Abstract
A previously unknown chemolithoautotrophic arsenite-oxidizing bacterium has been isolated from a gold mine in the Northern Territory of Australia. The organism, designated NT-26, was found to be a gram-negative motile rod with two subterminal flagella. In a minimal medium containing only arsenite as the electron donor (5 mM), oxygen as the electron acceptor, and carbon dioxide-bicarbonate as the carbon source, the doubling time for chemolithoautotrophic growth was 7.6 h. Arsenite oxidation was found to be catalyzed by a periplasmic arsenite oxidase (optimum pH, 5.5). Based upon 16S rDNA phylogenetic sequence analysis, NT-26 belongs to the Agrobacterium/Rhizobium branch of the α-Proteobacteria and may represent a new species. This recently discovered organism is the most rapidly growing chemolithoautotrophic arsenite oxidizer known.
Arsenic is found in many environments and is toxic to life in some forms. When present in insoluble forms, which are nontoxic, arsenic is frequently found as a mineral in combination with sulfur (e.g., orpiment [As2S3] and realgar [AsS]) and especially with iron and sulfur (e.g., arsenopyrite [FeAsS]). The oxidation of these insoluble forms, chemically and/or microbiologically, results in the formation of arsenite (AsIII [H3AsO3]). In environments such as acid mine drainage, arsenite concentrations can be extremely high (2 to 13 mg/liter) (22). The arsenite formed in various environments can then be oxidized to arsenate (AsV [H2AsO4− + H+]). Both of these soluble forms of arsenic, arsenite and arsenate, are toxic to living organisms, with arsenite considered more toxic than arsenate (5, 22).
Interestingly, the chemical oxidation of arsenite to arsenate is slow. Most arsenite is, therefore, oxidized to arsenate microbiologically (22). Arsenite-oxidizing bacteria were first described in 1918 (7). These organisms, as well as a number of others that have been isolated more recently, are almost all heterotrophic arsenite-oxidizing bacteria, as they require the presence of organic matter for growth (17, 18, 23, 24). The most common organism found has been Alcaligenes faecalis (7, 17, 18, 23, 24). For these heterotrophic bacteria, the oxidation (equation 1) is considered a detoxification mechanism rather than one that can support growth, despite the fact that the reaction is exergonic.
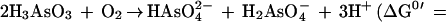 |
1 |
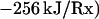 |
Only one bacterium has been described that is able to use the energy gained from this reaction for growth. This organism, Pseudomonas arsenitoxidans, was found to grow chemolithoautotrophically with arsenite, oxygen, and carbon dioxide. The fastest doubling time reported for growth with arsenite was, however, in the order of 2 days (8).
This report describes a new bacterium, isolated from a gold mine in the Northern Territory of Australia, that can also grow chemolithoautotrophically with arsenite as the electron donor, oxygen as the electron acceptor, and carbon dioxide (CO2) or bicarbonate (HCO3−) as the carbon source. Growth was rapid, with a doubling time of 7.6 h for chemolithoautotrophic growth. Preliminary studies with the enzyme that catalyzes arsenite oxidation are also described.
MATERIALS AND METHODS
Growth and media conditions.
The minimal salt enrichment medium used was essentially that described by Ilyaletdinov and Abdrashitova (8), except that also included in the medium were Na2SO4 · 10H2O (0.07 g/liter), trace elements SL8 (14), Na2SeO3 · 5H2O (0.017 mg/liter), Na2WO4 · 2H2O (0.03 mg/liter), and vitamins (14). The minimal salt growth medium contained Na2SO4 · 10H2O, 0.07 g/liter; KH2PO4, 0.17 g/liter; KCl, 0.05 g/liter; MgCl2 · 6H2O, 0.04 g/liter; CaCl2 · 2H2O, 0.05 g/liter; KNO3, 0.15 g/liter; (NH4)2SO4, 0.1 g/liter; NaHCO3, 0.5 g/liter; and the trace elements and vitamins cited above. The concentration of the electron donor, arsenite (NaAsO2), included in the medium was 5 mM. The final pH of both media was about 8. All incubations were carried out at 28°C.
Isolation of arsenite oxidizers.
For enrichments, samples of arsenopyrite (FeAsS) rock were obtained from moist locations of a gold mine in the Northern Territory of Australia. Small portions of the rock were placed in Macartney bottles containing minimal enrichment medium with no arsenite. These bottles were then transported back to the laboratory and were incubated for 7 days at 28°C. The enrichments were subcultured twice into minimal enrichment medium containing 10 mM arsenite. After the second subculture, each enrichment was serially diluted and spread onto minimal agar (2%) medium containing 10 mM arsenite. After growth, a number of different colonies were purified and then tested for their ability to grow in minimal enrichment medium containing 10 mM arsenite.
Growth on various substrates.
The ability of isolate NT-26 to grow on various carbon sources in the minimal salts growth medium described above was tested. These tests included (i) media containing, at final concentrations of 10 mM, sodium acetate, sodium succinate, sodium fumarate, sodium pyruvate, sodium lactate, sodium malate, and sodium citrate, and (ii) media containing, at final concentrations of 5 mM, salicin, glycerol, inositol, mannitol, sucrose, glucose, arabinose, fructose, trehalose, raffinose, lactose, maltose, xylose, galactose, sorbitol, and rhamnose. All substrates were sterilized separately and were added to the medium before inoculation. Substrates in list (i) were sterilized by filtration; all other substrates were sterilized by autoclaving. Minimal salt media, containing one of the various substrates, were inoculated (1% inoculum) with an overnight culture of bacteria grown with 5 mM acetate. Prior to inoculation, cells were harvested (21,000 × g, 20 min), were resuspended in minimal growth medium, and were centrifuged again (21,000 × g, 20 min). Cells were then suspended in minimal growth medium before inoculation. Following growth, or lack of growth, on a given substrate, organisms were subcultured (1% inoculum) twice to be certain of the result. If growth had not occurred in the first culture, growth in subsequent cultures was not observed.
PCR amplification of 16S rRNA genes.
A bacterial suspension (108 CFU/ml) of isolates NT-25 and NT-26 was boiled for 10 min to release the DNA and was centrifuged for 5 min in a microcentrifuge, and the supernatant was used as the DNA template for PCR amplification of the 16S rRNA genes. PCR amplification was performed in a 100-μl reaction volume containing PCR buffer [67 mM Tris-HCl (pH 8.8), 16.6 mM (NH4)2SO4, 0.45% (vol/vol) Triton X-100, 200 μg ml of gelatin−1] (Biotech International Ltd., Perth, Australia), 1.5 mM MgCl2, each deoxynucleotide phosphate at a concentration of 200 μM, 0.25 μM primer 27f (12), 0.25 μM primer 1525r (12), 5 μl of lysed cells, and 2 U of Tth Plus DNA polymerase (Biotech International Ltd.). All PCR amplifications were performed in a Perkin-Elmer Cetus model 480 thermal cycler (Applied Biosystems, Foster City, Calif.). The PCR conditions consisted of an initial denaturation step at 96°C for 5 min, 28 cycles of 48°C for 1 min, 72°C for 2 min, and 94°C for 1 min and one additional cycle at 48°C for 1 min and 72°C for 5 min to allow all extension products to be completed. The PCR products were purified by using the Promega Wizard Minipreps DNA purification system according to the manufacturer's instructions (Promega Corporation, Annadale, Australia).
16S rDNA sequencing.
The purified PCR product was used as the template for sequencing. A Taq DyeDeoxy Terminator Cycle sequencing kit (Applied Biosystems) or the ABI PRISM Dye Terminator Cycle Sequencing kit (Applied Biosystems) was used following procedures recommended by the manufacturer. The following nine 16S rDNA sequencing primers were used in the sequencing reactions: 27f, 342r, 357f, 519r, 530f, 907r, 1114f, 1525r (12), and 803f (20). The sequencing products were purified according to the manufacturer's instructions. The sequences were determined on an Applied Biosystems 373A DNA sequencer.
Phylogenetic analysis.
The near-full-length 16S rDNA sequences determined for strains NT-25 and NT-26 were aligned with the sequences of Escherichia coli and reference sequences of members of the α-Proteobacteria using the FAST Aligner (version 1.02) within the ARB EDIT tool of the ARB software for the analysis of sequence data (Department of Microbiology, Technische Universität München, Munich, Germany [http://www.mikro.biologie.tumuenchen.de]). The alignment was checked by eye, and some adjustments were made manually. Evolutionary similarities and distances were calculated using the Felsenstein correction (6), and a phylogenetic tree was constructed using the neighbor-joining method of Saitou and Nei (19). Bootstrap analysis of 100 data resamplings was performed with SEQBOOT and CONSENSE (6) to determine the statistical confidence of branch points in the tree. The sequence of Sinorhizobium meliloti was used as the outgroup in phylogenetic analyses.
The following sequences of bacterial strains with strain numbers where available were obtained from GenBank and the Ribosomal Database Project (15) for inclusion in the phylogenetic analyses: “Acinetobacter” sp. strain IF-19 (accession no. X86602), Agrobacterium rhizogenes IFO 13257 (accession no. D14501), Agrobacterium rubi LMG156 (accession no. X67228), Agrobacterium tumefaciens NCPPB 2437 (accession no. D14500), Agrobacterium vitis NCPPB 3354 (accession no. D14502), Rhizobium etli CFN 42 (accession no. U28916), Rhizobium galgae ATCC 43677 (accession no. D11343), Rhizobium gallicum R602 (accession no. U86343), Rhizobium giardinii H152 (accession no. U86344), Rhizobium huatlense S02 (accession no. AF025852), Rhizobium leguminosarum USDA 3270 (accession no. U299386), Rhizobium mongolense USDA 1844 (accession no. U89817), Rhizobium tropici USDA 9030 (accession no. U89832), Sinorhizobium fredii ATCC 35423 (accession no. D14516), Sinorhizobium meliloti LMG 6133 (accession no. X67222), and Sinorhizobium saheli LMG 7837 (accession no. X68390).
Growth experiments.
NT-26 was grown in minimal growth medium containing 5 mM arsenite. Cultures, grown overnight under experimental conditions, were inoculated (5%) into the experimental medium (200 ml). Samples were taken periodically, and either total cell numbers (14) or optical density (600 nm) were determined. Portions of the samples were also centrifuged (15,000 × g, 10 min) in a microfuge to remove cells, and the supernatant was frozen until analyses could be performed.
Analytical methods.
Arsenite was determined using a Varian Spectra AA20 atomic absorption spectrophotometer, with a VGA 76 hydride generator, as described previously (14). Arsenate was determined using high-pressure liquid chromatography (HPLC) (13) and inductively coupled plasma (ICP) (Jobin Yvon 24; France).
Electron microscopy.
Microorganisms were visualized by transmission negative-contrast electron microscopy. Cells, grown in minimal growth medium containing 5 mM arsenite, were harvested (as for preparation of cell extracts) and were then suspended in 50 mM 2-[N-morpholino]ethanesulfonic acid (MES) (pH 7). Small amounts of the suspension were applied to copper grids that had been coated with formvar followed by carbon. Excess preparation was removed, and the grids were stained with one-tenth saturated ammonium molybdate. Grids were viewed using a JEOL JEM100S transmission electron microscope operating at an accelerating voltage of 60,000.
Preparation of cell extracts and spheroplasts.
For preparation of cell extracts, NT-26 was grown to late log phase (i.e., 40 h for minimal arsenite, 18 h for minimal arsenite–0.004% yeast extract, 14 h for minimal arsenite–0.04% yeast extract), with shaking, in 5 liters of minimal growth medium (5 mM arsenite). Cells were harvested by centrifugation for 20 min at 21,000 × g (4°C) and were resuspended in ice-cold MES (pH 5.5) (1 g/10 ml). Cells were disrupted by a single passage through a French press (1.4 × 108 lb/in2) (11). Unbroken cells were removed by centrifugation at 30,000 × g (4°C). The supernatant was desalted by passage through a Sephadex G-25 column.
For spheroplast formation, NT-26 was grown in minimal growth medium containing arsenite (5 mM) and yeast extract (0.004% or 0.04%), for 16 or 13 h, respectively. Cells were harvested by centrifugation for 20 min at 21,000 × g (4°C). Cells (0.5 g/ml) were suspended in ice-cold 10 mM Tris/HCl (pH 8) and were centrifuged at 21,000 × g (4°C) for 20 min. The pellet was suspended in ice-cold 750 mM sucrose 30 mM Tris/HCl pH 8 (0°C). Spheroplasts were formed as described previously (11), with the following modifications: (i) the concentration of lysozyme added was 2.2 mg/ml, and (ii) after the addition of the lysozyme, and before the addition of the EDTA, cells were incubated for 20 min at 0°C and then 50 min at room temperature, with gentle stirring, to permit formation of spheroplasts. For certain experiments (indicated below), the periplasmic fraction was desalted by passage through a Sephadex G-25 column before measurement of enzyme activity.
Enzyme assays.
Malate dehydrogenase was measured as described previously (11). The arsenite oxidase (Aro) activity was measured with an assay based upon that of Anderson et al. (1). The reduction of the artificial electron acceptor 2,4-dichlorophenolindophenol (DCIP) by arsenite, as catalyzed by the Aro, was measured. The concentration of arsenite included in the assay was 2.5 mM, and the concentration of DCIP was 0.25 to 0.4 mM (depending upon the assay buffer pH). The reaction was started by the addition of arsenite. A number of different buffers (50 mM), including MES, 3-[N-morpholino], propanesulfonic acid (MOPS), and sodium acetate were tested at different pHs, indicated below.
Protein determination.
Bradford reagent (3) was used to determine protein concentrations. Bovine serum albumin served as the standard.
Nucleotide sequence accession numbers.
The sequences determined in this study for strains NT-25 and NT-26 have been deposited in GenBank under accession no. AF159452 and AF159453, respectively.
RESULTS
Isolation of new chemolithoautotrophic arsenite oxidizers.
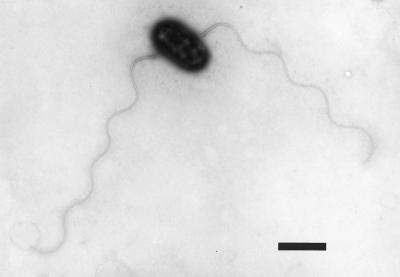
Moist samples of arsenopyrite from a gold mine in the Northern Territory of Australia were inoculated into minimal enrichment medium (without arsenite). Once samples had been brought back to the laboratory, organisms were isolated as described in Materials and Methods. Two organisms were isolated, and these were designated NT-25 and NT-26. Both organisms were found to be small (0.5 by 1.0 μm), motile, gram-negative rods (Fig. 1). The organisms were motile by means of two subterminal flagella (Fig. 1). NT-26 was chosen for further physiological study, as it grew slightly more rapidly than NT-25 with arsenite, oxygen, and CO2-HCO3−.
FIG. 1.
Electron micrograph of NT-26 grown in minimal medium plus arsenite (5 mM). The bar represents 1 μm.
Phylogenetic characterization of new arsenite oxidizers.
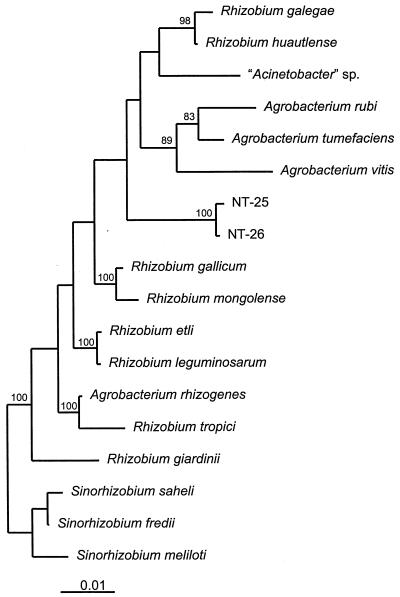
The phylogenetic analysis of almost complete 16S rDNA sequences (>1,476 nucleotides) of the two arsenite-oxidizing isolates NT-25 and NT-26 showed that the two strains belonged to a separate line of descent in the α-Proteobacteria (Fig. 2). The analysis of 1,332 unambiguous nucleotide positions showed that the two strains belonged to a poorly supported subbranch of the Agrobacterium-Rhizobium branch. The sequence similarity of the two strains is 99.8%, and their nearest known phylogenetic relatives are R. gallicum (97.4% sequence similarity) and a misidentified “Acinetobacter” strain IF-19 (97.4% sequence similarity) isolated from a deep subsurface mine gallery (2). On the basis of their phylogenetic position and differing growth and energy requirements when compared with their nearest phylogenetic relatives in the Agrobacterium-Rhizobium subbranch, it is likely that strains NT-25 and NT-26 belong to a new species. However, as the sequence similarity with their nearest described relative, R. gallicum, is >97%, DNA-DNA hybridization of genomic DNA would be required to confirm their separate species status (21).
FIG. 2.
Neighbor-joining tree showing the phylogenetic relationship of arsenite-oxidizing isolates NT-25 and NT-26 with species belonging to the α-Proteobacteria. The sequence of S. meliloti was used as the outgroup. The analysis included data from 1,332 unambiguous nucleotide positions. Significant bootstrap values from 100 analyses are shown at the branch points of the trees. The scale bar represents 1 nucleotide substitution per 100 nucleotides of 16S rRNA sequence.
Growth of strain NT-26 in minimal medium with arsenite.
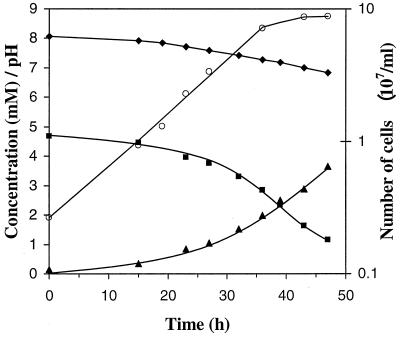
As can be seen in Fig. 3, NT-26 grew aerobically in minimal medium with arsenite (5 mM) as the electron donor and CO2-HCO3− as the carbon source. The experiment presented in Fig. 3 represents the results from one of two separate experiments done (i.e., on separate days); the results of both experiments were essentially the same. Thereafter, NT-26 was subcultured at least 30 times in arsenite minimal medium, and then another two separate experiments were carried out (i.e., on separate days); the results were again essentially the same as those presented in Fig. 3. The doubling time for all experiments was 7.6 ± 0.2 h (average of four experiments). As can be seen in Fig. 3, the growth rate, rate of arsenite utilization, and rate of arsenate formation were proportional during logarithmic growth. For example, during logarithmic growth, the relationship between arsenite oxidized and increase in cell numbers per milliliter was linear; the average of two experiments is 3.6 × 107 cells formed per mM arsenite oxidized. It should be noted that cell numbers are reported, because the optical density (600 nm) of the cultures increased from 0.000 to only 0.006 when cell numbers had increased to 8.7 × 107 per ml.
FIG. 3.
Growth of NT-26 in minimal medium containing arsenite (5 mM). Arithmetic plot: ■, concentration of arsenite; ▴, concentration of arsenate; ⧫, pH. Logarithmic plot: ○, cell number per milliliter.
In all cultures, when the pH reached 7.3 to 7.4, the rate of growth began to slow, although arsenite oxidation continued (Fig. 3). For instance, in the culture shown in Fig. 3, between hours 36 (thereafter growth began to slow; pH 7.3) and 64, cell numbers had only increased from 7.1 × 107 per ml to 10.6 × 107 per ml, while a further 2.7 mM arsenite was oxidized. This observation is consistent with the finding that when NT-26 was inoculated into arsenite (5 mM) minimal medium with an initial pH of 7.4, growth was slowed significantly (i.e., the estimated doubling time increased to greater than 15 h [J. M. Santini and J. M. Macy, unpublished data]).
Inoculation of NT-26 into minimal medium without arsenite resulted in no growth (date not shown). This experiment was carried out four times, with the same results.
The effect of small amounts of organic matter on the growth and arsenite oxidation rates of NT-26.
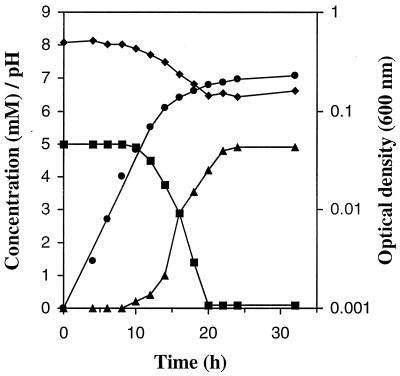
Growth of NT-26 and arsenite oxidation are not inhibited by the presence of small amounts of organic matter (Table 1). Instead, the presence of yeast extract stimulated both the rate of growth and arsenite oxidation; the doubling time was decreased from 7.6 h (in minimal medium plus 5 mM arsenite) to 2.8 ± 0.4 h (average of three experiments) (0.004% yeast extract plus 5 mM arsenite) or 2.7 ± 0.3 h (average of three experiments) (for 0.04% yeast extract plus 5 mM arsenite) (Table 1, Fig. 4). The yeast extract was not used preferentially, as arsenite was oxidized to equimolar amounts of arsenate throughout growth (e.g., Fig. 4). It is also clear that energy was gained from arsenite oxidation during growth in the presence of yeast extract, as the final optical density of NT-26 was greater when arsenite was present (e.g., 0.026 and 0.228 for 0.004% and 0.04% yeast extract plus arsenite, respectively; see Table 1 for results of all experiments) than in its absence (e.g., 0.011 and 0.167 for 0.004% and 0.04% yeast extract only, respectively; see Table 1 for results of all experiments).
TABLE 1.
Growth rate, final OD600, concentration of arsenite used, and medium pH change of strain NT-26 grown in minimal medium containng 0.004% or 0.04% yeast extract, with and without arsenitea
| Medium additions | Generation time (h) | Final OD600 | Concn of arsenite (initial → final) (mM)b | pH (initial → final) |
|---|---|---|---|---|
| AsIII, 0.004% YEc | 3.4 | 0.026 | 5.3 → 0.1 | 8.15 → 6.37 |
| 3.0 | 0.035 | 5.7 → 0.1 | 8.17 → 6.50 | |
| 3.0 | 0.035 | 4.0 → 0.1 | 8.17 → 6.61 | |
| 0.004% YE | 3.6 | 0.011 | 8.02 → 8.28 | |
| 3.0 | 0.015 | 8.12 → 8.39 | ||
| 2.8 | 0.017 | 8.09 → 8.38 | ||
| AsIII, 0.04% YE | 2.8 | 0.210 | 5.3 → 0.1 | 8.07 → 6.44 |
| 2.4 | 0.224 | 4.9 → 0.1 | 8.04 → 6.54 | |
| 3.0 | 0.192 | 4.8 → 0.1 | 8.09 → 6.58 | |
| 0.04% YE | 2.4 | 0.167 | 7.91 → 8.06 | |
| 2.4 | 0.165 | 8.11 → 8.16 | ||
| 2.5 | 0.157 | 8.06 → 8.19 |
Results of three separate experiments.
YE, yeast extract.
The arsenite was oxidized to an equivalent amount of arsenate.
FIG. 4.
Growth of NT-26 in minimal medium plus yeast extract (0.04%) and arsenite (5 mM). Arithmetic plot: ■, concentration of arsenite; ▴, concentration of arsenate; ⧫, pH. Logarithmic plot: ●, optical density.
Growth of NT-26 in a rich medium, such as nutrient broth, was slow; at least 1 week of incubation was required before a significant level of turbidity was observed in cultures, and the cells present were pleomorphic.
Growth on other substrates.
In addition to chemolithoautotrophic growth on arsenite, oxygen, and bicarbonate, NT-26 also grew (within 24 h of inoculation) in minimal medium containing the following carbon and energy sources: acetate, succinate, fumarate, pyruvate, malate, mannitol, sucrose, glucose, arabinose, fructose, trehalose, raffinose, maltose, xylose, or galactose. NT-26 also grew (within 48 h of inoculation) in minimal medium containing the following carbon and energy sources: lactate, salicin, glycerol, lactose, or inositol. Growth was not observed on citrate, sorbitol, or rhamnose.
The Aro of strain NT-26—optimum conditions.
Cells grown in minimal medium containing arsenite (5 mM) and yeast extract (0.004%) were harvested, broken in a French press, and tested for Aro activity. It was found that the pH optimum for Aro activity (average of triplicate measurements) was at pH 5.5 in the presence of 2 mM magnesium (0.14 μmol · min−1 · mg of protein−1). Specific activity in the absence of magnesium was 40% of this maximum, but the pH optimum was the same. Magnesium could be replaced by calcium (2 mM) and manganous (2 mM), but not by either potassium or sodium.
The effect of growth with yeast extract on Aro activity.
The highest Aro activity was measured (average of triplicate measurements) in cells grown in minimal medium with arsenite only (i.e., no yeast extract) (0.24 μmol · min−1 · mg of protein−1). The presence of 0.004% yeast extract in minimal arsenite medium resulted in a decrease in Aro activity (0.14 μmol · min−1 · mg of protein−1) to 58% of chemolithoautotrophic activity. The activity was decreased to 25% of chemolithoautotrophic activity when NT-26 was grown with arsenite plus 0.04% yeast extract (0.06 μmol · min−1 · mg of protein−1). Clearly, in the presence of yeast extract, energy was also gained from the oxidation of yeast extract as well as arsenite.
The cellular location of Aro.
The Aro is located in the periplasm of NT-26, as 88% of the Aro activity was found in the periplasmic fraction of the cell, while 12% remained with the spheroplasts. The opposite was found for the cytoplasmic enzyme malate dehydrogenase; 95% of the malate dehydrogenase activity was associated with the spheroplasts, while only 5% of the activity was detected in the periplasmic fraction.
It should be noted that the activity of the Aro in periplasmic fractions (i.e., in the presence of EDTA [5 mM]) was not stimulated by the presence of 2, 4, or 12 mM magnesium. However, when the protein fraction was separated from the EDTA by desalting (see Materials and Methods), the activity was stimulated (40%) by the addition of 2 mM magnesium, calcium, or manganous. The specific activity of the purified Aro was, however, not stimulated by magnesium (J. M. Santini and J. M. Macy, unpublished data).
DISCUSSION
The first identified arsenite-oxidizing bacterium, named Bacillus arsenoxydans, was isolated from an arsenical cattle dip in South Africa and was described in 1918 by Green (7). The isolation and growth medium contained both arsenite (0.1%) and organic matter (in the form of dung extract). Growth in the absence of organic matter was not tested, and the organism has since been lost. Subsequently, Turner, also working with cattle dips, but in Australia, described the isolation of 15 arsenite-oxidizing bacterial strains; all were isolated by including organic matter in the medium, and were, therefore, heterotrophic arsenite oxidizers (23, 24). Turner studied one isolate (presumably the most rapid arsenite oxidizer) in depth. This organism, Pseudomonas arsenoxydans-quinque, is considered synonymous with Alcaligenes faecalis (5).
Since the work of Turner, a number of other workers have also isolated heterotrophic arsenite-oxidizing bacteria, and most of the isolates have been strains of A. faecalis, found in environments such as raw sewage (18) and soil (17). None of these organisms have been reported as able to grow in the absence of organic matter (i.e., chemolithoautotrophically). Neither the strain isolated by Phillips and Taylor (18), nor that isolated by Osborne and Ehrlich (17), oxidized arsenite at a significant rate until the late exponential phase or stationary phase of growth was reached in batch culture (4, 18). It has, therefore, been suggested that arsenite oxidation by these organisms is a detoxification mechanism (17, 22). Although there is evidence that A. faecalis may be able to derive some energy from arsenite oxidation (4, 5) from an electron transport chain consisting of an arsenite oxidase, azurin, a specific cytochrome c, and presumably a cytochrome c oxidase (1, 16). Certainly, there is a significant amount of energy available from the oxidation of arsenite with oxygen (see equation 1).
The inducible enzyme that catalyzes arsenite oxidation in A. faecalis strain NCIB 8687 was purified from the outer surface of the cytoplasmic membrane of this organism (1). It was found to be a monomeric (85 kDa) oxomolybdoenzyme containing three redox-active centers, including a molybdopterin cofactor and two different iron-sulfur clusters (1, 16).
Prior to the present report, the only described chemolithoautotrophic arsenite-oxidizer (i.e., able to grow in a minimal medium with arsenite, oxygen, and CO2-HCO3−) was P. arsenitoxidans (8). Unfortunately, this organism has been lost, so it was not possible to directly compare it to NT-26. In their report, however, Ilyaletdinov and Abdrashitova reported that P. arsenitoxidans grew chemolithoautotrophically with arsenite more slowly than did NT-26. Their experiments usually lasted about 30 days, and under optimal conditions the fastest doubling time reported was at best 2 days (8). This is considerably slower than the 7.6 h doubling time of NT-26.
Unlike the A. faecalis heterotrophic arsenite oxidizers described above, NT-26 can gain energy for growth from arsenite oxidation. Not only is the organism able to grow chemolithoautotrophically with arsenite, oxygen, and CO2-HCO3−, but when organic matter is present arsenite is oxidized throughout growth, and not only during late log or stationary phase. In addition, with different amounts of yeast extract present in the medium, the amount of growth is greater when arsenite is present than when it is absent—clearly indicating that energy is gained by NT-26 from arsenite oxidation, as well as from the oxidation of yeast extract.
NT-26 is, therefore, a true arsenite-oxidizing bacterium that can gain energy from arsenite oxidation. The yield of cells determined, when NT-26 was grown chemolithoautotrophically with arsenite, oxygen, and CO2/HCO3−, was similar to that observed for growth of P. arsenitoxidans (8).
Phylogenetically, NT-26 (and NT-25) is not related to A. faecalis, which is a member of the β-Proteobacteria (J. M. Santini, P. Durand, L. I. Sly, D. Comrie, B. V. Pauling, and J. M. Macy, unpublished data). Instead, NT-26 and NT-25 are members of the Agrobacterium-Rhizobium branch of the α-Proteobacteria. The carbon/energy substrates that can support the heterotrophic growth of NT-26 are consistant with those used by Agrobacterium and Rhizobium species (9, 10). There is, however, no reported evidence of arsenite oxidation in Agrobacterium or Rhizobium, and the heterotrophic growth of NT-25 and NT-26 is poorer, supporting the phylogenetic evidence that these strains belong to a novel species.
The finding that NT-26 and NT-25 have evolved divergently from A. faecalis also supports the suggestion that the arsenite-oxidizing enzyme of NT-26 may be different from that of A. faecalis. Preliminary work with the Aro of NT-26 has shown that the enzyme is located in the periplasm, while that of A. faecalis (1) is located on the outer surface of the cytoplasmic membrane (i.e., facing the periplasm). The specific activity of the NT-26 enzyme in crude extracts of cells grown with 0.04% yeast extract is almost threefold greater than that of A. faecalis (and 10-fold greater when cells were grown in minimal arsenite medium). In crude extracts, the enzyme of NT-26 is stimulated by magnesium (2 mM) and other divalent cations such as calcium or manganous, while this is not the case with the purified protein (J. M. Santini and J. M. Macy, unpublished data), suggesting that another electron-carrying protein present in the cell extracts may be involved in transferring electrons to DCIP. The pH optimum of the NT-26 Aro is slightly lower (i.e., 5.5 compared to 6.0) than the A. faecalis enzyme (1).
In conclusion, NT-26 is unique because of its ability to grow so rapidly with arsenite chemolithoautotrophically, oxidizing the arsenite with an enzyme (Aro) that is present in the periplasmic space. To gain insight into how the Aro of NT-26 might be involved in energy conservation, the enzyme which has recently been purified (J. M. Santini and J. M. Macy, unpublished data) is being characterized, and the gene(s) encoding the enzyme is being cloned and sequenced.
ACKNOWLEDGMENTS
This work was supported by an Australian Research Council Grant (A09925054) and two Central Large La Trobe University Grants to J.M.M.
We thank Derek Flood for technical assistance and Andy Purvis and Michelle Lau for help in obtaining samples. We thank Aimin Wen, Department of Microbiology and Parasitology, University of Queensland, for DNA sequencing and Mark Fegan, Cooperative Research Centre for Tropical Plant Pathology, for assistance with phylogenetic analyses.
REFERENCES
- 1.Anderson G L, Williams J, Hille R. The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J Biol Chem. 1992;267:23674–23682. [PubMed] [Google Scholar]
- 2.Boivin-Jahns V, Bianchi A, Ruimy R, Garcin J, Daumas S, Christen R. Comparison of phenotypical and molecular methods for the identification of bacterial strains isolated from a deep subsurface environment. Appl Environ Microbiol. 1995;61:3400–3406. doi: 10.1128/aem.61.9.3400-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Ehrlich H L. Inorganic energy sources for chemolithotrophic and mixotrophic bacteria. Geomicrobiol J. 1978;1:65–83. [Google Scholar]
- 5.Ehrlich H L. Geomicrobiology. New York, N.Y: Marcel Dekker, Inc.; 1996. [Google Scholar]
- 6.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 7.Green H H. Description of a bacterium which oxidizes arsenite to arsenate, and of one which reduces arsenate to arsenite, isolated from a cattle-dipping tank. S Afr J Sci. 1918;14:465–467. [Google Scholar]
- 8.Ilyaletdinov A N, Abdrashitova S A. Autotrophic oxidation of arsenic by a culture of Pseudomonas arsenitoxidans. Mikrobiologiya. 1981;50:197–204. [PubMed] [Google Scholar]
- 9.Jordan D C. Family III. Rhizobiaceae. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Baltimore, Md: Williams and Wilkins; 1984. pp. 234–242. [Google Scholar]
- 10.Kersters K, De Ley J. Genus III. Agrobacterium. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Baltimore, Md: Williams and Wilkins; 1984. pp. 244–254. [Google Scholar]
- 11.Krafft T, Macy J M. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur J Biochem. 1998;255:647–653. doi: 10.1046/j.1432-1327.1998.2550647.x. [DOI] [PubMed] [Google Scholar]
- 12.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons; 1991. pp. 115–163. [Google Scholar]
- 13.Macy J M, Michel T A, Kirsch D G. Selenate reduction by a Pseudomonas species: a new mode of anaerobic respiration. FEMS Microbiol Lett. 1989;61:195–198. doi: 10.1016/0378-1097(89)90195-x. [DOI] [PubMed] [Google Scholar]
- 14.Macy J M, Nunan K, Hagen K D, Dixon D R, Harbour P J, Cahill M, Sly L. Chrysiogenes arsenatis gen. nov., sp. nov., a new arsenate-respiring bacterium isolated from gold mine wastewater. Int J Syst Bacteriol. 1996;46:1153–1157. doi: 10.1099/00207713-46-4-1153. [DOI] [PubMed] [Google Scholar]
- 15.Maidak B L, Olsen G L, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:198–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNellis L, Anderson G L. Redox-state dependent chemical inactivation of arsenite oxidase. J Inorganic Biochem. 1998;69:253–257. [Google Scholar]
- 17.Osborne F H, Ehrlich H L. Oxidation of arsenite by a soil isolate of Alcaligenes. J Appl Bacteriol. 1976;41:295–305. doi: 10.1111/j.1365-2672.1976.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 18.Phillips S E, Taylor M L. Oxidation of arsenite to arsenate by Alcaligenes faecalis. Appl Environ Microbiol. 1976;32:392–399. doi: 10.1128/aem.32.3.392-399.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Stackebrandt E, Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol. 1990;136:37–43. doi: 10.1099/00221287-136-1-37. [DOI] [PubMed] [Google Scholar]
- 21.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriol. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 22.Tamaki S, Frankenberger W T., Jr Environmental biochemistry of arsenic. Rev Environ Contam Toxicol. 1992;124:79–110. doi: 10.1007/978-1-4612-2864-6_4. [DOI] [PubMed] [Google Scholar]
- 23.Turner A W. Bacterial oxidation of arsenite. Nature (London) 1949;164:76–77. doi: 10.1038/164076a0. [DOI] [PubMed] [Google Scholar]
- 24.Turner A W. Bacterial oxidation of arsenite. I. Description of bacteria isolated from arsenical cattle-dipping fluids. Aust J Biol Sci. 1954;7:452–478. doi: 10.1071/bi9540452. [DOI] [PubMed] [Google Scholar]