Abstract
Cryptosporidium parvum is a parasitic zoonotic pathogen responsible for diarrheal illness in humans and animals worldwide. We report an investigation of a cryptosporidiosis outbreak in raccoons and wildlife rehabilitation workers at a Virginia facility. Fifteen (31%) of 49 facility personnel experienced symptoms meeting the case definition, including four laboratory-confirmed cases. Seven juvenile raccoons were reported to have diarrhoea; six had laboratory-confirmed cryptosporidiosis. Cryptosporidium parvum of the same molecular subtype (IIaA16G3R2) was identified in two human cases and six raccoons. Raccoon illness preceded human illness by 11 days, suggesting possible zoonotic transmission from raccoons to humans. This appears to be the first report of a human cryptosporidiosis outbreak associated with exposure to raccoons infected with C. parvum. Raccoons might be an under-recognized reservoir for human C. parvum infections. Further study is needed to explore the prevalence of cryptosporidial species in raccoons and their role as a wildlife reservoir.
Keywords: Cryptosporidium parvum, outbreak, Procyon lotor, raccoons
1 |. INTRODUCTION
Cryptosporidium parvum is a protozoan parasite causing diarrheal illness in both humans and animals worldwide. Infection occurs through ingestion of infectious oocysts; human transmission can occur directly from person-to-person or animal-to-person, or through ingestion of contaminated food or water (Ryan et al., 2014). Illness is characterized by watery diarrhoea and abdominal cramping beginning 7–10 days after exposure and lasting 7–14 days, but shedding of oocysts might continue after symptoms resolve (Chen et al., 2002; Hunter et al., 2004).
On 19 June 2019, a wildlife rehabilitation facility notified the Lord Fairfax Health District (LFHD) in Virginia of an outbreak of gastrointestinal illness (vomiting and diarrhoea) affecting 10 (42%) of 24 full-time staff and interns. Twenty-five volunteers worked part-time at the facility; none were known to be ill at the time of the initial report. The wildlife facility cared for a variety of animal species native to the area. When the human outbreak was identified, the facility also reported juvenile raccoons with intermittent diarrhoea, but other animals were described as healthy. The raccoons arrived at the facility during May 13–June 7, 2019 and the first documented diarrhoea amongst raccoons occurred on May 28. Illness onset in the first human case occurred on June 8. Based on the human symptoms reported (vomiting and diarrhoea) and negative raccoon faecal floatation testing for ova and parasites (O&P), the LFHD recommended norovirus testing and infection control measures. On June 20, 2019, the facility notified the LFHD that one person tested positive for Cryptosporidium antigen at a private laboratory. LFHD conducted an investigation to confirm the etiologic agent, determine the outbreak source, identify risk factors for illness and prevent additional transmission.
2 |. MATERIALS AND METHODS
2.1 |. Epidemiologic study
A retrospective cohort study was designed to better understand clinical illness and possible exposures; a questionnaire was developed and distributed to all 49 facility personnel, including seven staff, 17 interns and 25 volunteers. The questionnaire captured data on demographic characteristics, illness, healthcare-seeking behaviours, contact with animals (by species) and food and water exposure. As the investigation progressed, a supplemental questionnaire was created to collect additional detail about the type of animal care each person provided, personal protective equipment used and adherence to recommended cleaning protocols. Information about opportunities for person-to-person exposure and animal care, housing and feeding was gathered during a site visit. The facility provided a list of dates each person worked from May 25 to June 26, 2019, as well as data on ill raccoons at the facility, including intake date, location where the animal was found and onset date of diarrhoea.
For the purposes of the outbreak investigation, a probable case of human illness was defined as gastrointestinal illness occurring after June 6, 2019 in a person who worked or volunteered at the wildlife facility, characterized by diarrhoea and one or more of the following: abdominal cramping, vomiting, anorexia or diarrhoea lasting 72 hr or more. A confirmed case met probable case criteria and had laboratory evidence of Cryptosporidium infection.
Survey data were entered and stored in REDCap® and analysis was performed in Epi Info™ 7. The study population was characterized by age, sex and role (i.e. staff, intern, volunteer). Attack rates, risk ratios and 95% confidence intervals were calculated for specific animal exposures associated with the wildlife facility, contact with animals outside the facility, handling of produce for feeding the animals, drinking facility well water, contact with recreational water and use of personal protective equipment. Fisher’s exact test was used to identify statistically significant exposures (p < .05). Multivariate analyses were performed to assess the association between relevant exposures and illness. This study did not involve the use of human or animal research subjects but was conducted as routine public health practice in accordance with Virginia Department of Health outbreak investigation policy. As such, ethical review from either an Institutional Review Board or Institutional Animal Care and Use Committee was not sought.
2.2 |. Laboratory investigation
Stool specimens from four ill persons were collected for real-time polymerase chain testing at the Virginia Division of Consolidated Laboratory Services (DCLS) for norovirus and Cryptosporidium. Two additional human stool specimens were tested via clinical laboratories; one for Cryptosporidium and one for a general O&P. The Virginia Department of Agriculture and Consumer Services tested six raccoon stool specimens using a modified Kinyoun stain and Pathasure® enzyme-linked immunosorbent assay (ELISA).1 Human and raccoon specimens were forwarded to the CryptoNet Reference Laboratory at the Centers for Disease Control and Prevention (CDC) for genotyping (Roellig & Xiao, 2020). One raccoon was euthanized for reasons unrelated to this outbreak; a small intestinal tissue specimen from this animal was also forwarded to CDC CryptoNet for Cryptosporidium testing.
2.3 |. Environmental investigation
The facility was served by a private well. The Virginia Department of Health Office of Drinking Water collected three water samples from pretreatment taps inside and outside the building; these were tested by DCLS for bacterial indicators of faecal contamination (total coliforms and Escherichia coli) with the IDEXX Colilert test detection and quantification by most probable number (MPN). Facility records were reviewed to assess the geographic locations where raccoons were found prior to facility intake.
3 |. RESULTS
3.1 |. Epidemiologic study
Of 49 facility workers, 43 (88%) completed the initial survey. Fifteen respondents (35%) experienced illness meeting the case definition, including four confirmed cases (Table 1). Four people, including the person with the earliest symptom onset date (June 6), experienced one or more symptoms but did not meet the outbreak case definition and were excluded from further analysis. Amongst cases, illness was characterized by diarrhoea (15; 100%), abdominal cramps (13; 87%), headache (11; 73%) and vomiting (8; 53%). Fewer than half of ill persons reported fever (6; 40%) and no one reported blood in the stool. Illness duration ranged from 1 to 13 days (median = 4 days). Five persons sought healthcare; no one was hospitalized. Most respondents (36; 92%) reported performing adequate hand hygiene whilst at the facility. Contact with raccoons (risk ratio [RR] = 4.4; 95% confidence interval [CI] = 1.7–11.3), contact with foxes (RR = 4.0; 95% CI = 1.5–10.2), contact with cottontails (RR = 2.8; 95% CI = 0.9–8.3) and drinking facility tap water (RR = 3.8; 95% CI = 1.5–10.0) were significantly associated with illness (Table 2). Of 15 persons with raccoon contact, 14 (93%) also reported contact with foxes (p < .01). Because fox and raccoon contact were not independent (only those vaccinated against rabies could care for these species) and foxes were not demonstrating clinically significant illness, fox contact was not included in the multivariate logistic regression model. In the multivariate model, contact with raccoons (p < .01) and drinking tap water (p < .05) remained significantly associated with illness.
TABLE 1.
Characteristics of individuals working or volunteering at wildlife facility associated with cryptosporidiosis outbreak, Virginia, June 2019
| Ill n (%) | Not Ill n (%) | Total n (%) | |
|---|---|---|---|
| Total | 15 (38) | 24 (62) | 39 (100) |
| Case status | |||
| Confirmed | 4 (27) | 0 | 4 (10) |
| Probable | 11 (73) | 0 | 11 (28) |
| Not a case | 0 | 24 (100) | 24 (62) |
| Role | |||
| Full-time employee | 3 (20) | 0 | 3 (8) |
| Intern | 9 (60) | 6 (25) | 15 (38) |
| Volunteer | 3 (20) | 18 (75) | 21 (54) |
| Sex | |||
| Female | 14 (93) | 19 (79) | 33 (85) |
| Male | 1 (7) | 5 (21) | 6 (15) |
| Age in years: median (range) | 21 (18–55) | 26 (19–74) | 25 (18–74) |
| Days worked between 5/25–6/26: median (range) | 9 (3–20) | 4 (1–13) | 5 (1–20) |
TABLE 2.
Cryptosporidiosis attack rates and risk ratios, by exposure, Virginia, June 2019
| Bivariate exposure analysis | Exposed | Unexposed | |||||
|---|---|---|---|---|---|---|---|
| Ill/total | Attack rate | Ill/total | Attack rate | Risk ratio | 95% CI | p-value | |
| Wildlife facility | |||||||
| Cottontails | 12/23 | 52.2 | 3/16 | 18.8 | 2.8 | 0.9–8.3 | <.05 |
| Foxes | 11/16 | 68.8 | 4/23 | 17.4 | 4.0 | 1.5–10.2 | <.01 |
| Frogs | 4/13 | 30.8 | 11/26 | 42.3 | 0.7 | 0.3–1.8 | .7 |
| Opossums | 14/35 | 40.0 | 1/4 | 25.0 | 1.6 | 0.3–9.2 | 1 |
| Raccoons | 11/15 | 73.3 | 4/24 | 16.7 | 4.4 | 1.7–11.3 | <.01 |
| Raptors | 8/24 | 33.3 | 7/15 | 46.7 | 0.7 | 0.3–1.6 | .3 |
| Snakes | 5/15 | 33.3 | 10/24 | 41.7 | 0.8 | 0.3–1.9 | .7 |
| Songbirds | 12/30 | 40.0 | 3/9 | 33.3 | 1.2 | 0.4–3.3 | 1 |
| Squirrels | 6/17 | 35.3 | 9/22 | 40.9 | 0.9 | 0.4–1.5 | .8 |
| Turtles | 12/33 | 36.4 | 3/6 | 50.0 | 0.7 | 0.3–1.8 | .7 |
| Handle animal produce | 13/31 | 41.9 | 1/7 | 14.3 | 2.9 | 0.5–18.8 | .2 |
| Drink well water | 10/15 | 66.7 | 4/23 | 17.4 | 3.8 | 1.5–10.0 | <.01 |
| Wear PPE | 14/31 | 45.2 | 0/7 | 0.0 | Undefined | ||
| Use hose to clean | 10/20 | 50.0 | 1/6 | 16.7 | 3.0 | 0.5–18.9 | 0.2 |
| Clean cages | 11/23 | 47.8 | 0/3 | 0.0 | Undefined | ||
| Feed animals | 11/23 | 47.8 | 0/3 | 0.0 | Undefined | ||
| Restrain animals | 8/18 | 44.4 | 3/8 | 37.5 | 1.2 | 0.4–3.3 | 1 |
| Nonfacility exposures | |||||||
| Animals outside facility | 12/32 | 37.5 | 3/6 | 50.0 | 0.8 | 0.3–1.9 | .7 |
| Recreational water | 4/7 | 57.1 | 9/30 | 30.0 | 1.9 | 0.8–4.4 | .2 |
| Unpasteurized dairy | 3/5 | 60.0 | 7/20 | 35.0 | 1.7 | 0.7–4.3 | .4 |
| Multivariate analysis | Adjusted odds ratio | 95% CI | p-value |
|---|---|---|---|
| Cottontails | 2.5 | 0.3–18.2 | .4 |
| Raccoons | 12.0 | 1.9–76.4 | <.01 |
| Drink well water | 7.6 | 1.2–48.8 | <.05 |
Seven raccoons had documented diarrhoea. The median number of days between intake and diarrhoea onset was 3 days (range: 2–42 days). Onset of diarrhoea in the first raccoon occurred on May 28, followed 5 days later (June 2) by onset in its cage-mate and 8 days later (June 5) by a third raccoon in another cage. The first human illness meeting the outbreak case definition was on June 8 in an intern who cared for raccoons. Additional raccoon and human cases of illness followed (Figure 1).
FIGURE 1.
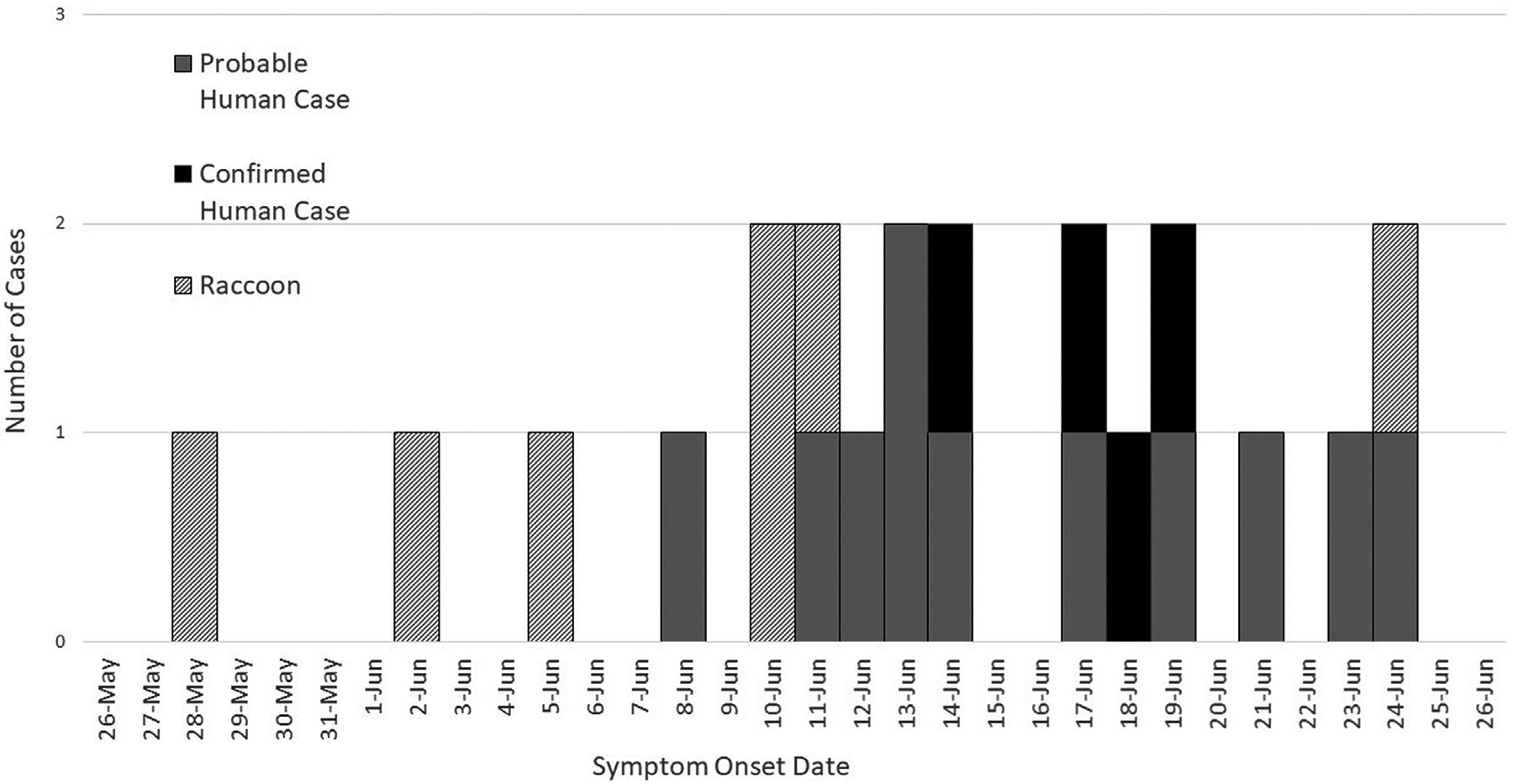
Wildlife facility cryptosporidiosis outbreak epidemiologic curve, Virginia, June 2019
3.2 |. Laboratory testing
Four human stool specimens tested negative for norovirus; three specimens tested positive for Cryptosporidium at DCLS. Of the two human stool specimens tested at clinical laboratories, one was positive for Cryptosporidium via antigen testing; the other was negative on O&P, but cryptosporidia testing was not performed on this specimen.
All six raccoon faecal specimens were acid-fast positive and positive for Cryptosporidium via ELISA. Molecular typing revealed C. parvum subtype (IIaA16G3R2) in six raccoon faecal specimens, one raccoon tissue specimen and two human stool specimens.
3.3 |. Environmental investigation
All three water samples had detectable levels of total coliforms (3.0, 10.9 and 13.5 MPN/100 ml). One of three samples was positive for E. coli (1 MPN/100 ml). Raccoons were found in a variety of locations between 8–46 miles from the facility. At the facility, raccoons were housed with their littermates in a dedicated room with no other animal species.
4 |. DISCUSSION
During 2009–2017, there were 444 outbreaks of cryptosporidiosis reported in the U.S., of which 86 (19%) were associated with animal contact, most frequently cattle (n = 65) (Gharpure et al., 2019; Xiao & Feng, 2008). Wild animals were not identified as a source of exposure in these outbreaks. Whilst raccoon infections with Cryptosporidium species and genotypes are known to occur in the U.S. and elsewhere, these infections have not been reported in association with human illness (Gonzalez-Astudillo et al., 2021; Hattori et al., 2018; Perz & Le Blancq, 2001; Snyder, 1988). To the authors’ knowledge, this is the first report of human illness after exposure to raccoons infected with C. parvum.
In this outbreak, raccoon illness preceded human illness by 11 days. The first ill raccoon came to the wildlife facility only 2 days prior to its onset of diarrhoea. Though the incubation period for C. parvum in raccoons is unknown, incubation periods of approximately 7 days have been documented in other species including cattle (Uga et al., 2000), sheep (Bukhar & Smith, 1997), pigs (Pereira et al., 2002) and mice (Tarazona et al., 1998). The timing of illness amongst raccoons and humans suggests that one or more raccoons could have been infected prior to intake, with subsequent transmission occurring between raccoons and from raccoons to humans (Figure 1).
Four exposures (contact with foxes, raccoons and cottontails and drinking facility well water) were shared by a majority of ill persons and were significantly associated with human illness. Of these, contact with raccoons had the highest attack rate (73%) and risk ratio (4.4; CI = 1.7–11.3) and was significantly associated with illness (p < .01) in multivariate modelling. Because vaccination against rabies is required to care for both raccoons and foxes, most individuals with raccoon contact also had fox contact (14 of 15); two persons had fox contact but no raccoon contact. The lack of clinically significant symptoms in foxes or cottontails compared to the profuse diarrhoea observed in raccoons made these animal exposures appear less plausible as sources of human infection. However, a limitation to this investigation is that foxes and cottontails were not tested for Cryptosporidium and could have had asymptomatic or mild infections. Drinking facility well water remained significantly associated with illness in multivariate analysis (p < .05). The water was tested for total coliforms and E. coli, but not for Cryptosporidium, which is a second limitation to this investigation. However, the water was used for all animals and no other species exhibited notable diarrheal illness. Bearing in mind the timing of illness onset for the first ill raccoon relative to facility intake, the well water at the facility was considered less likely to be a source of Cryptosporidia in this outbreak. Based on the clinical, epidemiologic and laboratory findings of this investigation, raccoons were considered to be the probable source of exposure for human illness.
The outbreak investigation revealed that standard cleaning and disinfection protocols at the facility were ineffective against Cryptosporidium oocysts. Once the outbreak aetiology was confirmed, protocols were changed to include disinfection with hydrogen peroxide to disrupt further transmission. This investigation also prompted further training of personnel, especially interns and volunteers, about effective infection control practices and proper use of personal protective equipment.
Molecular typing identified C. parvum IIaA16G3R2 in tested outbreak specimens rather than the dominant C. parvum IIaA15G2R1 found in dairy calves from the United States (Santín et al., 2008); the IIa subtype family causes zoonotic infections in most industrialized countries (Feng et al., 2018). The outbreak subtype, C. parvum IIaA16G3R2, was previously reported in four humans from Wisconsin (Feltus et al., 2006) and detected in 16 sporadic human cases as part of CryptoNet surveillance from Wisconsin, Nebraska and Tennessee from 2012 to 2019. The outbreak subtype has not been previously reported in raccoons.
Routine testing for Cryptosporidium might not be conducted at facilities caring for wild animals due to limited resources, a low index of suspicion for this pathogen and a low perceived benefit from testing. Still, raccoons and other wildlife species capable of infection with C. parvum (including opossums, foxes, lagomorphs and squirrels) might be an under-recognized reservoir for human infection. Expanded animal testing for Cryptosporidium could provide human health benefits by informing diagnostic and treatment choices for patients with exposure to infected animals. Human healthcare providers should consider cryptosporidiosis as a differential diagnosis in persons who have contact with raccoons (and potentially, other wildlife species susceptible to C. parvum) and present with gastrointestinal illness. Diagnostic testing for Cryptosporidium no longer requires microscopy; molecular detection is increasingly available through multiplex PCR panels. CryptoNet aims to systematically collect exposure and molecular characterization data to fill knowledge gaps about wildlife-associated zoonotic cryptosporidiosis, including identifying under-recognized reservoirs and risk factors.
Impacts.
Cryptosporidium parvum of the same molecular subtype (IIaA16G3R2) was identified in stool specimens from humans and raccoons associated with the same wildlife rehabilitation facility.
Raccoon illness preceded human illness by 11 days, indicating possible zoonotic transmission from raccoons to humans.
Raccoons might be an under-recognized reservoir for human C. parvum infections.
ACKNOWLEDGEMENTS
Dr. Riley and Dr. Darby contributed equally to this manuscript. The authors would like to thank the many individuals who contributed to this outbreak investigation: P. Bair, C.M. Greene, W.J. Davis, C. Bonnefond, M. Allen, A. Moore, M. Perry, from the Virginia Department of Health; J. Weisman from the Virginia Department of Agriculture and Consumer Services; E. Craig and K. Niedzielska-Hessler from the Virginia Division of Consolidated Laboratory Services; and M. Hlavsa from the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
CONFLIC T OF INTEREST
None of the authors of this study have a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Pathasure test is validated for bovine species and was used off-label in this investigation.
DATA AVAIL ABILIT Y STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- Bukhar Z, & Smith HV (1997). Cryptosporidium parvum: Oocyst excretion and viability patterns in experimentally infected lambs. Epidemiology and Infection, 119, 105–108. 10.1017/s0950268897007590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-M, Keithly JS, Paya CV, & LaRusso NF (2002). Cryptosporidiosis. New England Journal of Medicine, 346, 1723–1731. 10.1056/NEJMra013170 [DOI] [PubMed] [Google Scholar]
- Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, & McEvoy JM (2006). Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. Journal of Clinical Microbiology, 44, 4303–4308. 10.1128/JCM.01067-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Ryan UM, & Xiao L (2018). Genetic diversity and population structure of Cryptosporidium. Trends in Parasitology, 34, 997–1011. 10.1016/j.pt.2018.07.009 [DOI] [PubMed] [Google Scholar]
- Gharpure R, Perez A, Miller AD, Wikswo ME, Silver R, & Hlavsa MC (2019). Cryptosporidiosis outbreaks – United States, 2009–2017. MMWR Morbidity and Mortality Weekly Report, 68, 568–572. 10.15585/mmwr.mm6825a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Astudillo V, Sheley MF, Uzal FA, & Navarro MA (2021). Pathology of cryptosporidiosis in raccoons: Case series and retrospective analysis, 1990–2019. Journal of Veterinary Diagnostic Investigation, 33(4), 721–727, 10.1177/10406387211011949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori K, Donomoto T, Manchanayake T, Shibahra T, Sasai K, & Matsubayashi M (2018). First surveillance and molecular identification of the Cryptosporidium skunk genotype and Cryptosporidium parvum in wild raccoons (Procyon lotor) in Osaka, Japan. Parasitology Research, 117, 3669–3674. 10.1007/s00436-018-6089-y [DOI] [PubMed] [Google Scholar]
- Hunter PR, Hughes S, Woodhouse S, Syed Q, Verlander NQ, Chalmers RM, Morgan K, Nichols G, Beeching N, & Osborn K (2004). Sporadic cryptosporidiosis case-control study with genotyping. Emerging Infectious Diseases, 10, 1241–1249. 10.3201/eid1007.030582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SJ, Ramirez NE, Xiao L, & Ward LA (2002). Pathogenesis of human and bovine Cryptosporidium parvum in Gnotobiotic pigs. The Journal of Infectious Diseases, 186, 715–718. 10.1086/342296 [DOI] [PubMed] [Google Scholar]
- Perz JF, & Le Blancq SM (2001). Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Applied and Enviromental Microbiology, 67, 1154–1162. 10.1128/AEM.67.3.1154-1162.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roellig DM, & Xiao L (2020). Cryptosporidium genotyping for epidemiology tracking. Methods in Molecular Biology, 2052, 103–116. 10.1007/978-1-4939-9748-0_7 [DOI] [PubMed] [Google Scholar]
- Ryan U, Fayer R, & Xiao L (2014). Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology, 141, 1667–1685. 10.1017/S0031182014001085 [DOI] [PubMed] [Google Scholar]
- Santín M, Trout JM, & Fayer R (2008). A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Veterinary Parasitology, 155, 15–23. 10.1016/j.vetpar.2008.04.018 [DOI] [PubMed] [Google Scholar]
- Snyder DE (1988). Indirect immunofluorescent detection of oocysts of Cryptosporidium parvum in the feces of naturally infected raccoons (Procyon lotor). Journal of Parasitology, 74, 1050–1052. 10.2307/3282233 [DOI] [PubMed] [Google Scholar]
- Tarazona R, Blewett DA, & Carmona MD (1998). Cryptosporidium parvum infection in experimentally infected mice: Infection dynamics and effect of immunosuppression. Folia Parasitologica, 45, 101–107. [PubMed] [Google Scholar]
- Uga S, Matsuo J, Kono E, Kimura K, Inoue M, Rai K, & Ono K (2000). Prevalence of Cryptosporidium parvum infection and pattern of oocyst shedding in calves in Japan. Veterinary Parasitology, 194, 27–32. 10.1016/s0304-4017(00)00338-1 [DOI] [PubMed] [Google Scholar]
- Xiao L, & Feng Y (2008). Zoonotic cryptosporidiosis. FEMS Immunology and Medical Microbiology, 52, 309–323. 10.1111/j.1574-695X.2008.00377.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


