ncbi.nlm.nih.gov/sra/PRJNA772550.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6(12):3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol. 1996;3(2):178–187. doi: 10.1038/nsb0296-178. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149(2):981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C, Gu J, Chopra S, Gu X, Peterson T. Ordered origin of the typical two- and three-repeat Myb genes. Gene. 2004;326:13–22. doi: 10.1016/j.gene.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 6.Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol. 1998;46(1):74–83. doi: 10.1007/PL00006285. [DOI] [PubMed] [Google Scholar]
- 7.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, Yunping S, Li Z, Xiaohui D, Jingchu L, Xing-Wang D, Zhangliang C, Hongya G, Li-Jia Q. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 8.Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13(2):67–73. doi: 10.1016/S0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 9.Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41(5):577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- 10.Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. Control of cell and petal morphogenesis by R2R3-MYB transcription factors. Development. 2007;134(9):1691–1701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- 11.Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 2008;148(3):1583–1602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Cao G, Qu LJ, Gu H. Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genet Genomics. 2009;36(2):99–107. doi: 10.1016/S1673-8527(08)60096-X. [DOI] [PubMed] [Google Scholar]
- 13.Cone KC, Cocciolone SM, Burr FA, Burr B. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell. 1993;5(12):1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du H, Feng BR, Yang SS, Huang YB, Tang YX. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012;7(6):e37463. doi: 10.1371/journal.pone.0037463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ampomah-Dwamena C, Thrimawithana AH, Dejnoprat S, Lewis D, Espley RV, Allan AC. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019;221(1):309–325. doi: 10.1111/nph.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Ren G, Guirgis A, Thornburg RW. The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell. 2009;21(9):2672–2687. doi: 10.1105/tpc.108.060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onkokesung N, Reichelt M, van Doorn A, Schuurink RC, van Loon JJ, Dicke M. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: role of kaempferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J Exp Bot. 2014;65(8):2203–2217. doi: 10.1093/jxb/eru096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y, Bao X, Zhi Y, Wu Q, Guo Y, Yin X, Zeng L, Li J, Zhang J, He W, Liu W, Wang Q, Jia C, Li Z, Liu K. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic Rice. Front Plant Sci. 2019;10:168. doi: 10.3389/fpls.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Q, Liu R, Zeng L, Wu Y, Jiang Y, Yang Q, Nie Q. Isolation and molecular characterization of NtMYB4a, a putative transcription activation factor involved in anthocyanin synthesis in tobacco. Gene. 2020;760:144990. doi: 10.1016/j.gene.2020.144990. [DOI] [PubMed] [Google Scholar]
- 20.Song Z, Luo Y, Wang W, Fan N, Wang D, Yang C, Jia H. NtMYB12 positively regulates Flavonol biosynthesis and enhances tolerance to low pi stress in Nicotiana tabacum. Front Plant Sci. 2020;10:1683. doi: 10.3389/fpls.2019.01683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen WK, Yu KJ, Liu B, Lan YB, Sun RZ, Li Q, He F, Pan QH, Duan CQ, Wang J. Comparison of transcriptional expression patterns of carotenoid metabolism in 'Cabernet Sauvignon' grapes from two regions with distinct climate. J Plant Physiol. 2017;213:75–86. doi: 10.1016/j.jplph.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Hörtensteiner S. Chlorophyll degradation during senescence. Annu Rev Plant Biol. 2006;57:55–77. doi: 10.1146/annurev.arplant.57.032905.105212. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Wang L, Liu H, Zhao R, Liu B, Fu Q, Zhang Y. The antioxidative defense system is involved in the premature senescence in transgenic tobacco (Nicotiana tabacum NC89) Biol Res. 2016;49(1):30. doi: 10.1186/s40659-016-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Zhai J, Shao L, Lin W, Peng C. Accumulation of Anthocyanins: an adaptation strategy of Mikania micrantha to low temperature in winter. Front Plant Sci. 2019;10:1049. doi: 10.3389/fpls.2019.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4(5):447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 26.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 27.Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics. 2012;13:544. doi: 10.1186/1471-2164-13-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Liu Y, Chen XL, Kong QS. Characterization and divergence analysis of duplicated R2R3-MYB genes in watermelon. J Am Soc Hortic Sci. 2020;145(5):281. doi: 10.21273/JASHS04849-19. [DOI] [Google Scholar]
- 30.Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. 2014;5:3833. doi: 10.1038/ncomms4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaul S, Koo HL, Jenkins J, et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 32.Burr B. Mapping and sequencing the rice genome. Plant Cell. 2002;14(3):521–523. doi: 10.1105/tpc.140310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 34.Schmutz J, Cannon SB, Schlueter J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 35.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedorova L, Fedorov A. Introns in gene evolution. Genetica. 2003;118(2–3):123–131. doi: 10.1023/A:1024145407467. [DOI] [PubMed] [Google Scholar]
- 37.Jiang C, Gu X, Peterson T. Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 2004;5(7):R46. doi: 10.1186/gb-2004-5-7-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Islam S, Sajib SD, Jui ZS, Arabia S, Ghosh A. Genome-wide identification of glutathione S-transferase gene family in pepper, its classification, and expression profiling under different anatomical and environmental conditions. Sci Rep. 2019;9:15. doi: 10.1038/s41598-018-36763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei Q, Chen R, Wei X, Liu Y, Zhao S, Yin X, Xie T. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genomics. 2020;21(1):792. doi: 10.1186/s12864-020-07175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63(7):2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15(13):1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 42.Lee SB, Suh MC. Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 2015;56(1):48–60. doi: 10.1093/pcp/pcu142. [DOI] [PubMed] [Google Scholar]
- 43.Zhu F, Luo T, Liu C, Wang Y, Yang H, Yang W, Zheng L, Xiao X, Zhang M, Xu R, Xu J, Zeng Y, Xu J, Xu Q, Guo W, Larkin RM, Deng X, Cheng Y. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017;216(1):178–192. doi: 10.1111/nph.14684. [DOI] [PubMed] [Google Scholar]
- 44.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 45.Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, Allen F, Hurst R, White B, Kernodle SP, Bromley JR, Sanchez-Tamburrino JP, Lewis RS, Mueller LA. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics. 2017;18(1):448. doi: 10.1186/s12864-017-3791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, Bombarely A, Fisher-York T, Pujar A, Foerster H, Yan A, Mueller LA. The Sol Genomics Network (SGN)--from genotype to phenotype to breeding. Nucleic Acids Res. 2015;43:D1036–D1041. doi: 10.1093/nar/gku1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 49.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu GQ, Li ZQ, Cao H, Wang JL. Genome-wide identification and expression analysis of the WRKY genes in sugar beet (Beta vulgaris L.) under alkaline stress. PeerJ. 2019;7:e7817. doi: 10.7717/peerj.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, Cui X, Peng L. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010;10:282. doi: 10.1186/1471-2229-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin M, Zhang B, Gu G, Yuan J, Yang X, Yang J, Xie X. Genome-wide analysis of the G2-like transcription factor genes and their expression in different senescence stages of tobacco (Nicotiana tabacum L.) Front Genet. 2021;12:626352. doi: 10.3389/fgene.2021.626352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B, Yang J, Gu G, Jin L, Chen C, Lin Z, Song J. Xie X. integrative analyses of biochemical properties and transcriptome reveal the dynamic changes in leaf senescence of tobacco (Nicotiana tabacum L.) Front Genet. 2021;12:790167. doi: 10.3389/fgene.2021.790167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 60.Xu J, Chen Q, Liu P, Jia W, Chen Z, Xu Z. Integration of mRNA and miRNA analysis reveals the molecular mechanism underlying salt and alkali stress tolerance in tobacco. Int J Mol Sci. 2019;20:2391. doi: 10.3390/ijms20102391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin J, Zhang H, Zhang J, Liu P, Chen X, Li Z, Xu Y, Lu P, Cao P. Integrated transcriptomics and metabolomics analysis to characterize cold stress responses in Nicotiana tabacum. BMC Genomics. 2017;18:496. doi: 10.1186/s12864-017-3871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song Z, Pan F, Yang C, Jia H, Jiang H, He F, Li N, Lu X, Zhang H. Genome-wide identification and expression analysis of HSP90 gene family in Nicotiana tabacum. BMC Genet. 2019;20:35. doi: 10.1186/s12863-019-0738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the the NCBI Sequence Read Archive repository, https://www.ncbi.nlm.nih.gov/sra/PRJNA772550.