Abstract
Our skin is the interface through which we mediate lifelong interactions with our surrounding environment. Initial development of the skin’s epidermis, adnexal structures and barrier function are necessary for normal cutaneous microbial colonization, immune development, and prevention of disease. Early life microbial exposures can have unique and long-lasting impacts on skin health. The identity of neonatal skin microbes and the context in which they are first encountered, i.e. through a compromised skin barrier or in conjunction with cutaneous inflammation, can have additional short- and long-term health consequences. Here, we discuss key attributes of infant skin and endogenous and exogenous factors that shape its relationship to the early life cutaneous microbiome, with a focus on their clinical implications.
Introduction
The skin and its adnexal structures constitute the body’s external epithelial surface. Rather than an impermeable barrier, our skin serves as a dynamic interface that integrates signals from the surrounding environment (Gallo, 2017). The cutaneous host-microbe relationship is initiated moments after birth, marked by the skin’s colonization with a diverse community of commensal microbes, including bacteria, fungi, viruses and even parasites (Byrd et al., 2018; Chen et al., 2018; Dominguez-Bello et al., 2010). Key age-dependent differences in cutaneous physiology and immune function influence host-microbe interactions in skin during the neonatal window. These early life interactions remain relatively understudied as compared to those in adulthood. However, an emerging body of evidence suggests that the postnatal period is characterized by unique, non-redundant mechanisms that shape our relationship with the skin microbiome. These microbe-skin interactions support cutaneous immune homeostasis in infancy and may impact longer-term skin health. Here we review the unique physiologic and microbial features of infant skin, discuss skin-microbe crosstalk during this developmental window, and identify contexts in which this relationship can be pathologically altered.
Human skin microbiome: topographical and ecological considerations
Compared to other microbial niches on the human body, such as the nasopharynx, intestines or other mucosal surfaces, the skin is cool, acidic, dry, and relatively nutrient sparse. It also produces salty sweat and antimicrobial peptides (AMPs) that limit bacterial growth (Belkaid and Segre, 2014; Byrd et al., 2018; Grice and Segre, 2011). Despite this, human skin houses a rich and diverse microbial community (Byrd et al., 2018; Oh et al., 2016). Whereas intestinal bacteria feed off of a steady stream of ingested dietary nutrients, skin microbes must derive their sustenance from the skin’s epidermis and the secretions coating its surface. Fluctuations in the relative density of skin adnexal structures, i.e. sweat ducts, hair follicles and sebaceous glands, across the body’s surface contribute to a range of skin microenvironments that vary with regard to temperature, pH, moisture, AMP and lipid content. These sub-niches preferentially support distinct groups of commensal microbes that are more or less adept at deriving nutrients from various skin sources and surviving in the face of cutaneous host defenses (Belkaid and Segre, 2014; Oh et al., 2014). In adult skin, these niches are categorized according to dry, moist or sebaceous sites. Sebaceous sites, such as the face and chest, are enriched for lipid-loving Cutibacterium, whereas the phyla Staphylococcus and Corynebacteria are found more so on moist or dry sites, such as the arms, groin or feet (Byrd et al., 2018; Grice et al., 2009).
While a planar surface macroscopically, skin topography at the microscopic level is highly three dimensional. In fact, when considering invaginations such as hair follicles as part of the skin’s surface area, it exceeds that of the intestines (Gallo, 2017). The relatively protected and microaerophilic environment of the hair follicle provides a unique, densely occupied niche for bacteria on skin (Hall et al., 2018). It also represents an important cutaneous immune microenvironment, where chemokines attract and influence behavior of skin resident immune cells (Adachi et al., 2015; Nagao et al., 2012). Thus, the hair follicle is thought to be a central location for microbe-immune interactions during homeostasis and in disease (Constantinou et al., 2021; Harries and Paus, 2010). Skin bacteria and their products are also found to extend into the interfollicular dermis and dermal adipose tissue. This suggests other important avenues through which they can influence skin biology (Nakatsuji et al., 2013).
The skin is a rich immunological organ, containing numerous tissue-resident immune cell types. These immune cells sample and respond to microbial products (Byrd et al., 2018; Chen et al., 2018), limit deeper penetration of skin commensal organisms (Shen et al., 2014), and contribute to the cutaneous pool of antimicrobial peptides (AMPs) (Takahashi and Gallo, 2017). Whereas in the intestine, B cell secretion of IgA is thought to help curate composition of luminal microbes (Fadlallah et al., 2018), deficiencies in lymphocytes or Langerhans cells have not been shown to strongly impact skin microbial ecology (Scholz et al., 2014). Certain skin immune cells, such as innate lymphoid cells (ILCs), have been shown to indirectly affect skin bacteria, i.e. via their effects on sebaceous gland biology (Kobayashi et al., 2019). However, the structural and chemical considerations discussed above remain paramount in dictating skin microbiome composition.
The cutaneous immune system displays many age-specific features. Recent studies using high-parameter cytometry and single cell RNA sequencing have revealed that T cells in human fetal skin are relatively naïve as compared to those in adult skin, but do contain activated memory-type regulatory T cells (Tregs) and subsets of CD45RO+ T effector cells (Dhariwala et al., 2020; Mishra et al., 2021; Xu et al., 2021; Yona et al., 2013). Distinct immune signatures have also been demonstrated by bulk RNA sequencing of neonatal, pediatric or adult skin (Brunner et al., 2018; Visscher et al., 2021). The composition of immune cells in pediatric skin is not yet as well defined. However, one study used flow cytometry to demonstrate a higher percentage of Tregs in healthy pediatric versus adult skin (Cordoro et al., 2017), akin to what has been shown for other early life human tissues such as the lung and intestines (Thome et al., 2015). Collectively, these considerations provide an important backdrop for examining the development of the early life skin microbiome and its relationship to infant skin heath.
Composition and evolution of the infant skin microbiome
The presence of live microbes in utero remains an active area of investigation, as does their potential contribution to immune education, with some recent studies suggesting a role in priming of lymphocytes in human fetal tissues (Mishra et al., 2021; Rackaityte et al., 2020). While debate continues regarding the fetal microbiome (Kennedy et al., 2021a, 2021b), live microbes, when detected, are found in very limited abundance in utero (Kennedy et al., 2021a; Rackaityte et al., 2020). Thus, while pre-natal microbial encounters are possible, our skin’s first substantial introduction to microbes occurs at birth. Within minutes, bacteria can be cultured from healthy infant skin (Sarkany and Gaylarde, 1967). Sampling of the neonatal skin microbiome by 16S rRNA sequencing has shown that its composition during the first hours of life is very similar to the microbiome at other body sites. In vaginally delivered infants, this largely reflects composition of the maternal vaginal community (Dominguez-Bello et al., 2010, 2016). By 6 weeks of age, the skin microbiome signature has diverged from that of the stool, nares, or oral cavity (Chu et al., 2017). By 3 months of age, regional specificity within the skin microbiome also emerges, with distinct communities found on the arm, forehead and buttock (Capone et al., 2011). A great deal of longitudinal fluctuation in the bacterial and fungal skin communities persists throughout the first 6 months of life (Capone et al., 2011; Zhu et al., 2020), likely due to progressive changes in infant skin physiology (Stamatas et al., 2011; Visscher et al., 2015) as well as interspecies interactions that are recognized as part of normal complex microbial community establishment (Coyte et al., 2021).
By 6 to 12 months of age, infant skin acquires a more stable microbial community, the composition of which is notably distinct from that seen in adults. Multiple studies have confirmed that the infant skin microbiome contains relatively more Firmicutes, such as Staphylococci and Streptococci, and fewer Actinobacteria, such as Cutibacteria and Corynebacteria (Oh et al., 2012). Bacteria from the Proteobacteria and Bacteroidetes phyla are also present (Capone et al., 2011). Of note, Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes have been renamed Bacillota, Actinomycetoma, Pseudomonadata, and Bacteriodata, in the newest nomenclature guidelines from NCBI (Oren and Garrity, 2021); however, the former names will be used throughout this review.
Fewer studies have focused on the infant skin mycobiome, but longitudinal examination of 110 infants less than 6 months of age found Malassezia to be the most abundant fungi across three distinct skin sites, followed, at several fold lower abundance, by Pleosporales and Saccharomycetales (Zhu et al., 2020). Notably Malassezia burden decreases on infant skin between 3 and 6 months of age (Zhu et al., 2020) and is thought to remain lower throughout early childhood as compared to teens or adults (Findley et al., 2013; Jo et al., 2017; Park et al., 2021).
Early life factors influencing skin microbiome composition
Published studies examining microbiota on infant and pediatric skin have relied largely on 16S and 18S rRNA sequencing rather than metagenomic sequencing. This means we have limited insight into strain level data or potential age-dependent differences in microbial pathway abundance on skin during early life. Even so, there is an emerging consensus regarding factors that help shape genera and species-level composition of microbial communities on infant skin.
Infant skin physiology
As in adults, the composition and physiology of infant skin is thought to be a driving force in curating what microbes establish early residency in this niche. At birth, human skin is coated by the vernix caseosa (VC), a protective layer produced in utero by fetal sebaceous glands. The VC contains lipids (10%), protein (10%), and water (80%) (Nishijima et al., 2019; Visscher et al., 2011). While its function is not completely understood, the VC has been proposed to aid the transition from an aqueous intrauterine to a dry extrauterine environment (King et al., 2013; Nishijima et al., 2019). Its hydrophobic lipid content may hasten development of a fully functional stratum corneum towards the end of in utero gestation (Visscher et al., 2005; Youssef et al., 2001). Postnatally, longer retention of the VC is associated with higher infant skin hydration 24 hours after birth (Visscher et al., 2005).
The VC likely also contributes to initial seeding of microbes both on skin and in the gut by providing microbial prebiotics in the form of saturated branched fatty acids, squalene and other lipids (Houghteling and Walker, 2015; Ran-Ressler et al., 2008; Wang et al., 2018). Meanwhile it is also provides early antimicrobial defense to infant skin both by serving as a physical obstruction for bacterial colonization (Bautista et al., 2000; Okah et al., 1994) as well as a source of innate immune cytokines, e.g. interleukin (IL)-1α and IL-1β, IL-6, IL-8, and tumor necrosis factor alpha (Narendran et al., 2010; Walker et al., 2008; Yoshio et al., 2003) and antimicrobial peptides (AMPs), e.g. LL-37, lysozyme, psoriasin, lactorferrin and alpha defensins (Yoshio et al., 2003; Marchini et al., 2002). Previous studies have found that the VC or its subcomponents can limit growth of S. aureus, Klebsiella, Bacillus megaterium and Candida albicans (Yoshio et al., 2003), suggesting that the VC may favor colonization by particular bacterial or fungal species over other (Yoshio et al., 2003). The first bath of a newborn removes the VC and is sometimes delayed rather than performed immediately at birth due to observed benefits on infant health, such as temperature regulation (Visscher et al., 2015). Further studies are needed, however, to understand how timing of infant bathing and removal of the VC affect early postnatal development of the human skin microbiome (Lund and Zukowsky, 2016).
Compositional differences between the microbial communities on infant versus adult skin are thought to be driven in large part by age-dependent changes in skin physiology. Human skin development begins in utero by 4 weeks gestation at which point multi-layer specialization of the epidermis and keratinization, i.e. formation of the outermost stratum corneum, are seen (Jö et al., 1999; King et al., 2013). Adnexal structures such as hair follicles and sweat glands appear at nine weeks and further mature throughout the second and early third trimesters (Jö et al., 1999; King et al., 2013). Despite developing in an aquatic environment, full-term, healthy infant skin is considered to be fully functional at birth (Visscher et al., 2015). For example, transepidermal water loss (TEWL), a proximal measurement for skin barrier function, is generally comparable between infants and adults (Fluhr et al., 2000, 2012; Kelleher et al., 2013; Kikuchi et al., 2006). Compared with adult skin, however, infant skin is moister and more alkaline and lower in lipid and sebaceous content (Moskovicz et al., 2020). Firmicutes, which are overrepresented on infant skin, generally have higher salt tolerance and can thrive in moist, alkaline environments. In contrast, underrepresented genera such Cutibacteria from the Actinobacteria phylum thrive on lipid-rich skin secretions that are not as abundant on infant skin (Byrd et al., 2018; Scharschmidt and Fischbach, 2013). Consistent with this paradigm, significant associations have been observed in the relative abundance of certain bacterial species and locally measured skin pH or moisture content in 1 year old infants (Zhu et al., 2019). Hormonally-driven changes in sebum production that occur in late puberty are accompanied by a shift towards a more Actinobacteria-dominant and Malassezia-rich skin microbiome (Oh et al., 2012; Park et al., 2021). Thus, chemical and physical properties help dictate which microbes establish residency on skin through life, including in infancy (Figure 1).
Figure 1: Physiologic Features of Infant skin.
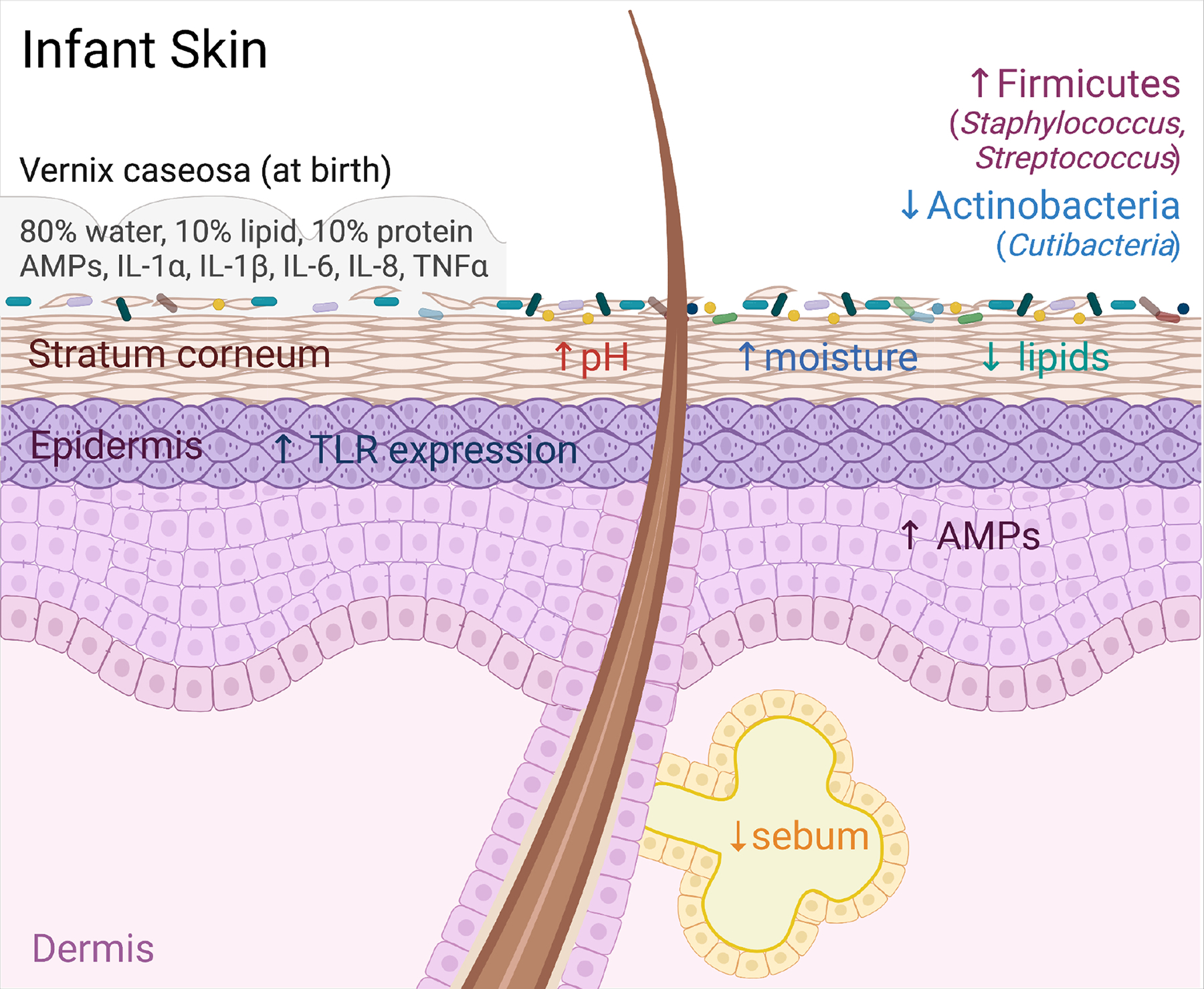
Infant skin at birth is covered in the vernix caseosa (VC). The VC is made up of 80% water, 10% lipid, 10% protein, and contains various antimicrobial peptides (AMPs) and cytokines. The VC likely influences initial microbial colonization of the skin via its role as both a physical and chemical barrier as well as a nutrient substrate for microbes. Compared to adults, infant skin has increased expression of AMPs and toll like receptors (TLRs), an elevated pH, increased moisture content, and decreased lipid and sebum production. Collectively these features shape early life skin microbial ecology, which is distinguished by relatively increased colonization by Firmicutes, including members of Staphylococcus and Streptococcus, and decreased colonization by Actinobacteria, including Cutibacteria.
Maternal factors
As is true of the gut microbiome, seeding of skin microbes is thought to occur largely via vertical colonization from the mother. It is not surprising then that the skin microbiome of infants is dominated by strains found on their mothers immediately after birth (Dominguez-Bello et al., 2010). Perhaps more surprising is the durability of this maternal influence. Even at 10 years of age, the relative abundance of bacteria at several skin sites more closely resembles that of their mother compared to unrelated women (Zhu et al., 2019). The extent to which this is due to first priority effects of microbes transferred at birth (Fukami, 2015; Nayfach et al., 2016) versus continued microbial transfer from mother to child during early life remains to be determined, but likely both are at play. These maternal microbiome seeding effects suggest some level of heritability in the skin microbiome and imply that dramatic shifts in the maternal microbiome, on skin or other body sites, during the perinatal window may impact this process.
Birth mode
It has been clearly demonstrated that mode of delivery plays a large role in shaping the composition of the infant skin microbiome immediately after birth. One seminal study used 16S rRNA sequencing to sample the maternal skin, oral, and vaginal microbiota just before delivery in tandem with the infants’ skin, oral, and nasopharyngeal microbiota within minutes of birth and meconium within 1 day of life (Dominguez-Bello et al., 2010). This revealed that vaginally-delivered infants had skin bacterial communities resembling their mother’s vaginal microbiota (Lactobacillus, Prevotella, and Sneathia spp.), whereas infants delivered by Cesarean section (C-section) had skin bacterial communities most similar to maternal skin (Staphylococcus, Corynebacterium, and Cutibacterium spp.) (Dominguez-Bello et al., 2010). Subsequent studies have largely confirmed these findings with regard to the immediate infant skin microbiome signature following vaginal versus C-section delivery (Capone et al., 2011; Chu et al., 2017). However, they have also shown that these delivery mode signatures are fairly transient and abate significantly or disappear entirely by 1–3 months of life (Capone et al., 2011). One study did find subtle associations between delivery mode and composition of the skin microbial community on the volar forearm in 1 year old children and on the face of 10-year-old children, driven respectively by slightly more Streptococcus and Chryseobacterium in the C-section delivered children (Zhu et al., 2019). However, differences were not seen at other body sites, nor at four other intervening sampling time points. The significance of such subtle differences in the skin and gut microbial communities remains an area of investigation with regard to certain clinical associations with C-section birth, such as higher rates of asthma and allergies (Bager et al., 2008; Negele et al., 2004).
Other environmental exposures
While initial seeding of the infant skin microbiome is likely of seminal importance due to enduring priority effects of founder species (Sprockett et al., 2018), there is also evidence that the infant’s environment influences composition of the skin microbial community. For example, bacteria from the hospital environment can been found on the skin of full term and preterm infants during the first weeks of life (Younge et al., 2018). Looking at later time points, there is also evidence that residency in an urban versus rural environment or in a larger household may subtly shift the diversity and composition of skin bacteria (Lehtimäki et al., 2017). Although limited by its small sample size, a study of infants born in Illinois versus three different areas of Mexico demonstrated higher alpha diversity on the skin of infants raised in a rural environment, in a larger household size, and by a larger total number of caregivers (Manus et al., 2020). The relative contribution of these various factors remains unclear but these types of data suggest that further work examining environmental factors associated with atopy or other outcomes, for example the presence of pets in the home, will be important to study with respect to skin microbiome composition as has been done for the infant gut (Tun et al., 2017).
Microbial influences on neonatal skin function
Of obvious interest and clinical significance are the potential effects of the cutaneous microflora on physiologic and immune functions of infant skin. Assessing many of these requires full thickness skin biopsies, a minimally invasive procedure but still one largely avoided in healthy children for understandable reasons. We therefore have limited human data on functional microbe-skin interactions in this developmental window. However, studies in mice have created a framework for thinking about the likely effects of cutaneous microbes on infant skin physiology.
One key area for functional interactions is development of the physical and antimicrobial skin barrier. Commensal microbes produce their own AMPs (Nakatsuji et al., 2017) and can also augment expression of cutaneous host-derived AMPs (Lai et al., 2010). Studies in gnotobiotic (i.e. germ-free mice), conventionalized and antibiotic-treated mice, have shown that bacteria also promote terminal differentiation of keratinocytes and a fully functional stratum corneum (Uberoi et al., 2021). Although not examined specifically in neonatal animals, these results would suggest that initial colonization of skin by bacteria and fungi may help bolster infant skin defenses against more pathogenic organisms via multiple mechanisms. Whether the greater expression of toll-like receptors (TLRs) (Iram et al., 2012) and AMPs (Nishijima et al., 2019; Walker et al., 2008) in infant versus adult skin reflects innate immune activation in response to initial microbial colonization is not known, but of potential relevance.
Skin bacteria may also influence infant skin physiology via effects on the number or function of skin immune cells. Information is again limited due to challenges associated with the study of human neonates and the fact that most murine studies have been performed in adult animals. Even so, it worth highlighting a few emerging principles of microbial-mediated effects on cutaneous immunity that inform our thinking about the early life window.
Microbes have been shown in multiple contexts to alter the abundance and function of lymphocytes in murine skin. This has been rigorously demonstrated with regard to effects of Staphylococcus epidermidis on skin CD4+, CD8+ and mucosal-associated invariant T (MAIT) cells (Belkaid and Naik, 2013; Chen et al., 2019; Harrison et al., 2019; Linehan et al., 2018). Analogous effects have been seen for Corynebacterium accolens on Vγ4+ dermal γδ T cells (Ridaura et al., 2018), and for cutaneous fungi such as Candida albicans and Malessazia furfur on IL-17-producing CD4+ T cells (Hurabielle et al., 2020; Kashem et al., 2015). Comparatively minimal effects have been shown in adult mice with respect to the impact of microbes on the numbers of innate lymphoid cells (Ricardo-Gonzalez et al., 2018) or myeloid cells (Tamoutounour et al., 2013). Consistent with immunological dogma implicating type 17 responses in immunity to extracellular microbes, these studies have largely demonstrated that skin commensal bacteria and fungi bolster homeostatic type 17 immune responses through both antigen dependent and independent mechanisms. In certain more pathological contexts, however, skin bacteria have also been shown to trigger a type 2 immune signature (Byrd et al., 2017; Harrison et al., 2019). Although not studied specifically in neonatal mice or human infant skin, this body of work suggests that skin commensals have the potential to optimally “tune” the cutaneous type 17 immune responses in a manner that could accelerate this arm of skin immune development post birth.
As discussed above, human skin and other pediatric tissues tend to be relatively enriched in Tregs (Cordoro et al., 2017; Thome et al., 2015). Studies of human skin have also revealed increased tolerogenic capacity among dendritic cells isolated from fetal versus adult skin (McGovern et al., 2017). The skin of neonatal mice also contains higher numbers of Tregs, with an influx specifically between postnatal days 6 and 13 (Scharschmidt et al., 2015). Experiments in gnotobiotic mouse pups and models of impaired hair follicle development showed that this neonatal Treg wave into murine skin is at least in part directed by commensal-induced expression of the chemokine Ccl20 in developing hair follicles (Scharschmidt et al., 2017). Studies of adult gnotobiotic mice have not revealed persistent differences in skin Treg numbers (Naik et al., 2012), however, suggesting that other mechanisms can help compensate for early commensal-facilitated recruitment. Tregs appear much earlier in developing human skin. Although absolute numbers of Tregs remain low in fetal skin as compared to adult skin, by the late second trimester the percentage of Tregs among skin CD4+ T cells, as well as their activation and memory status, start to resemble that seen in adult skin (Dhariwala et al., 2020). Preferential fetal skin Treg accumulation at sites of higher hair density suggest that hair follicles may help facilitate early Treg accumulation in human skin as is seen in the mouse (Dhariwala et al., 2020). Whether postnatal interactions with colonizing skin microbes further recruit, activate, or expand human skin Tregs remains an open question.
Unique contributions of early life skin-microbe interactions
While many aspects of skin-microbe crosstalk likely retain equal relevance and potency throughout the lifespan, there are already two examples from animal studies where early life microbial interactions have unique effects on cutaneous immune function not replicated by later life microbial exposure. The first relates to MAIT cells, which are a type of nonconventional lymphocyte enriched in skin and other body barrier tissues. These cells express semi-invariant T cell receptors, which recognize microbial molecules, in particular riboflavin derivatives. In mice, it has been shown that trafficking of these microbially-produced riboflavin derivatives from the intestine to the thymus regulates MAIT development (Legoux et al., 2019), and that this microbially-dependent process occurs preferentially during the first few weeks of life (Constantinides et al., 2019) (Figure 2A). MAIT cells accumulate in mouse skin between two and three weeks of age and can be further expanded and activated by the presence of skin bacteria that produce riboflavin derivatives (Constantinides et al., 2019). MAIT cells are present in adult human skin (Li et al., 2017) but the timing of their appearance and expansion in infant skin has not yet been explicitly studied. Activation of MAIT in human blood occurs soon after birth followed by a gradual expansion over the next five to ten years (Dusseaux et al., 2011; Pellicci et al., 2020; ben Youssef et al., 2018). Skin bacteria fall along a wide continuum with regard to their capacity for riboflavin metabolism and MAIT stimulation (Tastan et al., 2018), suggesting that composition of the infant skin microbiome might impact MAIT recruitment and activation, thereby influencing local skin immune function.
Figure 2: Early life microbial-immune interactions.
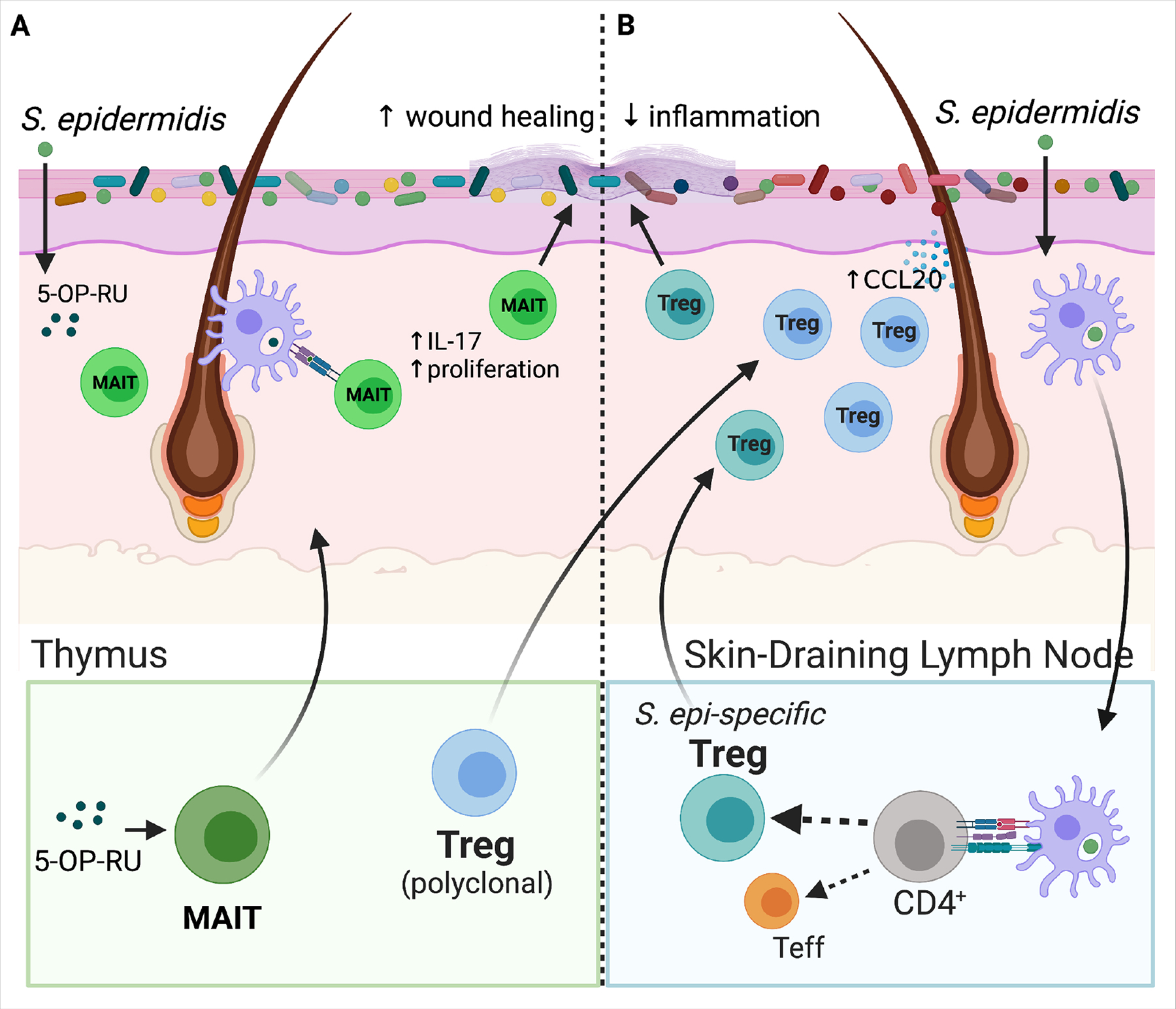
(A) 5-OP-RU, a microbially-produced riboflavin-derived antigen stimulates production of MAIT cells specifically in the neonatal versus adult thymus. As shown in mice, inadequate microbial exposure during this early window results in a lost opportunity to expand this lymphocyte population. MAIT cells travel from the thymus to skin where subsequent local production of 5-OP-RU by S. epidermidis and other skin bacteria promotes proliferation of MAITs and stimulates their IL-17 production. These cutaneous MAIT cells contribute to skin homeostasis for example by augmenting local would healing. (B) A polyclonal wave of regulatory T cells (Tregs) migrates from the thymus and secondary lymphoid organs into murine skin between the first and second week of postnatal life. Microbial colonization of developing hair follicles during this postnatal window in mice augments production of the ligand Ccl20 in the hair follicle infundibulum, which helps draw this polyclonal Ccr6+ Treg population into the tissue. Cutaneous exposure to commensal antigens in this early window, for example via neonatal S. epidermidis colonization, leads to a persistent enrichment of Tregs among commensal-specific CD4+ T cells in skin. They help to limit skin inflammation upon subsequent re-exposure to S. epidermidis under an inflammatory context. Expansion of these commensal-specific Tregs is dependent on dendritic-cell mediated uptake of bacterial antigens and their trafficking to the skin-draining lymph nodes (SDLN) for presentation to naïve CD4+ T cells. When initial skin exposure to S. epidermidis is delayed until adulthood, the repertoire of CD4+ T cells specific for that bacteria is shifted instead towards effector CD4+ T cells (Teffs). The mechanistic basis for this neonatal capacity to establish commensal-specific tolerance is likely multifactorial, but the enriched presence of Tregs in neonatal skin has been shown to be one contributing factor.
The repertoire of conventional αβ T cells that recognize and respond to skin bacteria is also influenced by the timing of bacterial exposure. Studies in mice have revealed that neonatal colonization with S. epidermidis (S. epi) supports development of a S. epi-specific CD4+ compartment that is rich in Tregs (Scharschmidt et al., 2015). By comparison, exposure to the same bacteria in an adult mouse led predominantly to generation of effector CD4+ T cells (Teff) (Figure 2B). Generation of these S. epi-specific Tregs in neonatal life was closely linked to protection against skin tissue inflammation when the mice were re-exposed to S. epi as adults, indicating functional immune tolerance (Scharschmidt et al., 2015). The mechanistic basis of this early life predisposition for tolerance to skin bacteria remains incompletely understood but likely relates to a combination of factors. These may include the heighted early life propensity for naive T cells to become Tregs (Mold et al., 2010), augmented Tregpromoting capacity among skin dendritic cells (McGovern et al., 2017), and an increased abundance of polyclonal Tregs in the skin of young mice (Scharschmidt et al., 2015) and humans (Cordoro et al., 2017) that may affect the quality of antigen presentation. It will be difficult to test whether humans experience similar age-dependency in their establishment of tolerance to skin commensal bacteria. However, data from the literature on peanut allergy (Toit et al., 2015) and oral as well as epicutaneous antigen exposure to prevent this allergy in children (Kim and Burks, 2020) suggest that a preferential early window for immune tolerance also exists in human. Notably, antigen-specific Tregs are not indiscriminately generated to any bacteria present on neonatal skin, as mice colonized with the pathobiont Staphylococcus aureus generate relatively fewer S. aureus-specific versus S. epi-specific Tregs. This brake on tolerance to a prototypical skin pathogen, is due in part to heightened IL-1R1 signaling elicited by S. aureus alpha toxin (Leech et al., 2019). The percentage of Tregs generated neonatally to S. aureus, however, still exceeded that generated to S. epi in adult mice suggesting that early exposure even to pathogenic bacteria in the early window may be more tolerizing than in later life.
Infant Skin Microbiome in Health & Disease
‘Physiologic’ Infant Skin-Microbe Interactions
When early life skin-microbe interactions go smoothly, they are clinically ‘invisible’. However, some very common childhood dermatoses likely reflect low levels of cutaneous inflammation elicited by microbe-immune crosstalk. Erythema toxicum neonatorum (ETN) is a cutaneous eruption seen during the first week of life in about half of healthy newborns (Harris and Schick, 1956). It is typified by the appearance of small splotchy areas of redness that may contain central papules or pustules (Chadha and Jahnke, 2019). Microscopic examination of these ETN lesions show infiltration of eosinophils, neutrophils and myeloid cell types especially around hair follicles. This is found in conjunction with increased expression of innate immune mediators such as IL-1, IL-8, nitric oxide synthases and antimicrobial peptides (Marchini et al., 2001, 2003). Coagulase-negative Staphylococci as well as other Firmicutes that are prevalent on infant skin have been cultured from ETN lesions, and electron microscopy has revealed cocci-shaped bacteria within hair follicle epithelium and phagocytosed by nearby immune cells (Marchini et al., 2005). Collectively, these findings have led to the yet unproven hypothesis that ETN results from an early innate immune response to colonization of infant skin by healthy commensal bacteria driven both by keratinocytes and macrophages (Marchini et al., 2005, 2007).
Similarly, fungi or yeast on neonatal skin may contribute to rashes seen in healthy infants, for example infantile seborrheic dermatitis (ISD). This condition presents as waxy and sometimes erythematous areas of scale, referred to commonly as “cradle cap” when seen on the scalp (Chadha and Jahnke, 2019). ISD most frequently affects skin sites rich in sebaceous activity and has been associated with increased frequency of Malassezia furfur (Broberg and Faergemann, 1989; Tollesson et al., 1997), raising the possibility of yeast-driven biology in its pathogenesis. Neonatal cephalic pustulosis (NCP) is another condition seen in up to 20% of healthy infants and typified by facial pustules starting in the first month of life. NCP has been associated in some studies but not others with increased rates of Malassezia colonization and is sometime treated, as is ISD, with topical anti-fungal creams (Ayhan et al., 2007; Bernier et al., 2002). However, mechanistic links between Malassezia and ISD or NCP have not yet been firmly established. The fact that ETN, ISD and NCP generally self-resolve, are seen in otherwise healthy infants, and do not generally predict risk of long-term skin disease suggests that these cutaneous eruptions represent visible manifestations of early life microbe-immune interactions that remain within the normal physiologic spectrum.
‘Pathologic’ Contexts for Infant Skin-Microbe Interaction
Preterm birth
Preterm birth (PTB) represents the seminal risk factor for morbidity and mortality in the first year of life (Callaghan et al., 2006; Kramer et al., 2000). It is thought that disrupted microbe-immune interactions may contribute to the inflammatory neonatal pathologies associated with PTB as well as the differential risk of later life diseases, such as increased rates of asthma (Arrieta et al., 2015; Fujimura and Lynch, 2015) and food allergies (Feehley et al., 2019) or reduced rates of atopic dermatitis (Schoch et al., 2021). The structural and functional immaturity of skin in preterm versus full term birth (FTB) infants (Narendran et al., 2010) does appear to affect establishment of the skin microbiome and the host immune response to these organisms (Schoch et al., 2019) (Figure 3A). In general, diversity of the neonatal skin microbiome has been shown to positively correlate with gestational age at birth (Pammi et al., 2017). One cross-sectional study of 129 infants documented lower alpha diversity and greater enrichment for Staphylococcus and Escherichia at multiple skin body sites in PTB versus FTB infants (Younge et al., 2018). Fungal species on the skin of PTB infants are notably influenced by antibiotic exposure, body site, NICU environment, mode of delivery, and diet (Paul et al., 2019).
Figure 3: Early life pathologies associated with changes in skin physiology and colonization.
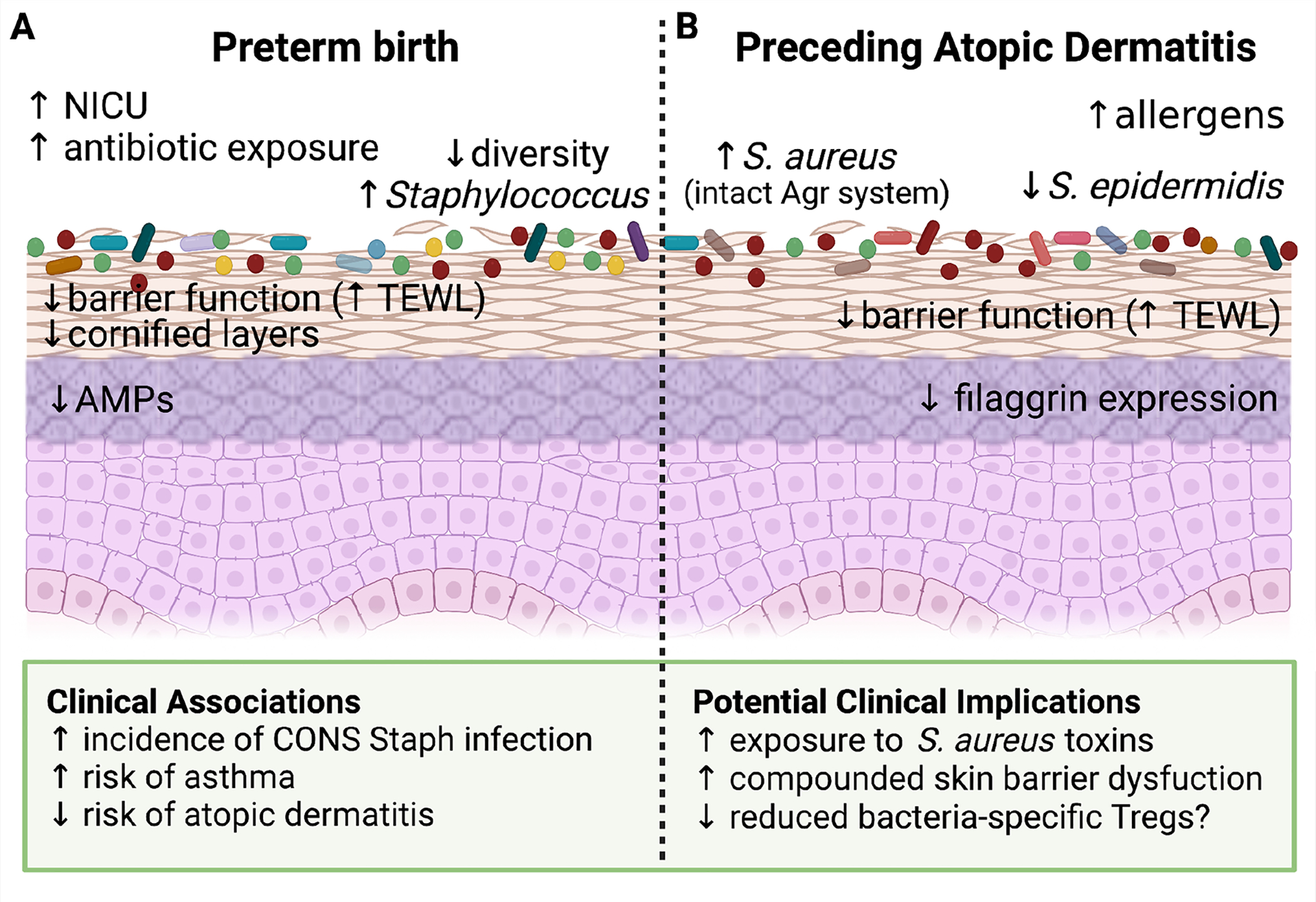
(A) Preterm birth (PTB) has a significant impact on neonatal skin. These infants are often exposed to the NICU environment as well as more frequent topical and systemic antibiotics. Because full maturation of the epidermis occurs during the final weeks of normal in utero gestation, the stratum corneum in PTB infants has fewer cornified layers, reduced antimicrobial peptide (AMP) expression and decreased barrier function as evidenced by increased transepidermal water loss (TEWL). These changes in skin physiology are accompanied by reduced diversity of the PTB skin microbiome, with a notable increase in Staphylococcus prevalence. If and how these altered host and microbial features in preterm infants contribute to the clinical associations seen in this population, such as increased opportunistic infections and decreased later risk of atopic dermatitis, remains an open area of investigation. (B) Blooms of S. aureus and S. epidermidis are well documented during flares of established atopic dermatitis (AD). However, there is also evidence for skin microbial changes that precede disease onset. Specifically, increased colonization by S. aureus, especially stains with a preserved Agr quorum sensing system, and a relative decrease in the prevalence of S. epidermidis have both been observed among infants that go on to develop AD. These ecological differences are accompanied by distinct skin physiological features, namely decreased filaggrin expression and skin barrier function (increased TEWL) that could alter the host response to skin microbes. Collectively, this suggests that an altered relationship with commensal microbes may be established prior to AD onset and contribute to its early pathogenesis alongside other key environmental or allergic exposures.
These changes in skin microbial composition between PTB and FTB infants may relate both to endogenous differences in their skin physiology as well as exogeneous environmental factors. The human skin barrier functionally matures by 34 weeks gestational age (Casterline and Paller, 2020; Evans and Rutter, 1986; King et al., 2013). The skin of infants born before 34 weeks demonstrates an underdeveloped epidermis with fewer cornified layers, reduced VC levels, and lower expression of key dermal proteins (Visscher et al., 2015). This collectively results in compromised skin barrier integrity and increased skin fragility (Evans and Rutter, 1986). PTB infants can also demonstrate cutaneous innate immune dysfunction, for example producing lower levels of antimicrobial proteins but increased amounts of certain proinflammatory cytokines, such as IL-1β, IL-6, IL-8 and monocyte chemoattractant protein-1 (MCP-1) (Visscher et al., 2021).
Systemic infections in PTB infants, many of which are caused by skin-resident bacteria, remain a major source of morbidity and mortality in the first year of life (Steiner et al., 2019). For example, rates of methicillin-resistant S. aureus (MRSA) infections as well as neonatal sepsis induced by S. epidermidis (Dong et al., 2018; Kitajima, 2003) are elevated in PTB infants. This heightened infection risk stems from a combination of both endogenous and exogenous factors. Reduced skin barrier function (Evans and Rutter, 1986) and altered systemic immune function (Melville and Moss, 2013) are clear precipitants. The microbial skin ecology of PTB infants, marked by an increased relative abundance of Staphylococcus spp (Younge et al., 2018) including some strains that contain antibiotic resistance and other virulence genes (Hellmann et al., 2021), may further compound the risk.
How altered composition of the PTB infant skin microbiota relates to longer-term risk of disease remains less clear. Preterm infants have lower rates of atopic dermatitis (AD) compared to full term infants (Schoch et al., 2021), and recent studies have found that longer NICU stays are inversely correlated with later development of AD (Schoch et al., 2021). AD is generally considered an early step in the “atopic march,” subsequently followed by development of food allergies, asthma, and allergic rhinitis (Lowe et al., 2018). Risk of allergic rhinitis was found to be positively correlated with gestational age at birth in a large cohort, i.e. PTB was protective (Crump et al., 2011). Conversely, PTB infants have higher rates of asthma (Zhang et al., 2018). How development of these atopic disorders relates to alterations in the infant skin and gut microbiomes among PTB infants has yet to be determined. Some hypothesize, however, that the altered composition of skin microbes or the fact that microbial exposure occurs earlier in life might functionally contribute to the protection against cutaneous atopy associated with PTB (Crump et al., 2011; Schoch et al., 2021).
Early life antibiotics
Systemic antibiotics during early childhood can be life-saving but have also been associated with increased risk of immune, metabolic and neurobehavioral conditions of pediatric onset (McKeever et al., 2002; Russell et al., 2012). A recent study confirmed an increased risk of asthma, allergic rhinitis, and AD in a population-based cohort study of 14,572 children who received one or more antibiotic prescriptions before the age two (Aversa et al., 2021). Murine studies likewise document that early antibiotic administration can alter severity of later life pathologies (Roubaud-Baudron et al., 2019; Russell et al., 2012; Wlodarska et al., 2011) including susceptibility to models of skin inflammation (Zanvit et al., 2015). The relative role of the intestinal versus cutaneous microbes in these effects however have yet to be teased apart. Studies in adults suggest that oral antibiotics can shift skin microbiota composition (Chien et al., 2019; Marples and Kligman, 1971). Analogous studies have not yet been performed in healthy infants, but documentation of lower skin microbiome diversity in preterm infants that received intravenous antibiotics (Pammi et al., 2017) would indicate that at least temporary changes in the skin microbiota might be expected following early life antibiotics, as is seen for the intestinal microbiome (Moskovicz et al., 2020; Yallapragada et al., 2015). Further work is needed to understand the significance of any changes to the skin microbiome community induced by oral antibiotics or topical antiseptics (Lund and Zukowsky, 2016; Strzępa et al., 2018) used in early life and their relationship to clinical associations with later life skin disease.
Atopic Dermatitis
Atopic dermatitis (AD), often referred to as eczema, is an inflammatory skin condition seen in 15–30% of infants in industrialized countries (Silverberg, 2017), which manifests as chronic, recurrent flares of scaling red patches on the skin. While AD can start in adulthood, most patients present within the first year of life (Chadha and Jahnke, 2019). Flares of disease have long been known to often be accompanied by blooms of S. aureus on affected skin (Geoghegan et al., 2018). More recent work using genomic methods to study the skin microbiome in pediatric AD patients have confirmed this, and also found the relative skin abundance of other coagulase-negative Staphylococci (CoNS) increases during AD flares (Byrd et al., 2017; Key et al., 2021; Kong et al., 2012). S. aureus is thought to contribute to AD pathogenesis through various mechanisms, such as production of proteases, toxins, pruritogens, and superantigens, which collectively elicit and perpetuate atopic skin inflammation (Geoghegan et al., 2018; Kobayashi et al., 2015; Nakatsuji et al., 2016; Ogonowska et al., 2021; Paller et al., 2019; Towell et al., 2021).
We understand less about the role of the skin microbiome in the initial development of AD. However, several studies have tracked early skin microbial composition in young infants revealing microbiome features that associate with subsequent AD development. These include increased skin colonization by S. aureus prior to AD onset, especially by strains that retain a functional quorum sensing system, and a decreased relative abundance of S. epidermidis (Kennedy et al., 2017; Meylan et al., 2017; Nakamura et al., 2020). Additional research is needed to validate that these early microbiome signatures robustly correlate with subsequent AD development, to tease apart the endogenous vs. exogenous factors that lead to early alterations of the skin microbiome in AD patients and to understand the functional contribution of these early microbial shifts towards early events in AD pathogenesis and the atopic march (Figure 3B).
Looking forward
The early life window is replete with microbial and other environmental exposures that have lasting impacts on human health. As one of the body’s largest barrier tissues and a site of rich microbial interactions, skin is a key site for these early formative events. As discussed herein, recent work has shed light on the nature of some of these interactions, but much remains to be understood. With regard to neonatal skin microbial ecology, metagenomic sequencing will help elucidate the influence of various endogenous and exogenous factors on the genomic content and functional capacity of the infant skin microbial community. Likewise, additional work is needed to track individual strains on infant skin and determine the relative contributions of initially seeded versus newly acquired strains in longitudinal shifts in composition of the skin microbiota throughout childhood. Work from the adult skin microbiome (Zhou et al., 2020) would suggest that on-person evolution of strains occurs at least for some species, such as S. epidermidis and C. acne (Conwill et al., 2022). Understanding the timing of strain acquisition and evolution may provide insight into the role of specific strains on pediatric skin health as well as inform potential strategies to manipulate early life skin microbial communities for therapeutic benefit.
On the host side of the equation, a more granular understanding of the immune cell populations in healthy neonatal and pediatric skin would provide a helpful landscape on which to layer observations from in vitro and ex vivo studies that can more explicitly test microbial effects on these cells’ phenotype and function. Pairing such translational work with continued and expanded studies that use murine systems to tease apart unique early life skin-microbiome interactions will be an important strategy for the field. Although skin microbial transfer may face different challenges than fecal transplantation, there is a growing momentum and solid science behind the use of commensal bacteria to improve skin health, specifically for treatment of inflammatory skin diseases such as atopic dermatitis or acne (Nakatsuji et al., 2021). Taking cues from the gut microbiome field (Durack and Lynch, 2019) and the general principle that prevention of disease is often more efficient than cure, it seems a matter of time before we see a push to optimize composition of the neonatal skin microbial community to maximize health and limit disease risk. We have a journey ahead before that can be safely realized, but it seems a path very worthy of rigorous pursuit.
The skin barrier continually mediates interactions with our surrounding microbial environment. In this context, early life microbial-immune crosstalk is central to establishing skin health and homeostasis. Conversely, when disrupted, it can contribute to increased disease risk. Here, we discuss the factors that shape this relationship during a pivotal developmental window.
Acknowledgements
We thank our colleagues in the fields of skin microbiome and early life skin immunity whose work inspired this review piece and apologize to any whose contributions we could not adequately highlight due to space constrains. Figures were generated using Biorender. LRD is supported by 5T32AI007334-33 from NIAID. TCS is supported by a Sun Pharma Award from the Dermatology Foundation and grant DP2AI44968 from NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
TCS serves as a member of the Scientific Advisory Board of Concerto Biosciences.
References
- Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, Saya H, Amagai M, and Nagao K (2015). Hair follicle–derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nature Medicine 21, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M-C, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science Translational Medicine 7. [DOI] [PubMed] [Google Scholar]
- Aversa Z, Atkinson EJ, Schafer MJ, Theiler RN, Rocca WA, Blaser MJ, and LeBrasseur NK (2021). Association of Infant Antibiotic Exposure With Childhood Health Outcomes. Mayo Clinic Proceedings 96, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayhan M, Sancak B, Karaduman A, Arikan S, and Sahin S (2007). Colonization of neonate skin by Malassezia species: relationship with neonatal cephalic pustulosis. Journal of the American Academy of Dermatology 57, 1012–1018. [DOI] [PubMed] [Google Scholar]
- Bager P, Wohlfahrt J, and Westergaard T (2008). Caesarean delivery and risk of atopy and allergic disesase: Meta-analyses. Clinical and Experimental Allergy 38, 634–642. [DOI] [PubMed] [Google Scholar]
- Bautista MIB, Wickett RR, Visscher MO, Pickens WL, and Hoath SB (2000). Characterization of Vernix Caseosa as a Natural Biofilm: Comparison to Standard Oil-Based Ointments. Pediatric Dermatology 17, 253–260. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, and Naik S (2013). Compartmentalized and systemic control of tissue immunity by commensals. Nature Immunology 14, 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Segre JA. (2014). Dialogue between skin microbiota and immunity. Science 346, 950–954. [DOI] [PubMed] [Google Scholar]
- Bernier V, Weill F, Hirigoyen V, Elleau C, Feyler A, Labrèze V, Sarlangue J, Chène G, Couprie B, and Taïeb A (2002). Skin colonization by Malassezia species in neonates: a prospective study and relationship with neonatal cephalic pustulosis. Archives of Dermatology 138, 215–218. [DOI] [PubMed] [Google Scholar]
- Broberg A, and Faergemann J (1989). Infantile seborrhoeic dermatitis and Pityrosporum ovale. The British Journal of Dermatology 120, 359–362. [DOI] [PubMed] [Google Scholar]
- Brunner PM, Israel A, Zhang N, Leonard A, Wen HC, Huynh T, Tran G, Lyon S, Rodriguez G, Immaneni S, et al. (2018). Early-onset pediatric atopic dermatitis is characterized by TH2/TH17/TH22-centered inflammation and lipid alterations. Journal of Allergy and Clinical Immunology 141, 2094–2106. [DOI] [PubMed] [Google Scholar]
- Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng W-I, Conlan S, Program NCS, Belkaid Y, Segre JA, and Kong HH (2017). Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Science Translational Medicine 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AL, Belkaid Y, and Segre JA (2018). The human skin microbiome. Nature Reviews Microbiology 16, 143–155. [DOI] [PubMed] [Google Scholar]
- Callaghan WM, MacDorman MF, Rasmussen SA, Qin C, and Lackritz EM (2006). The Contribution of Preterm Birth to Infant Mortality Rates in the United States. Pediatrics 118, 1566–1573. [DOI] [PubMed] [Google Scholar]
- Capone KA, Dowd SE, Stamatas GN, and Nikolovski J (2011). Diversity of the human skin microbiome early in life. Journal of Investigative Dermatology 131, 2026–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casterline BW, and Paller AS (2020). Early development of the skin microbiome: therapeutic opportunities. Pediatric Research 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha A, and Jahnke M (2019). Common neonatal rashes. Pediatric Annals 48, e16–e22. [DOI] [PubMed] [Google Scholar]
- Chen YE, Fischbach MA, and Belkaid Y (2018). Skin microbiota–host interactions. Nature 553, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YE, Bouladoux N, Hurabielle C, Mattke AM, Belkaid Y, and Fischbach MA (2019). Decoding commensal-host communication through genetic engineering of Staphylococcus epidermidis. BioRxiv 3, 664656. [Google Scholar]
- Chien AL, Tsai J, Leung S, Mongodin EF, Nelson AM, Kang S, and Garza LA (2019). Association of Systemic Antibiotic Treatment of Acne With Skin Microbiota Characteristics. JAMA Dermatology 155, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, and Aagaard KM (2017). Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nature Medicine 23, 314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han S-J, Chen YE, Li K, Farhat S, Weckel A, et al. (2019). MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science (New York, NY) 366, eaax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Kanti V, Polak-Witka K, Blume-Peytavi U, Spyrou GM, and Vogt A (2021). The Potential Relevance of the Microbiome to Hair Physiology and Regeneration: The Emerging Role of Metagenomics. Biomedicines 9, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conwill A, Kuan AC, Damerla R, Poret AJ, Baker JS, Tripp AD, Alm EJ, and Lieberman TD (2022). Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host & Microbe 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoro KM, Hitraya-Low M, Taravati K, Sandoval PM, Kim E, Sugarman J, Pauli ML, Liao W, and Rosenblum MD (2017). Skin-infiltrating, interleukin-22-producing T cells differentiate pediatric psoriasis from adult psoriasis. Journal of the American Academy of Dermatology 77, 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte KZ, Rao C, Rakoff-Nahoum S, and Foster KR (2021). Ecological rules for the assembly of microbiome communities. PLOS Biology 19, e3001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump C, Sundquist K, Sundquist J, and Winkleby MA (2011). Gestational age at birth and risk of allergic rhinitis in young adulthood. Journal of Allergy and Clinical Immunology 127, 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwala MO, Karthikeyan D, Vasquez KS, Farhat S, Taravati K, Leitner EG, Pauli M, Lowe MM, Rosenblum MD, and Scharschmidt TC (2020). Developing human fetal skin demonstrates a unique lymphocyte signature. BioRxiv. [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, and Knight R (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America 107, 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, de Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, et al. (2016). Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nature Medicine 2016 22:3 22, 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Speer CP, and Glaser K (2018). Beyond sepsis: Staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity. Virulence 9, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J, and Lynch S. v. (2019). The gut microbiome: Relationships with disease and opportunities for therapy. The Journal of Experimental Medicine 216, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, Milder M, le Bourhis L, Soudais C, Treiner E, et al. (2011). Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17–secreting T cells. Blood 117, 1250–1259. [DOI] [PubMed] [Google Scholar]
- Evans NJ, and Rutter N (1986). Development of the Epidermis in the Newborn. Biol. Neonate 49, 74–80. [DOI] [PubMed] [Google Scholar]
- Fadlallah J, el Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, et al. (2018). Microbial ecology perturbation in human IgA deficiency. Science Translational Medicine 10, 1217. [DOI] [PubMed] [Google Scholar]
- Feehley T, Plunkett C, Bao R, Choi Hong S, Culleen E, Belda-Ferre P, Campbell E, Aitoro R, Nocerino R, Paparo L, et al. (2019). Healthy infants harbor intestinal bacteria that protect against food allergy. Nature Medicine 25, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Program, N.I.H.I.S.C.C.S., et al. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr J, Pfisterer S, and Gloor M (2000). Direct comparison of skin physiology in children and adults with bioengineering methods. Pediatric Dermatology 17, 436–439. [DOI] [PubMed] [Google Scholar]
- Fluhr J, Darlenski R, Lachmann N, Baudouin C, Msika P, de Belilovsky C, and Hachem J (2012). Infant epidermal skin physiology: adaptation after birth. The British Journal of Dermatology 166, 483–490. [DOI] [PubMed] [Google Scholar]
- Fujimura K, and Lynch S (2015). Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host & Microbe 17, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T (2015). Historical Contingency in Community Assembly: Integrating Niches, Species Pools, and Priority Effects. 10.1146/Annurev-Ecolsys-110411-160340 46, 1–23. [DOI] [Google Scholar]
- Gallo RL (2017). Human Skin Is the Largest Epithelial Surface for Interaction with Microbes. Journal of Investigative Dermatology 137, 1213–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JA, Irvine AD, and Foster TJ (2018). Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends in Microbiology 26, 484–497. [DOI] [PubMed] [Google Scholar]
- Grice EA, and Segre JA (2011). The skin microbiome. Nature Reviews Microbiology 9, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JB, Cong Z, Imamura-Kawasawa Y, Kidd BA, Dudley JT, Thiboutot DM, and Nelson AM (2018). Isolation and Identification of the Follicular Microbiome: Implications for Acne Research. Journal of Investigative Dermatology 138, 2033–2040. [DOI] [PubMed] [Google Scholar]
- Harries MJ, and Paus R (2010). The pathogenesis of primary cicatricial alopecias. American Journal of Pathology 177, 2152–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JR, and Schick B (1956). Erythema neonatorum. A.M.A. Journal of Diseases of Children 92, 27–32. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Linehan JL, Shih H-Y, Bouladoux N, Han S-J, Smelkinson M, Sen SK, Byrd AL, Enamorado M, Yao C, et al. (2019). Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science (New York, N.Y.) 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann KT, Tuura CE, Fish J, Patel JM, and Robinson DA (2021). Viability-Resolved Metagenomics Reveals Antagonistic Colonization Dynamics of Staphylococcus epidermidis Strains on Preterm Infant Skin. MSphere 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghteling P, and Walker WA (2015). Why is initial bacterial colonization of the intestine important to infants’ and children’s health? Journal of Pediatric Gastroenterology and Nutrition 60, 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurabielle C, Link VM, Bouladoux N, Han S-J, Merrill ED, Lightfoot YL, Seto N, Bleck CKE, Smelkinson M, Harrison OJ, et al. (2020). Immunity to commensal skin fungi promotes psoriasiform skin inflammation. Proceedings of the National Academy of Sciences 117, 16465–16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iram N, Mildner M, Prior M, Petzelbauer P, Fiala C, Hacker S, Schöppl A, Tschachler E, and Elbe-Bürger A (2012). Age-related changes in expression and function of Toll-like receptors in human skin. Development (Cambridge, England) 139, 4210–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jö J, Ersch J, and Stallmach T (1999). Assessing Gestational Age From Histology of Fetal Skin: An Autopsy Study of 379 Fetuses. [DOI] [PubMed]
- Jo J-H, Kennedy EA, and Kong HH (2017). Topographical and physiological differences of the skin mycobiome in health and disease. Virulence 8, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashem SW, Igyártó BZ, Gerami-Nejad M, Kumamoto Y, Mohammed J, Jarrett E, Drummond RA, Zurawski SM, Zurawski G, Berman J, et al. (2015). Candida albicans Morphology and Dendritic Cell Subsets Determine T Helper Cell Differentiation. Immunity 42, 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher MM, O’Carroll M, Gallagher A, Murray DM, Galvin AD, Irvine AD, and Hourihane JO (2013). Newborn Transepidermal Water Loss Values: A Reference Dataset. Pediatric Dermatology 30, 712–716. [DOI] [PubMed] [Google Scholar]
- Kennedy EA, Connolly J, Hourihane JO, Fallon PG, Mclean WHI, Murray D, Jo J-H, Segre JA, Kong HH, and Irvine AD (2017). Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol 139, 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy KM, Gerlach MJ, Adam T, Heimesaat MM, Rossi L, Surette MG, Sloboda DM, and Braun T (2021a). Fetal meconium does not have a detectable microbiota before birth. Nature Microbiology 2021 6:7 6, 865–873. [DOI] [PubMed] [Google Scholar]
- Kennedy KM, Bellissimo CJ, Breznik JA, Barrett J, Braun T, Bushman FD, de Goffau M, Elovitz MA, Heimesaat MM, Konnikova L, et al. (2021b). Over-celling fetal microbial exposure. Cell 184, 5839–5841. [DOI] [PubMed] [Google Scholar]
- Key FM, Khadka VD, Romo-González C, Blake KJ, Deng L, Lynn TC, Lee JC, Chiu IM, García-Romero MT, and Lieberman TD (2021). On-person adaptive evolution of Staphylococcus aureus during atopic dermatitis increases disease severity. BioRxiv 2021.03.24.436824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Kobayashi H, O’goshi K, and Tagami H (2006). Impairment of skin barrier function is not inherent in atopic dermatitis patients: a prospective study conducted in newborns. Pediatric Dermatology 23, 109–113. [DOI] [PubMed] [Google Scholar]
- Kim EH, and Burks AW (2020). Food allergy immunotherapy: Oral immunotherapy and epicutaneous immunotherapy. Allergy 75, 1337–1346. [DOI] [PubMed] [Google Scholar]
- King A, Balaji S, and Keswani SG (2013). Biology and Function of Fetal and Pediatric Skin. Facial Plastic Surgery Clinics of North America 21, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima H (2003). Prevention of methicillin-resistant Staphylococcus aureus infections in neonates. Pediatrics International : Official Journal of the Japan Pediatric Society 45, 238–245. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, Kong HH, Amagai M, and Nagao K (2015). Dysbiosis and Staphyloccus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity 42, 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo JH, Shih HY, Truong A, Doebel T, Sakamoto K, Cui CY, et al. (2019). Homeostatic Control of Sebaceous Glands by Innate Lymphoid Cells Regulates Commensal Bacteria Equilibrium. Cell 176, 982–997. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Program NCS, et al. (2012). Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Research 22, 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MS, Demissie K, Yang H, Platt RW, Sauvé R, Liston R, and for the Fetal and Infant Health Study Group of the Canadian Perinatal Surveillance System, for the F. and I.H.S.G. of the C.P.S.S. (2000). The Contribution of Mild and Moderate Preterm Birth to Infant Mortality. JAMA 284, 843–849. [DOI] [PubMed] [Google Scholar]
- Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, Ryan AF, di Nardo A, and Gallo RL (2010). Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. Journal of Investigative Dermatology 130, 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech JM, Dhariwala MO, Lowe MM, Chu K, Merana GR, Cornuot C, Weckel A, Ma JM, Leitner EG, Gonzalez JR, et al. (2019). Toxin-Triggered Interleukin-1 Receptor Signaling Enables Early-Life Discrimination of Pathogenic versus Commensal Skin Bacteria. Cell Host and Microbe 26, 795–809. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux F, Bellet D, Daviaud C, el Morr Y, Darbois A, Niort K, Procopio E, Salou M, Gilet J, Ryffel B, et al. (2019). Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science (New York, N.Y.) 366, 494–499. [DOI] [PubMed] [Google Scholar]
- Lehtimäki J, Karkman A, Laatikainen T, Paalanen L, von Hertzen L, Haahtela T, Hanski I, and Ruokolainen L (2017). Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Scientific Reports 2017 7:1 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Reantragoon R, Kostenko L, Corbett A, Varigos G, and Carbone F (2017). The frequency of mucosal-associated invariant T cells is selectively increased in dermatitis herpetiformis. The Australasian Journal of Dermatology 58, 200–204. [DOI] [PubMed] [Google Scholar]
- Linehan JL, Harrison OJ, Han S-J, Byrd AL, Vujkovic-Cvijin I, Villarino A. v, Sen SK, Shaik J, Smelkinson M, Tamoutounour S, et al. (2018). Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe AJ, Leung DYM, Tang MLK, Su JC, and Allen KJ (2018). The skin as a target for prevention of the atopic march. Annals of Allergy, Asthma & Immunology 120, 145–151. [DOI] [PubMed] [Google Scholar]
- Lund C, and Zukowsky K (2016). Bathing and Beyond: Current Bathing Controversies for Newborn Infants. Advances in Neonatal Care 16, S13–S20. [DOI] [PubMed] [Google Scholar]
- Manus MB, Kuthyar S, Perroni-Marañón AG, la Mora AN, and Amato KR (2020). Infant Skin Bacterial Communities Vary by Skin Site and Infant Age across Populations in Mexico and the United States. MSystems 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini G, Ulfgren A-K, Loré K, Ståbi B, Berggren V, and Lonne-Rahm S (2001). Erythema Toxicum Neonatorum: An Immunohistochemical Analysis. Pediatric Dermatology 18, 177–187. [DOI] [PubMed] [Google Scholar]
- Marchini G, Lindow S, Brismar H, Ståbi B, Berggren V, Ulfgren A-K, Lonne-Rahm S, Agerberth B, and Gudmundsson GH (2002). The newborn infant is protected by an innate antimicrobial barrier: peptide antibiotics are present in the skin and vernix caseosa. British Journal of Dermatology 147, 1127–1134. [DOI] [PubMed] [Google Scholar]
- Marchini G, Ståbi B, Kankes K, Lonne-Rahm S, Østergaard M, and Nielsen S (2003). AQP1 and AQP3, Psoriasin, and Nitric Oxide Synthases 1–3 are Inflammatory Mediators in Erythema Toxicum Neonatorum. Pediatric Dermatology 20, 377–384. [DOI] [PubMed] [Google Scholar]
- Marchini G, Nelson A, Edner J, Lonne-Rahm S, Stavréus-Evers A, and Hultenby K (2005). Erythema toxicum neonatorum is an innate immune response to commensal microbes penetrated into the skin of the newborn infant. Pediatric Research 58, 613–616. [DOI] [PubMed] [Google Scholar]
- Marchini G, Hultenby K, Nelson A, Yektaei-Karin E, Ståbi B, Lonne-Rahm S, Ulfgren A-K, and Brismar H (2007). Increased expression of HMGB-1 in the skin lesions of erythema toxicum. Pediatric Dermatology 24, 474–482. [DOI] [PubMed] [Google Scholar]
- Marples RR, and Kligman AM (1971). Ecological Effects of Oral Antibiotics on the Microflora of Human Skin. Archives of Dermatology 103, 148–153. [PubMed] [Google Scholar]
- McGovern N, Shin A, Low G, Low D, Duan K, Yao LJ, Msallam R, Low I, Shadan NB, Sumatoh HR, et al. (2017). Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature 546, 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeever TM, Lewis SA, Smith C, Collins J, Heatlie H, Frischer M, and Hubbard R (2002). Early exposure to infections and antibiotics and the incidence of allergic disease: A birth cohort study with the West Midlands General Practice Research Database. Journal of Allergy and Clinical Immunology 109, 43–50. [DOI] [PubMed] [Google Scholar]
- Melville JM, and Moss TJM (2013). The immune consequences of preterm birth. Frontiers in Neuroscience 0, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, Vial Y, Prod’hom G, Greub G, Kypriotou M, et al. (2017). Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. Journal of Investigative Dermatology 137, 2497–2504. [DOI] [PubMed] [Google Scholar]
- Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, Shepherdson E, Singh GSN, Pai R, Shanti A, et al. (2021). Microbial exposure during early human development primes fetal immune cells. Cell 184, 3394–3409. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mold JE, Venkatasubrahmanyam S, Burt TD, Michaëlsson J, Rivera JM, Galkina SA, Weinberg K, Stoddart CA, and McCune JM (2010). Fetal and Adult Hematopoietic Stem Cells Give Rise to Distinct T Cell Lineages in Humans. Science 330, 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovicz V, Gross A, and Mizrahi B (2020). Extrinsic factors shaping the skin microbiome. Microorganisms 8, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K, Kobayashi T, Moro K, Ohyama M, Adachi T, Kitashima DY, Ueha S, Horiuchi K, Tanizaki H, Kabashima K, et al. (2012). Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nature Immunology 13, 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. (2012). Compartmentalized Control of Skin Immunity by Resident Commensals. Science (New York, N.Y.) 337, 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, Shoji T, Takada S, Nakagawa S, Oguma R, et al. (2020). Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Science Translational Medicine 12, eaay4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, and Gallo RL (2013). The microbiome extends to subepidermal compartments of normal skin. Nature Communications 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, Hata TR, and Gallo RL (2016). Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. Journal of Investigative Dermatology 136, 2192–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik A. v, et al. (2017). Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Science Translational Medicine 9, eaah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Hata T, Tong Y, Cheng J, Shafiq F, Butcher A, Salem S, Brinton S, Rudman Spergel A, Johnson K, et al. (2021). Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nature Medicine 27, 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendran V, Visscher MO, Abril I, Hendrix SW, and Hoath SB (2010). Biomarkers of Epidermal Innate Immunity in Premature and Full-Term Infants. Pediatric Research 2010 67:4 67, 382–386. [DOI] [PubMed] [Google Scholar]
- Nayfach S, Rodriguez-Mueller B, Garud N, and Pollard K (2016). An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Research 26, 1612–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negele K, Heinrich J, Borte M, von Berg A, Schaaf B, Lehmann I, Wichmann H, and Bolte G (2004). Mode of delivery and development of atopic disease during the first 2 years of life. Pediatr Allergy Immunol. 15, 48–54. [DOI] [PubMed] [Google Scholar]
- Nishijima K, Yoneda M, Hirai T, Takakuwa K, and Enomoto T (2019). Biology of the vernix caseosa: A review. Journal of Obstetrics and Gynaecology Research 45, 2145–2149. [DOI] [PubMed] [Google Scholar]
- Ogonowska P, Gilaberte Y, Barańska-Rybak W, and Nakonieczna J (2021). Colonization With Staphylococcus aureus in Atopic Dermatitis Patients: Attempts to Reveal the Unknown. Frontiers in Microbiology 0, 3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Conlan S, Polley EC, Segre JA, and Kong HH (2012). Shifts in human skin and nares microbiota of healthy children and adults. Genome Medicine 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Segre JA, Barnabas B, Blakesley R, Bouffard G, Brooks S, et al. (2014). Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Park M, Program NCS, Kong HH, and Segre JA (2016). Temporal Stability of the Human Skin Microbiome. Cell 165, 854–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okah FA, Randall Wickett R, And KP, Hoath SB, and Heath SB (1994). Human Newborn Skin: The Effect of Isopropanol on Skin Surface Hydrophobicity. [PubMed]
- Oren A, and Garrity GM (2021). Notification that new names of prokaryotes, new combinations, and new taxonomic opinions have appeared in volume 70, part 11 of the IJSEM. International Journal of Systematic and Evolutionary Microbiology 71, 004645. [DOI] [PubMed] [Google Scholar]
- Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, Luger T, and Irvine AD (2019). The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol 143, 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammi M, O’Brien JL, Ajami NJ, Wong MC, Versalovic J, and Petrosino JF (2017). Development of the cutaneous microbiome in the preterm infant: A prospective longitudinal study. PLOS ONE 12, e0176669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Schwardt NH, Jo JH, Zhang Z, Pillai V, Phang S, Brady SM, Portillo JA, MacGibeny MA, Liang H, et al. (2021). Shifts in the Skin Bacterial and Fungal Communities of Healthy Children Transitioning through Puberty. Journal of Investigative Dermatology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AA, Hoffman KL, Hagan JL, Sampath V, Petrosino JF, and Pammi M (2019). Fungal cutaneous microbiome and host determinants in preterm and term neonates. Pediatric Research 2019 88:2 88, 225–233. [DOI] [PubMed] [Google Scholar]
- Pellicci DG, Koay H-F, and Berzins SP (2020). Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge. Nature Reviews Immunology 20, 756–770. [DOI] [PubMed] [Google Scholar]
- Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, Fujimura KE, Burt TD, and Lynch S. v. (2020). Viable bacterial colonization is highly limited in the human intestine in utero. Nature Medicine 2020 26:4 26, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran-Ressler RR, Devapatla S, Lawrence P, and Brenna JT (2008). Branched Chain Fatty Acids Are Constituents of the Normal Healthy Newborn Gastrointestinal Tract. Pediatric Research 2008 64:6 64, 605–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang H-E, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, et al. (2018). Tissue signals imprint ILC2 identity with anticipatory function. Nature Immunology 2018 19:10 19, 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Bouladoux N, Claesen J, Chen YE, Byrd AL, Constantinides MG, Merrill ED, Tamoutounour S, Fischbach MA, and Belkaid Y (2018). Contextual control of skin immunity and inflammation by Corynebacterium. The Journal of Experimental Medicine 215, 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubaud-Baudron C, Ruiz V, Swan A, Vallance B, Ozkul C, Pei Z, Li J, Battaglia T, Perez-Perez G, and Blaser M (2019). Long-Term Effects of Early-Life Antibiotic Exposure on Resistance to Subsequent Bacterial Infection. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet M-R, Mohn WW, McNagny KM, et al. (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Reports 13, 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkany I, and Gaylarde CC (1967). SKIN FLORA OF THE NEWBORN. The Lancet 289, 589–590. [DOI] [PubMed] [Google Scholar]
- Scharschmidt TC, and Fischbach MA (2013). What Lives On Our Skin: Ecology, Genomics and Therapeutic Opportunities Of the Skin Microbiome. Drug Discovery Today. Disease Mechanisms 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Truong H-A, Gearty S. v, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, et al. (2015). A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Pauli ML, Leitner EG, Chu K, Truong H-A, Lowe MM, Rodriguez RS, Ali N, Laszik ZG, et al. (2017). Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host and Microbe 21, 467–477. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch JJ, Monir RL, Satcher KG, Harris J, Triplett E, and Neu J (2019). The infantile cutaneous microbiome: A review. Pediatric Dermatology 36, 574–580. [DOI] [PubMed] [Google Scholar]
- Schoch JJ, Miranda N, Garvan CW, Monir RL, Neu J, and Lemas DJ (2021). Duration of neonatal intensive care unit exposure associated with decreased risk of atopic dermatitis. Pediatric Dermatology 38, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz F, Badgley BD, Sadowsky MJ, and Kaplan DH (2014). Immune Mediated Shaping of Microflora Community Composition Depends on Barrier Site. PLOS ONE 9, e84019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Li W, Hixon JA, Bouladoux N, Belkaid Y, Dzutzev A, and Durum SK (2014). Adaptive immunity to murine skin commensals. Proceedings of the National Academy of Sciences 111, E2977–E2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg JI (2017). Public Health Burden and Epidemiology of Atopic Dermatitis. Dermatologic Clinics 35, 283–289. [DOI] [PubMed] [Google Scholar]
- Sprockett D, Fukami T, and Relman DA (2018). Role of priority effects in the early-life assembly of the gut microbiota. Nature Reviews Gastroenterology & Hepatology 2018 15:4 15, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatas GN, Nikolovski J, Mack MC, and Kollias N (2011). Infant skin physiology and development during the first years of life: a review of recent findings based on in vivo studies. International Journal of Cosmetic Science 33, 17–24. [DOI] [PubMed] [Google Scholar]
- Steiner L, Diesner SC, and Voitl P (2019). Risk of infection in the first year of life in preterm children: An Austrian observational study. PLoS ONE 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzępa A, Lobo FM, Majewska-Szczepanik M, and Szczepanik M (2018). Antibiotics and autoimmune and allergy diseases: Causative factor or treatment? International Immunopharmacology 65, 328–341. [DOI] [PubMed] [Google Scholar]
- Takahashi T, and Gallo RL (2017). The Critical and Multifunctional Roles of Antimicrobial Peptides in Dermatology. Dermatologic Clinics 35, 39–50. [DOI] [PubMed] [Google Scholar]
- Tamoutounour S, Guilliams M, MontananaSanchis F, Liu H, Terhorst D, Malosse C, Pollet E, Ardouin L, Luche H, Sanchez C, et al. (2013). Origins and Functional Specialization of Macrophages and of Conventional and Monocyte-Derived Dendritic Cells in Mouse Skin. Immunity 39, 925–938. [DOI] [PubMed] [Google Scholar]
- Tastan C, Karhan E, Zhou W, Fleming E, Voigt AY, Yao X, Wang L, Horne M, Placek L, Kozhaya L, et al. (2018). Tuning of human MAIT cell activation by commensal bacteria species and MR1-dependent T-cell presentation. Mucosal Immunology 11, 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome JJC, Bickham KL, Ohmura Y, Kubota M, Matsuoka N, Gordon C, Granot T, Griesemer A, Lerner H, Kato T, et al. (2015). Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nature Medicine 22, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, Brough HA, Phippard D, Basting M, Feeney M, et al. (2015). Randomized Trial of Peanut Consumption in Infants at Risk for Peanut Allergy. The New England Journal of Medicine 372, 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollesson A, Frithz A, and Stenlund K (1997). Malassezia furfur in infantile seborrheic dermatitis. Pediatric Dermatology 14, 423–425. [DOI] [PubMed] [Google Scholar]
- Towell AM, Feuillie C, Vitry P, da Costa TM, Mathelié-Guinlet M, Kezic S, Fleury OM, McAleer MA, Dufrêne YF, Irvine AD, et al. (2021). Staphylococcus aureus binds to the N-terminal region of corneodesmosin to adhere to the stratum corneum in atopic dermatitis. Proceedings of the National Academy of Sciences 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, Guttman DS, Becker AB, Mandhane PJ, Turvey SE, et al. (2017). Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome 2017 5:1 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberoi A, Bartow-McKenney C, Zheng Q, Flowers L, Campbell A, Knight SAB, Chan N, Wei M, Lovins V, Bugayev J, et al. (2021). Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host & Microbe 29, 1235–1248. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher MO, Narendran V, Pickens WL, LaRuffa AA, Meinzen-Derr J, Allen K, and Hoath SB (2005). Vernix Caseosa in Neonatal Adaptation. Journal of Perinatology 2005 25:7 25, 440–446. [DOI] [PubMed] [Google Scholar]
- Visscher MO, Utturkar R, Pickens WL, Laruffa AA, Robinson M, Wickett RR, Narendran V, and Hoath SB (2011). Neonatal skin maturation-vernix caseosa and free amino acids. Pediatric Dermatology 28, 122–132. [DOI] [PubMed] [Google Scholar]
- Visscher MO, Adam R, Brink S, and Odio M (2015). Newborn infant skin: Physiology, development, and care. Clinics in Dermatology 33, 271–280. [DOI] [PubMed] [Google Scholar]
- Visscher MO, Carr AN, Winget J, Huggins T, Bascom CC, Isfort R, Lammers K, and Narendran V (2021). Biomarkers of neonatal skin barrier adaptation reveal substantial differences compared to adult skin. Pediatric Research 89, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker VP, Akinbi HT, Meinzen-Derr J, Narendran V, Visscher M, and Hoath SB (2008). Host Defense Proteins on the Surface of Neonatal Skin: Implications for Innate Immunity. Journal of Pediatrics 152, 777–781. [DOI] [PubMed] [Google Scholar]
- Wang DH, Ran-Ressler R, St Leger J, Nilson E, Palmer L, Collins R, and Brenna JT (2018). Sea Lions Develop Human-like Vernix Caseosa Delivering Branched Fats and Squalene to the GI Tract. Scientific Reports 2018 8:1 8, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Willing B, Keeney K, Menendez A, Bergstrom K, Gill N, Russell S, Vallance B, and Finlay B (2011). Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infection and Immunity 79, 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang J, Hu Y, Li X, Sun L, Peng Y, Sun Y, Liu B, Bian Z, and Rong Z (2021). Single-cell transcriptome analysis reveals the dynamics of human immune cells during early fetal skin development. Cell Reports 36. [DOI] [PubMed] [Google Scholar]
- Yallapragada SG, Nash CB, and Robinson DT (2015). Early-life exposure to antibiotics, alterations in the intestinal microbiome, and risk of metabolic disease in children and adults. Pediatric Annals 44, e265–e269. [DOI] [PubMed] [Google Scholar]
- Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshio H, Tollin M, Gudmundsson GH, Lagercrantz H, Jörnvall H, Marchini G, and Agerberth B (2003). Antimicrobial Polypeptides of Human Vernix Caseosa and Amniotic Fluid: Implications for Newborn Innate Defense. Pediatric Research 53, 211–216. [DOI] [PubMed] [Google Scholar]
- Younge NE, Araújo-Pérez F, Brandon D, and Seed PC (2018). Early-life skin microbiota in hospitalized preterm and full-term infants. Microbiome 2018 6:1 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef W, Wickett R, and Hoath S (2001). Surface free energy characterization of vernix caseosa. Potential role in waterproofing the newborn infant. Skin Research and Technology : Official Journal of International Society for Bioengineering and the Skin (ISBS) [and] International Society for Digital Imaging of Skin (ISDIS) [and] International Society for Skin Imaging (ISSI) 7, 10–17. [DOI] [PubMed] [Google Scholar]
- ben Youssef G, Tourret M, Salou M, Ghazarian L, Houdouin V, Mondot S, Mburu Y, Lambert M, Azarnoush S, Diana J-S, et al. (2018). Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. Journal of Experimental Medicine 215, 459–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanvit P, Konkel JE, Jiao X, Kasagi S, Zhang D, Wu R, Chia C, Ajami NJ, Smith DP, Petrosino JF, et al. (2015). Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nature Communications 6, 8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma C, Yang A, Zhang R, Gong J, and Mo F (2018). Is preterm birth associated with asthma among children from birth to 17 years old? -A study based on 2011–2012 US National Survey of Children’s Health. Italian Journal of Pediatrics 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Spoto M, Hardy R, Guan C, Fleming E, Larson PJ, Brown JS, and Oh J (2020). Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell 180, 454–470. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Liu X, Kong FQ, Duan YY, Yee AL, Kim M, Galzote C, Gilbert JA, and Quan ZX (2019). Age and Mothers: Potent Influences of Children’s Skin Microbiota. Journal of Investigative Dermatology 139, 2497–2505. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Duan YY, Kong FQ, Galzote C, and Quan ZX (2020). Dynamics of Skin Mycobiome in Infants. Frontiers in Microbiology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]


