Abstract
Neurons are the longest-lived cells in our bodies and lack DNA replication, which makes them reliant on a limited repertoire of DNA repair mechanisms to maintain genome fidelity. These repair mechanisms decline with age, but we have limited knowledge of how genome instability emerges and what strategies neurons and other long-lived cells may have evolved to protect their genomes over the human life span. A targeted sequencing approach in human embryonic stem cell–induced neurons shows that, in neurons, DNA repair is enriched at well-defined hotspots that protect essential genes. These hotspots are enriched with histone H2A isoforms and RNA binding proteins and are associated with evolutionarily conserved elements of the human genome. These findings provide a basis for understanding genome integrity as it relates to aging and disease in the nervous system.
Neurons are highly specialized postmitotic cells and comprise the major functional cell type of the nervous system. Although there is a limited capacity to generate new neurons throughout life, the majority of neurons age in parallel with the organism, making them especially susceptible to decline from age-related disruptions in cellular homeostasis (1). In total, neurons repair on the order of ~104 to 105 DNA lesions each day, or more than 1 billion repairs over the life span of humans (2). Deficiencies in DNA repair and genome instability have been linked to both developmental and age-associated neurodegenerative diseases (3, 4).
Studies of genome integrity in neurons suggest that their DNA repair efforts focus on transcribed genes at the expense of inactive regions of the genome (5). Accumulation of DNA lesions drives age-associated changes in transcription that lead to a decline in neuronal function (6, 7). Additionally, neuronal activity correlates with the generation of DNA double-strand breaks (DSBs), potentially contributing to genome instability (8, 9). Although genomics approaches have made the study of mitotic neural progenitor cells and the role of somatic mosaicism in neurons accessible, methods to detect DNA damage remain technically challenging, limiting their use (10, 11). Despite the link between genome maintenance and neuronal health, we know surprisingly little about the genome protection strategies with which neurons have evolved to ensure their distinctive longevity.
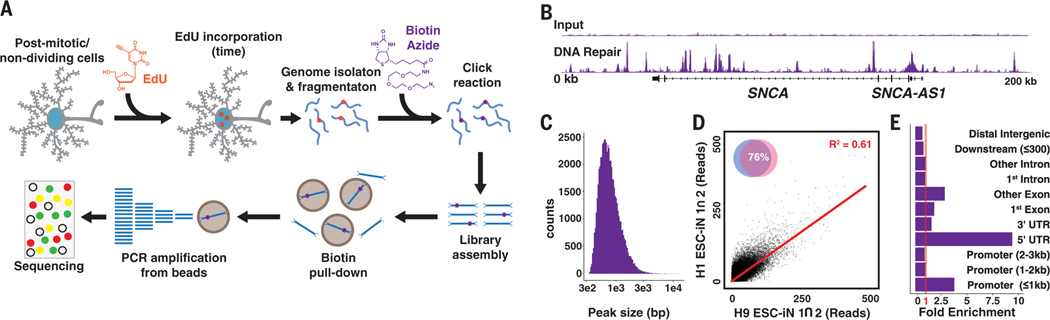
To better understand genome integrity in neurons, we developed a sequencing method capable of capturing a genomic distribution of all DNA repair by the nonreplicative incorporation of the nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU). We generated human embryonic stem cell–induced neurons (ESCiNs) that assume a postmitotic neuron identity after the addition of doxycycline through NEUROG2 expression (fig. S1) (12). ESC-iNs were labeled with EdU for 24 hours, and sites of DNA repair synthesis were identified by the enrichment of next-generation sequencing libraries containing EdU (Fig. 1A) (12). Our method, Repair-seq, revealed many sites enriched for EdU incorporation relative to whole-genome sequencing to the same depth and was relatively free of mitochondrial reads (Fig. 1B and figs. S2 and S3, A and B). EdU-enriched sites appeared as well-defined peaks of ~500 base pairs (Fig. 1C). We applied genome peak calling to our data and found 61,178 reproducible peaks, or DNA repair hotspots (DRHs), covering ~1.6% of the genome (Fig. 1D; fig. S3, C to E; and table S1). These DRHs were distributed throughout the genome on all chromosomes and were enriched in promoters of ≤1 kb, 5′-untranslated regions, and gene bodies (Fig. 1E and fig. S3, F to H).
Fig. 1. EdU incorporated into the genomes of postmitotic neurons by DNA repair can be mapped by next-generation sequencing.
(A) Repair-seq workflow: Neurons are cultured with EdU for 24 hours, the genomes are isolated and fragmented with sonication, a click reaction adds biotin to the EdU, and biotin-DNA fragments are enriched on streptavidin beads and subsequently amplified (12). PCR, polymerase chain reaction. (B) DNA repair peaks from the SNCA locus in EdU-fed neurons compared with input genomes sequenced to the same depth show a site with substantial enrichment. (C) Histogram of DNA repair hotspot peak widths. bp, base pairs. (D) Comparison of intersecting peaks in two H1 and two H9 ESC-iN samples. (E) Fold enrichment of DRHs over predicted genome distribution. UTR, untranslated region.
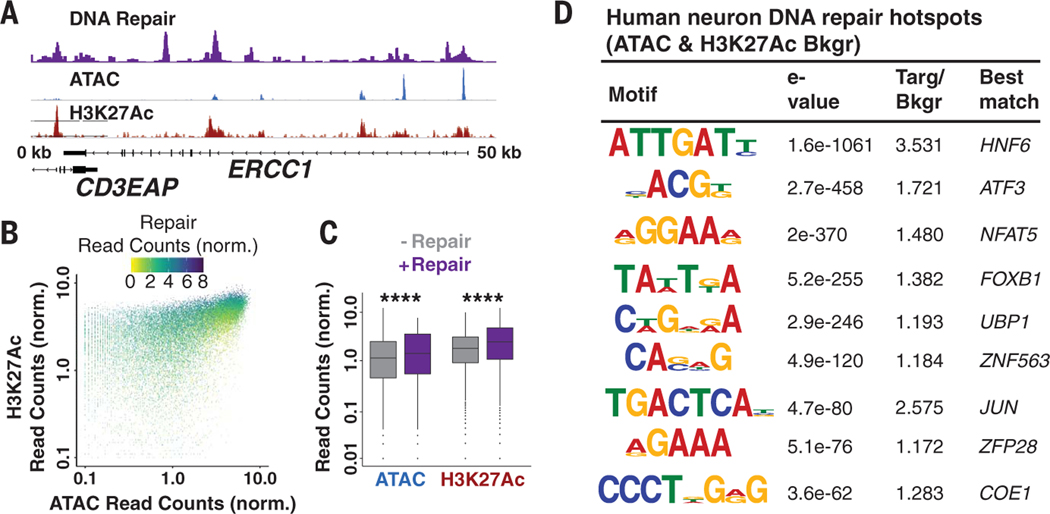
We compared the location of DRHs with open chromatin and active regulatory regions in neurons mapped by ATAC-seq (assay for transposase-accessible chromatin using sequencing) and histone 3 lysine 27 acetylation (H3K27Ac) ChIP-seq (chromatin immunoprecipitation sequencing), and we observed that ~23.5% of hotspots were located within these genomic regions (Fig. 2A and fig. S4, A to C). Intersecting peaks in open regions correlated with greater DNA repair signal strength (Fig. 2, B and C, and fig. S4D). Promoters were enriched for repair, ATAC, and H3K27Ac peak intersections, whereas DRHs not associated with open chromatin were predominantly located in intergenic and intronic elements of the genome (fig. S4E). De novo DNA sequence motif analysis identified significantly enriched sequences in DRHs when considering sequence bias and ATAC or H3K27Ac peaks as background to correct for the contributions of open chromatin (Fig. 2D, fig. S5, and table S2).
Fig. 2. Chromatin accessibility controls the placement of repair hotspots.
(A) Repair-seq, ATAC-seq, and H3K27Ac ChIP-seq data at the ERCC1 locus demonstrate overlap between DNA repair, chromatin accessibility, and histone acetylation. (B) Scatter plot of Repair-seq–normalized read counts compared to ATAC- and H3K27Ac-normalized read counts. (C) Box plots of ATAC and H3K27Ac peaks with and without DNA repair. The horizontal black lines represent the medians, whereas the whiskers are displayed at the largest and smallest values no more than 1.5 times the interquartile range from quartiles 3 and 1, respectively. ****P < 2.2 × 10−16 by Kruskal-Wallis test. (D) DNA sequence motifs identified de novo and predicted as enriched in DRHs relative to randomized sequence. Targ/Bkgr, target/background.
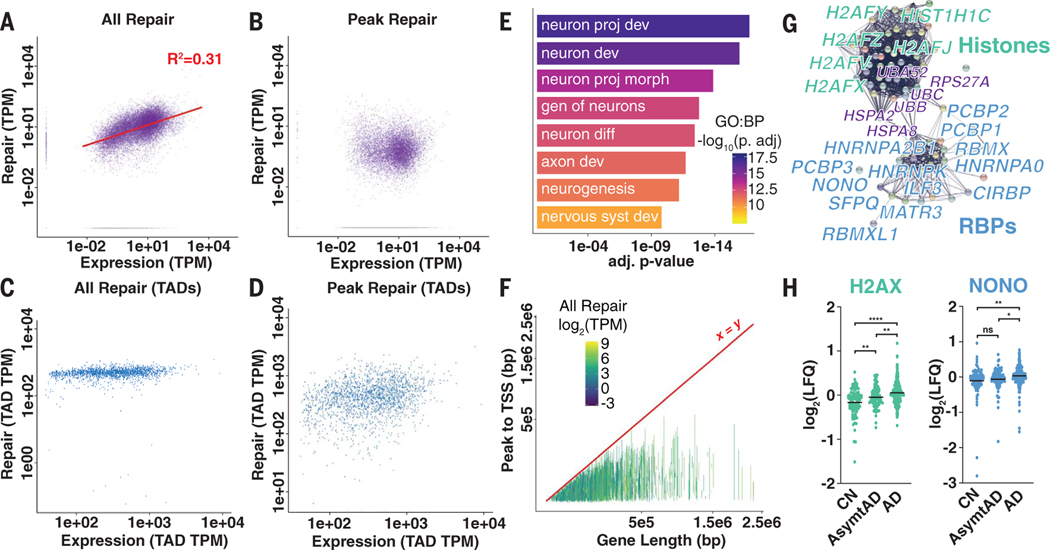
Repair-seq allowed us to compare all DNA repair- and transcription-associated reads.Most Repair-seq reads (~67%) could be assigned to genes, with the majority of the neuronal transcriptome exhibiting some level of maintenance that increased with expression levels (Fig. 3A and fig. S6, A and B). This finding corroborates prior work suggesting that in neurons, global DNA repair is attenuated and consolidated to actively transcribed genes, presumably to suppress the accumulation of lesions and mutations (5). However, when we examined DRH reads (~23% of all Repair-seq reads), we observed that many genes lacked recurrent DNA repair sites and showed no relationship with expression (Fig. 3B, fig. S6C, and table S3). Comparison of the locations of DRHs with transcribing RNA polymerases [global run-on sequencing (GRO-seq)] showed strong promoter enrichment (fig. S7). Almost one-third of DRHs were located in intergenic regions and could not be assigned to transcription of single genes.
Fig. 3. Transcriptional output correlates with total DNA repair in genes but not repair hotspots.
(A and B) All DNA repair–associated reads in genes (A) and repair peak–associated reads in genes (B) compared with RNA-associated reads from total RNA-seq. TPM, transcripts per kilobase million. (C and D) All DNA repair–associated reads in genes (C) and repair peak–associated reads in genes (D) compared with RNA-associated reads from total RNA-seq in length-normalized TADs. (E) Select biological process (BP) GO terms for genes containing DRHs. Terms are neuron projection development, neuron development, neuron projection morphogenesis, genesis of neurons, neuron differentiation, axon development, neurogenesis, and nervous system development. (F) Line plot of transcription start sites (TSSs) to DRHs in each gene compared with total gene length (colored by total DNA repair level). (G) String network representation of peptides enriched for histones (green), RNA binding proteins (RBPs; blue), and some chaperones and ubiquitin (purple). (H) LFQ proteomics data for H2AX and NONO abundance in cognitively normal (CN), asymptomatic Alzheimer’s disease (AsymtAD), and Alzheimer’s disease (AD). Horizontal black lines represent mean log2(LFQ). ns, not significant; *P < 0.5; **P < 0.01; and ****P < 0.0001 by analysis of variance (ANOVA) with Tukey’s multiple comparison test.
To address the potential contribution of these sites to transcription-associated repair, we generated Hi-C contact maps for ESC-iNs to assign intergenic peaks to genes using features of three-dimensional (3D) genome organization, such as topologically associating domains (TADs) (13). DNA repair levels in most TADs were uniform (Fig. 3C). Assignment of intergenic peaks did not substantially alter the finding that DRHs were not correlated with the level of gene transcription (Fig. 3D and fig. S8). A comparison of the distribution of either all DNA repair–associated reads or Repair-seq peaks with genome-wide features of 3D genome organization, such as A/B compartments, displayed an enrichment of DNA repair in the “active” A compartments (fig. S9).
We found that DRH genes were enriched for specific cellular processes irrespective of expression level, because they were correlated with genes essential for neuronal identity and function (Fig. 3E, fig. S10, and table S4). We explored whether gene length played a role in DRH density and found that both total repair and transcription were independent of gene length (fig. S11, A and B). However, when we examined reads that were only from DRHs in relationship to length, the total level of repair in these sites, as well as total peak density, paradoxically diminished in relationship with gene length (Fig. 3F and fig. S11, C and D). These findings suggest that DRHs in neuronal genes might arise from the requirements of maintaining transcriptional elongation and splicing in genes containing large introns (14)
To investigate if DRHs were linked to splicing in neurons, we performed rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) on chromatin that had undergone repair (12), and we detected 79 enriched proteins (table S5). Proteins identified by RIME were largely grouped into histone H2A isoforms and RNA binding proteins by network and gene ontology (GO) analysis (Fig. 3G, fig. S12, and table S6). The presence of the marker histone H2AZ was validated by ChIP-seq (fig. S13). We used the Consensus Brain Protein Coexpression Study dataset to compare protein abundance [label-free quantitation (LFQ)] in cognitive normal and asymptomatic or symptomatic Alzheimer’s disease patients (15). We found that 21 of the identified proteins showed differences in neurodegenerative disease (P < 2.67 × 10−10 by hypergeometric test), suggesting a role for changes to DNA repair in the etiology and progression of Alzheimer’s disease (Fig. 3H and table S7) (16).
In mice, neuronal activity generates DSBs and the damage marker γH2AX in select genes to initiate transcription for learning and memory (8, 9). Repair-seq was used on KCl-stimulated ESC-iNs to find activity-induced break sites in human neurons (12). No substantial changes were observed in DRHs after neuron depolarization, in contrast to cells where spontaneous activity was inhibited with tetrodotoxin (TTX) (fig. S14, A to D). Genes linked to activity-induced DSBs in mice showed minimal changes in DNA repair levels with either neuronal stimulation or inhibition (fig. S14E). Activity-induced breaks were linked to topoisomerase IIβ at CCCTC-binding factor (CTCF) sites; however, we found minimal intersection between CTCF ChIP-seq and Repair-seq peaks (fig. S14, F and G). This lack of increased DNA repair linked to neuronal stimulation suggests species-specific differences in how these genes are transcribed (17), that their repair might be highly reliable and not incorporate new nucleotides, or that the γH2AX that is associated with activity may not be a reliable marker of DSBs (18). However, given the ability of TTX to suppress many DRHs, we believe that a substantial fraction of DNA repair is linked to neuronal identity established by activity. Finally, we noted that FOS and NPAS4 contained predicted G-quadruplex structures, and we performed an analysis that suggested that these could be key regulatory features of neuron promoters that might be vulnerable to damage (fig. S15 and table S8) (19).
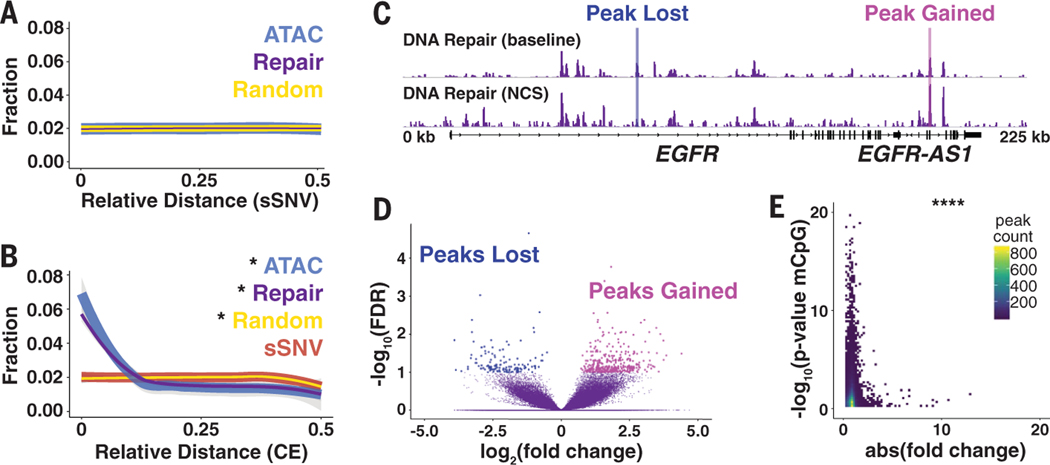
As cells age, the activity of DNA repair mechanisms declines, leading to an increase in somatic mutations and the accumulation of unrepaired lesions (20). Direct intersection and relative distance comparison between DRHs and somatic single-nucleotide variants (sSNVs) identified from single neurons isolated from postmortem human brains showed no proximal enrichment (Fig. 4A and fig. S16, A to C) (11), suggesting that mutations occurred randomly throughout the genome, irrespective of DRHs. We next used genomic evolutionary rate profiling (GERP)–defined constrained elements (CEs) in humans and compared the maximum GERP score and CE location with DRHs; the DRHs were enriched near CEs and more likely to have a single base under strong conservation, in contrast to sSNV sites (Fig. 4B; fig. S16, D to F; and fig. S17). These data suggest that DRHs might protect essential elements from both erroneous repairs and going unrepaired.
Fig. 4. Repair hotspots protect evolutionarily constrained regions of the human genome from epigenetic drift.
(A) Relative distance measurement from postmortem human neuron sSVNs to nearest DRH or randomly generated peaks. (B) Relative distance measurement from GERP CEs to nearest sSNV, DRH, ATAC, or random peaks. (C) Representative browser view of DRHs at baseline and 24 hours after 10 min of NCS treatment demonstrates that peaks are lost and gained. (D) Volcano plot for NCS differential peaks using a false discovery rate (FDR) of <0.1 from four samples. (E) Heat map of the DRH stability (absolute fold change after NCS treatment) compared with epigenetic clock mCpG sites from sorted human neurons. *P < 0.01 by Jaccard distance test;****P < 8.52 × 10−5 by hypergeometric test.
Aging drives fundamental changes in the epigenome (21), and biological age can be quantified quantified with epigenetic clock models created using changes in the methylation patterns on CpG dinucleotides (22). Despite the accuracy of such models, no satisfying biological explanation exists as to why these DNA modifications are linked to aging (22). We compared the locations and proximity of DRHs with CpG sites and methylated CpG dinucleotides (mCpG) statistically associated with aging in neurons from human prefrontal cortex (23), and we found that they were closely associated (fig. S18). Genome instability in the form of DSBs is a primary driver of biological aging (24). We treated ESC-iNs with the DNA-damaging agent neocarzinostatin (NCS) to assay injury-induced changes to DRHs. Acute NCS treatment triggered both the gain and loss of DRHs in neurons in a stochastic fashion, although at the dosage used, relatively few peaks were detected (Fig. 4, C and D, and table S9). In the context of aging, genome instability could potentially redistribute repair efforts away from hotspots to other locations in the genome, similar to NCS treatment. A comparison of absolute fold change for NCS and other DNA damage–treated samples with statistically significant mCpG sites indicated that the most stable DRHs were those associated with the epigenetic clock and CEs (Fig. 4E, figs. S19 and S20, and tables S10 to S15). Therefore, as DNA repair capacity declines with age and pathways become overtaxed, these sites could be susceptible to dysregulation.
Our results suggest that DRHs are established in neurons and play a key role in identity and function. Going forward, Repair-seq will be a powerful tool to explore how age and disease disrupt genome integrity in the nervous system. Finally, whether DRHs are specific to neurons, particular developmental lineages, or other nondividing cells or are found in only some long-lived species remains an open question. The discovery of these sites in other cell types might further aid in our understanding of how age-related changes in their organization could drive differential aging or the development of disease in other tissue types.
Note added in proof:
It was brought to our attention that a closely related paper by Wu et al. (25) is in press.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Moore, I. Guimont, K. E. Diffenderfer, W. Travis Berggren, J. Diedrich, and A. Pinto for technical assistance as well as M. L. Gage for editorial comments. We also acknowledge the Salk Institute Stem Cell Core, Next Generation Sequencing Core, and Mass Spectrometry Core for technical support.
Funding: D.A.R. is an Alzheimer’s Association Research Fellow (AARF-17-504089). This work was supported by an American Heart Association (AHA)-Allen Initiative in Brain Health and Cognitive Impairment award made jointly through the AHA and The Paul G. Allen Frontiers Group (19PABHI34610000); the JPB Foundation; the Dolby Foundation; the Helmsley Charitable Trust; NIH AG056306 to F.H.G.; NIH R01AG056511-02 to C.K.G.; NIH DP5OD023071-03 to J.R.D.; and P30 014195 to the Mass Spectrometry Core.
Footnotes
Competing interests: The Salk Institute for Biological Studies has filed a patent governing the use of Repair-seq for the detection of DNA repair in nondividing cells.
Data and materials availability: Primary data are available at www.ncbi.nlm.nih.gov/bioproject/PRJNA699309, and code is available at www.synapse.org/repairseq (26).
REFERENCES AND NOTES
- 1.Mattson MP, Magnus T, Nat. Rev. Neurosci 7, 278–294 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J, Nature 461, 1071–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow HM, Herrup K, Nat. Rev. Neurosci 16, 672–684 (2015). [DOI] [PubMed] [Google Scholar]
- 4.McKinnon PJ, Nat. Neurosci 16, 1523–1529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nouspikel T, Hanawalt PC, Mol. Cell. Biol 20, 1562–1570 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu T. et al. , Nature 429, 883–891 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Vermeij WP et al. , Nature 537, 427–431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suberbielle E. et al. , Nat. Neurosci 16, 613–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madabhushi R. et al. , Cell 161, 1592–1605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei PC et al. , Cell 164, 644–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lodato MA et al. , Science 359, 555–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Materials and methods are available as supplementary materials.
- 13.Dixon JR et al. , Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi A. et al. , Cell Rep. 23, 1326–1341 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Johnson ECB et al. , Nat. Med 26, 769–780 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swarup V. et al. , Cell Rep. 31, 107807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruunsil P, Bengtson CP, Bading H, Cell Rep. 18, 122–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanbhag NM et al. , Acta Neuropathol. Commun 7, 77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hänsel-Hertsch R. et al. , Nat. Genet 48, 1267–1272 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Maynard S, Fang EF, Scheibye-Knudsen M, Croteau DL, Bohr VA, Cold Spring Harb. Perspect. Med 5, a025130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-Otín C, Blasco MA, Partridge L, Serrano M,Kroemer G, Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath S, Raj K, Nat. Rev. Genet 19, 371–384 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Kozlenkov A. et al. , Genes (Basel) 8, 152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White RR et al. , Nat. Commun 6, 6790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu W. et al. , bioRxiv 2020.12.16.423085 [Preprint]. [Google Scholar]
- 26.Reed PJ, Code used in the analysis of Repair-seq data. Synapse (2021); 10.7303/syn24202004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.