Abstract
In order to evaluate the impact of an urban effluent on antibiotic resistance of freshwater bacterial populations, water samples were collected from the Arga river (Spain), upstream and downstream from the wastewater discharge of the city of Pamplona. Strains of Enterobacteriaceae (representative of the human and animal commensal flora) (110 isolates) and Aeromonas (typically waterborne bacteria) (118 isolates) were selected for antibiotic susceptibility testing. Most of the Aeromonas strains (72%) and many of the Enterobacteriaceae (20%) were resistant to nalidixic acid. Singly nalidixic acid-resistant strains were frequent regardless of the sampling site for Aeromonas, whereas they were more common upstream from the discharge for enterobacteria. The most common resistances to antibiotics other than quinolones were to tetracycline (24.3%) and beta-lactams (20.5%) for Enterobacteriaceae and to tetracycline (27.5%) and co-trimoxazole (26.6%) for Aeromonas. The rates of these antibiotic resistances increased downstream from the discharge at similar degrees for the two bacterial groups; it remained at high levels for enterobacteria but decreased along the 30-km study zone for Aeromonas. Genetic analysis of representative strains demonstrated that these resistances were mostly (enterobacteria) or exclusively (Aeromonas) chromosomally mediated. Moreover, a reference strain of Aeromonas caviae (CIP 7616) could not be transformed with conjugative R plasmids of enterobacteria. Thus, the urban effluent resulted in an increase of the rates of resistance to antibiotics other than quinolones in the riverine bacterial populations, despite limited genetic exchanges between enterobacteria and Aeromonas. Quinolone resistance probably was selected by heavy antibiotic discharges of unknown origin upstream from the urban effluent.
Antibiotic-resistant bacteria (2, 3, 9, 34) and antibiotics (22) are discharged in various amounts in the environment as a result of the increasing and often indiscriminate use of antibiotics in medical, veterinary, and agricultural practices. River waters are the main receptacle for these polluants, since they receive the sewage of urban effluents. As rivers are one of the major sources of water, directly or indirectly, for human and animal consumption, this pollution may contribute to the maintenance and even the spread of bacterial antibiotic resistance.
Most investigations on antibiotic resistance in the aquatic habitat have concerned bacteria of fecal origin, because they are used as pollution indicators and may be associated with infectious diseases. However, in many freshwater systems, fecal bacteria are of little numerical significance despite the fact that they are discharged into almost all inland waters (27). Thus, if the environmental pool of resistance is to be measured, bacteria other than those of fecal origin must be considered. Other studies were interested in whole bacterial populations (Enterobacteriaceae [18, 30], gram-negative bacteria [4, 28, 32], heterotrophic bacteria [14, 33, 34], or viable bacteria [11]) and dealt with global antibiotic resistance (the frequency of cells able to grow on antibiotic-supplemented media). Such studies cannot differentiate intrinsic resistance, i.e., species-specific resistance (17), from acquired resistance due to either chromosomal mutations or incoming and thus transferable genes, mainly carried by plasmids or transposable elements (36).
The aim of this study was to evaluate the impact of an urban effluent on the antibiotic resistance of riverine bacteria, namely, that of the wastewater discharge of the city of Pamplona on the Arga River (Spain). The rate of acquired resistance, upstream and downstream from the polluted discharge, was investigated for two bacterial groups whose wild-type antibiotic resistance patterns are well known: Enterobacteriaceae, most of which are of human or animal origin, and Aeromonas spp., which are typically waterborne bacteria. For strains exhibiting representative antibiotic resistance patterns, plasmid content was analyzed, and the location and transferability of resistance determinants were examined.
MATERIALS AND METHODS
Study area and sample collection.
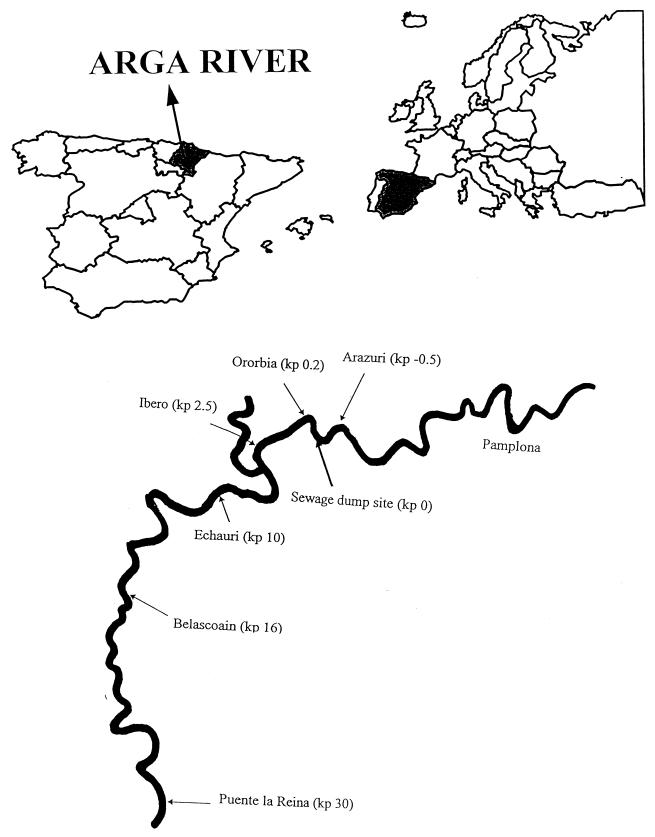
The river Arga (1.5 to 24 m3/s), located in northern Spain, receives the wastewater discharge (1 m3/s) from the sewage treatment plant of the city of Pamplona (200,000 inhabitants) (Fig. 1). Prior to discharge in the river, the urban sewage is filtered, and oil and suspended matter are removed by settling, but chlorination is not carried out. The sampling stations were chosen along the pollution gradient in the river, with one site upstream at Arazuri and five sites downstream at Ororbia (0.2 km from the sewage effluent), Ibero (2.5 km), Echauri (10 km), Belascoain (16 km), and Puente la Reina (30 km). Between Ibero and Echauri the river receives one additional unpolluted effluent (biotic and abiotic parameters comparable to those at Arazuri), which has approximately the same flow as the river. Water samples (500 ml) were collected at all sites in June, July, September, and October 1996, using sterile screw-capped bottles, and stored in cold bags at 4°C until analysis in the laboratory within 18 h of collection. The physicochemical and biological characteristics of water at the sampling sites during the sampling period were determined by the Mancomunidad de Aguas de la Comarca de Pamplona.
FIG. 1.
Locations of sampling sites in the Arga River. kp, kilometric point.
Isolation and identification of Enterobacteriaceae and Aeromonas spp.
The samples were plated on MacConkey agar. Representative colonies were purified on Trypticase soy agar. Preliminary identification of strains obtained in pure culture was based on Gram staining, respiration-fermentation tests, and oxidase reaction; biochemical tests were performed with the API bacterial identification system from bioMérieux (API 20E for enterobacteria and API 20NE for Aeromonas). Complete identification of enterobacteria was achieved by use of the tests in Bergey's Manual of Determinative Bacteriology (25) and the conventional methods described by Balows et al. (6). Final identification of Aeromonas was carried out according to the criteria of Popoff and Véron (35), as recommended by Austin et al. (5) for environmental isolates. Strains that could not be identified at the genus level were disregarded for this study.
Antibiotic susceptibility testing.
Antibiotic susceptibility was determined by the agar diffusion method (17), using 22 antibiotic disks (Sanofi Diagnostics Pasteur) corresponding to the drugs most commonly used in the treatment of human and animal infections caused by gram-negative bacilli (ampicillin, ticarcillin, cephalothin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, latamoxef, imipenem, amoxicillin-clavulanate, piperacillin-tazobactam, gentamicin, tobramycin, amikacin, netilmicin, nalidixic acid, ofloxacin, co-trimoxazole [trimethoprim-sulfamethoxazole], tetracycline, chloramphenicol, fosfomycin, and colistin). After 24 h of incubation at either 37°C (Enterobacteriaceae) or 30°C (Aeromonas), organisms were classified as sensitive, intermediate, or resistant according to French national guidelines (17). Acquired resistances were deduced from data on wild-type antibiotic susceptibility patterns characteristic of each species (17, 19, 37, 40). Strains with a decreased susceptibility were considered low-level resistant.
Plasmid DNA analysis.
Eighteen strains of Enterobacteriaceae and 16 strains of Aeromonas exhibiting representative antibiotic resistance patterns were selected for genetic analysis of their resistance determinants. Plasmid DNA was extracted by an alkaline lysis method and analyzed by electrophoresis on 0.9% (wt/vol) agarose gels in Tris-borate buffer, staining with ethidium bromide (2 mg/liter), and visualization under UV light (39). The molecular sizes of the plasmid bands were evaluated by comparison with reference plasmids pUC19 (2.6 kb) (41), pBR322 (4.3 kb) (10), pKK3535 (11.9 kb) (12), and PP4 (54.0 kb) (26).
Transformation and conjugation experiments.
The locations and the transferabilities of resistance determinants of the selected strains were further investigated by transformation and conjugation experiments. Transformation experiments with plasmid DNA extracts were performed by the high-voltage electroporation procedure (with Escherichia coli HB101 as the recipient) or by the heat shock method with competent cells prepared with calcium chloride (with Aeromonas caviae CIP 7616 as the recipient) (39). Conjugation experiments were carried out by a broth-mating procedure with a nalidixic acid-resistant (Nalr) mutant of E. coli K-12 as the recipient. Recipient strains and donor strains (resistant strains of enterobacteria or Aeromonas) were grown for 24 h in brain heart infusion broth. Equal volumes (2 ml) of parental strains were mixed and incubated with 100 μl of each conjugation mixture and then incubated for 1 to 3 days before they were examined for the presence of transconjugants. Incubation was performed at either 37°C (E. coli recipient) or 30°C (Aeromonas recipient). The selective antibiotic concentrations used for conjugation and transformation experiments were as follows: nalidixic acid, 50 mg/liter; ticarcillin, 100 mg/liter (Enterobacteriaceae) or 500 mg/liter (Aeromonas); tetracycline, 10 mg/liter; chloramphenicol, 15 mg/liter; sulfamethoxazole, 50 mg/liter (Enterobacteriaceae) or 500 mg/liter (Aeromonas); trimethoprim, 20 mg/liter; and tobramycin, 4 mg/liter.
RESULTS
As shown in Table 1, the indicators of pollution increased dramatically downstream from the wastewater discharge. Neither the pH nor the temperature had changed significantly after the discharge. However, the dissolved oxygen decreased from a range of 7 to 8.4 mg/liter at Arazuri to a range of 0.7 to 3.4 mg/liter at Ibero. Similarly, the chemical and biochemical oxygen demands increased from ranges of 9 to 32 and 1 to 2 mg of O2/liter at Arazuri to ranges of 45 to 146 and 25 to 79 mg of O2/liter at Ibero, respectively. Total and fecal coliforms also increased drastically; they were 100- to 1,000-fold greater at Ibero than at Arazuri. A total of 228 bacterial strains were isolated and could be identified to at least the genus level: 110 enterobacterial strains (23 E. coli, 23 Enterobacter, 22 Klebsiella, 19 Kluyvera, 15 Citrobacter, 3 Serratia, 3 indole-positive Proteus, and 2 Yersinia frederiksenii) and 118 Aeromonas strains (88 A. caviae, 19 Aeromonas sobria, and 11 Aeromonas hydrophila) (Table 2).
TABLE 1.
Biochemical characteristics of the sampling sites in May, June, July, August, and September 1996a
| Site | pH | Temp (°C) | SM (g/liter) | DO (mg/liter) | COD (mg of O2/liter) | BOD (mg of O2/liter) | TC (CFU/100 ml) | FC (CFU/100 ml) |
|---|---|---|---|---|---|---|---|---|
| Arazuri | 7.9–8.5 | 16–24 | 10–30 | 7–8.4 | 9–32 | 1–2 | 9 × 102–5 × 104 | 1 × 102–4 × 103 |
| Ororbia | 7.3–7.7 | 15–24 | 14–33 | 1–6 | 51–129 | 27–68 | 3 × 105–2 × 107 | 2 × 105–7 × 106 |
| Ibero | 7.3–7.6 | 13–24 | 14–31 | 0.7–3.4 | 45–146 | 25–79 | 4 × 105–4 × 107 | 2 × 105–1 × 107 |
| Echauri | 7.4–7.7 | 14–24 | 11–25 | 2–4 | 32–103 | 16–25 | 4 × 105–3 × 107 | 5 × 104–6 × 106 |
| Belascoain | 7.5–8.0 | 14–25 | 2–16 | 0.6–5.1 | 11–74 | 7–17 | 6 × 104–7 × 106 | 2 × 104–4 × 106 |
| Puente la Reina | 7.6–7.9 | 14–26 | 1–13 | 3–6 | 9–63 | 4–14 | 3 × 103–2 × 105 | 5 × 102–8 × 104 |
SM, suspended matter; DO, dissolved oxygen; COD, chemical oxygen demand; BOD, biological oxygen demand; TC, total coliforms; FC, fecal coliforms. Data are from the purifying station at Arazuri (Mancomunidad de la Comarca de Pamplona, Navarra, Spain). Maxima and minima for this period are shown.
TABLE 2.
Numbers of Aeromonas and Enterobacteriaceae strains isolated in the Arga River
| Site |
Aeromonas spp.
|
Enterobacteriaceae
|
||
|---|---|---|---|---|
| No. of strains | Identification | No. of strains | Identification | |
| Arazuri | 4 | A. caviae | 3 | Klebsiella spp. |
| 4 | A. sobria | 2 | E. coli | |
| 1 | Citrobacter freundii | |||
| Total | 8 | 6 | ||
| Ororbia | 26 | A. caviae | 8 | Klebsiella spp. |
| 3 | A. sobria | 9 | E. coli | |
| 3 | A. hydrophila | 3 | C. freundii | |
| 9 | Kluyvera spp. | |||
| 2 | Enterobacter cloacae | |||
| 1 | Morganella morganii | |||
| 1 | Serratia liquefaciens | |||
| 1 | Yersinia frederiksenii | |||
| Total | 32 | 34 | ||
| Ibero | 30 | A. caviae | 6 | Klebsiella spp. |
| 5 | A. sobria | 6 | E. coli | |
| 5 | A. hydrophila | 8 | Citrobacter spp. | |
| 4 | Kluyvera spp. | |||
| 9 | Enterobacter spp. | |||
| 1 | M. morganii | |||
| 2 | Serratia spp. | |||
| 1 | Y. frederiksenii | |||
| Total | 40 | 37 | ||
| Echauri | 14 | A. caviae | 4 | Klebsiella spp. |
| 1 | A. sobria | 4 | E. coli | |
| 1 | C. freundii | |||
| 4 | Kluyvera spp. | |||
| 7 | Enterobacter spp. | |||
| 1 | Providencia rettgeri | |||
| Total | 15 | 21 | ||
| Belascoain | 9 | A. caviae | 1 | Klebsiella spp. |
| 1 | A. sobria | 1 | E. coli | |
| 1 | A. hydrophila | 1 | C. freundii | |
| 2 | Kluyvera spp. | |||
| 1 | Enterobacter intermedius | |||
| Total | 11 | 6 | ||
| Puente la Reina | 5 | A. caviae | 1 | E. coli |
| 5 | A. sobria | 1 | C. freundii | |
| 2 | A. hydrophila | 4 | Enterobacter spp. | |
| Total | 12 | 6 | ||
| Total | 118 | 110 | ||
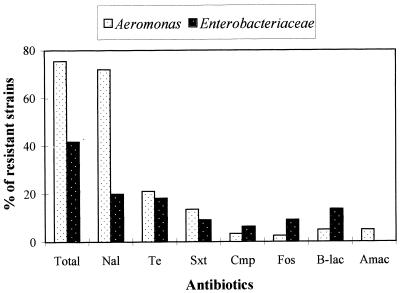
Most of the enterobacteria (58.2%) did not have any acquired resistance to the antibiotics tested; i.e., they presented a wild-type antibiotic resistance pattern (Fig. 2). Similar resistance rates were observed for nalidixic acid (20%) and tetracycline (18.2%); beta-lactam-resistant strains were also found (13.6%); fewer than 10% of the strains were resistant to co-trimoxazole, fosfomycin, and chloramphenicol; and all strains were uniformly susceptible to aminoglycosides. Only 21.8% of the isolates displayed a single acquired resistance, principally to nalidixic acid (6.3%), tetracycline (5.5%), or beta-lactams (4.5%) (Table 3). Multiple-antibiotic-resistant enterobacteria (isolates with more than three acquired resistances) were uncommon (5.5%).
FIG. 2.
Percentages of strains resistant to antibiotics. Total, strains showing at least one acquired resistance; Nal, nalidixic acid; Te, tetracycline; Sxt, co-trimoxazole; Cmp, chloramphenicol; Fos, fosfomycin, B-lac, beta-lactams; Amac, aminoglycosides.
TABLE 3.
Distribution of antibiotic resistance patterns in enterobacteria and locations of resistance determinants in representative strains
| Antibiotic resistance pattern
|
Representative strain studied | No. of plasmid bands (kb) | Transformation to E. coli HB101 | Conjugation of transformants to E. coli K-12 Nalr | Resistance determinant location
|
|||
|---|---|---|---|---|---|---|---|---|
| Resistance marker(s)a | No. of strains | Chromosome (pattern)a | Plasmid
|
|||||
| Patterna | Transferability (kb) | |||||||
| Nal | 7 | K. pneumoniae 487 | 0 | NDb | ND | Nal | ||
| Tic | 5 | E. coli 233 | 7 (54.0, 44.7, 33.9, 9.1, 3.5, 2.5, 1.9) | + | + | Tic | + (54.0) | |
| Tc | 6 | E. coli 163 | 1 (54.0) | + | + | Tc | + (54.0) | |
| Fos | 4 | E. coli 330 | 4 (>100, 54.0, 22.9, 6.0) | − | ND | Fos | ||
| Nal Tic | 3 | E. coli 463 | 7 (49.0, 21.9, 11.5, 9.1, 7.2, 4.0, 2.8) | + | − | Nal | Tic | − |
| Nal Fos | 4 | K. cryocrescens 499 | 4 (69.2, 49.0, 33.1, 8.7) | − | ND | Nal Fos | ||
| Tc Su | 2 | E. coli 149 | 5 (79.4, 49.0, 8.7, 3.6, 2.6) | − | ND | Tc Su | ||
| Su Tp | 1 | E. coli 368 | 7 (75.9, 41.7, 30.2, 15.8, 11.5, 10.7, 2.6) | − | ND | Su Tp | ||
| Nal Tc (C) | 1 | M. morganii 157 | 1 (49.0) | − | ND | Nal Tc (C) | ||
| Tic Tc (C) | 1 | C. freundii 266 | 1 (46.8) | − | ND | Tic Tc (C) | ||
| Tc Su Tp | 2 | E. coli 189 | 6 (79.4, 49.0, 18.2, 17.8, 12.6, 7.9) | + | ND | Tc Tp | Su | − |
| C (Su) (Tp) | 2 | E. cloacae 436 | 1 (49.0) | + | + | (Su) (Tp) | C | + (49.0) |
| Nal (Tc) Su Tp | 1 | Klebsiella 12 | 5 (>100, >100, 49.0, 42, 20.9) | + | − | Nal (Tc) Tp | Su | − |
| Tic Tc (Su) (Tp) | 1 | C. freundii 145 | 6 (44.7, 22.9, 12.0, 5.8, 5.2, 3.3) | + | ND | Tc (Su) (Tp) | Tic | − |
| Tc C Su Tp | 1 | M. morganii 467 | 1 (49.0) | − | ND | Tc C Su Tp | ||
| Nal Tic Tc Su Tp | 3 | E. coli 312 | 1 (49.0) | + | − | Nal Tc Su Tp | Tic | − |
| Nal Tc (C) Su Tp | 1 | E. coli 264 | 5 (>100, >100, 54.0, 41.7, 25.1) | + | − | Nal (C) Tp | Tc Su | − |
| Tic Tc C Su Tp | 1 | E. coli 173 | 1 (46.8) | + | + | Tic Tc C Su Tp | + (46.8) | |
Nal, nalidixic acid; Tic, ticarcillin; Tc, tetracycline; C, chloramphenicol; Su, sulfamethoxazole; Tp, trimethoprim; Fos, fosfomycin. Parentheses indicate low-level resistance.
ND, not determined.
Among the Aeromonas strains, 75% showed at least one acquired resistance (Fig. 2). Nalidixic acid resistance (72%) was much more frequent than tetracycline (21%) or co-trimoxazole (14%) resistance, resistances to other antimicrobial agents (chloramphenicol, fosfomycin, beta-lactams, and aminoglycosides) were scarce (≤5%). Strains with a single resistance (45.7%) were principally nalidixic acid resistant (43.2%) (Table 4). Of the 34 strains presenting more than one resistance, only one was sensitive to nalidixic acid. Multiple-antibiotic-resistant Aeromonas strains were rare (3.4%).
TABLE 4.
Distribution of antibiotic resistance patterns in Aeromonas and plasmid contents of representative strains
| Antibiotic resistance patterna
|
Representative strain studied | No. of plasmid bands (kb) | |
|---|---|---|---|
| Resistance marker(s) | No. of strains | ||
| Nal | 51 | A. caviae 100 | 11 (>100, 54.0, 39.8, 24.0, 9.5, 8.1, 7.6, 6.8, 4.6, 3.6, 2.4) |
| Tc | 1 | A. caviae 535 | 1 (24.0) |
| (Fos) | 1 | A. caviae 520 | 1 (21.9) |
| Nal Tic | 1 | A. caviae 218 | 0 |
| Nal Tc | 14 | A. caviae 28 | 2 (34.7, 22.9) |
| Nal Su | 3 | A. caviae 524 | 3 (>100, 43.7, 26.3) |
| Nal (Tm) | 2 | A. hydrophila 471 | 4 (>100, 50.1, 24.0, 18.2) |
| Nal Fos | 1 | A. caviae 495 | 1 (24.0) |
| Su (Tp) | 1 | A. caviae 210 | 1 (20.4) |
| Nal Tic (Tm) | 1 | A. caviae 94 | 0 |
| Su (Tp) Fos | 1 | A. caviae 248 | 2 (6.8, 3.0) |
| Nal Tc Su Tp | 6 | A. caviae 384 | 5 (>100, 46.8, 26.9, 15.8, 6.8) |
| Nal (Su) (Tp) (Tm) | 1 | A. caviae 224 | 2 (12.6, 3.1) |
| Nal Tc (C) Su Tp | 1 | A. caviae 2 | 1 (21.9) |
| Nal Tic (Tc) (C) Su | 1 | A. caviae 542 | 3 (>100, 30.9, 24) |
| Nal Tic Tc (C) Su (Tm) | 2 | A. hydrophila 34 | 7 (>100, 67.6, 46.8, 32.4, 21.9, 8.3, 6.5) |
Nal, nalidixic acid; Tic, ticarcillin; Tc, tetracycline; C, chloramphenicol; Su, sulfamethoxazole; Tm, tobramycin; Tp, trimethoprim; Fos, fosfomycin. Parentheses indicate low-level resistance.
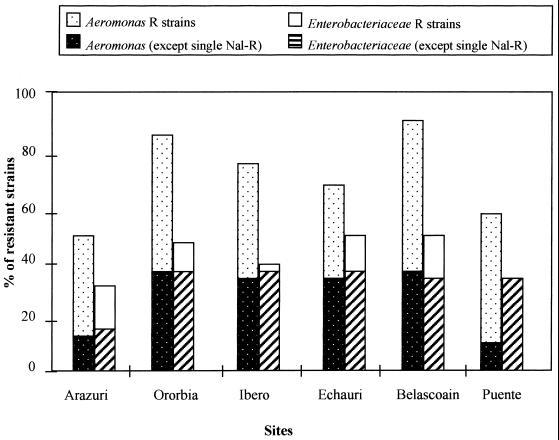
In both groups of bacteria, there was an increase in the percentages of resistant strains downstream from the wastewater discharge (Fig. 3): from 30% of enterobacteria and 50% of Aeromonas strains at Arazuri to 50% and more than 90%, respectively, at Belascoain. Enterobacteria were always less resistant than Aeromonas strains, at all sites. However, if we consider all antibiotic resistances except the single nalidixic acid resistances, the rates of resistant strains for the two groups were quite similar. For enterobacteria, singly nalidixic acid-resistant strains represented half of the resistant strains at Arazuri and fewer than one-third at the other sampling sites; at Puente la Reina, no singly nalidixic acid-resistant strain was isolated. In contrast, singly quinolone-resistant strains of Aeromonas represented more than half of the resistant strains at Ororbia (50.2%, with a total of 87.5% resistant strains), at Ibero (42.5%, with a total of 75% resistant strains), and at Echauri (33.3%, with a total of 66.6% resistant strains), and this rate increased upstream from the wastewater discharge (37.5% of the 50% resistant strains isolated at Arazuri) and far downstream, at Belascoain (54.7% of 91% resistant strains) and at Puente la Reina (50% of 58.3% resistant strains). For both bacterial groups, the rates of strains resistant to antibiotics other than quinolones increased downstream from the discharge. For Enterobacteriaceae, the greatest increase was found with beta-lactams (from 0% at Arazuri to 20.5% at Ororbia) and tetracycline (from 12.5% at Arazuri to 24.3% at Ibero). For Aeromonas, the greatest increase was found with tetracycline (from 0% at Arazuri to 27.5% at Ibero) and co-trimoxazole (from 0% at Arazuri to 26.6% at Echauri). At Puente la Reina (kp 30), enterobacteria resistant to tetracycline, co-trimoxazole, chloramphenicol, and beta-lactams were still encountered, whereas only one strain of Aeromonas was resistant to antibiotics other than quinolones.
FIG. 3.
Percentages of antibiotic-resistant strains by sampling site. R, resistant.
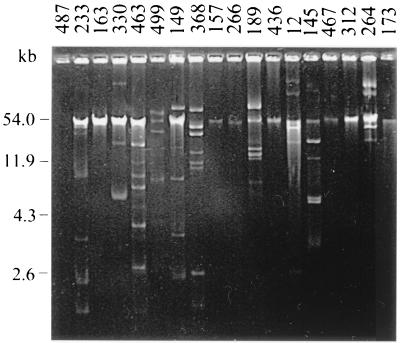
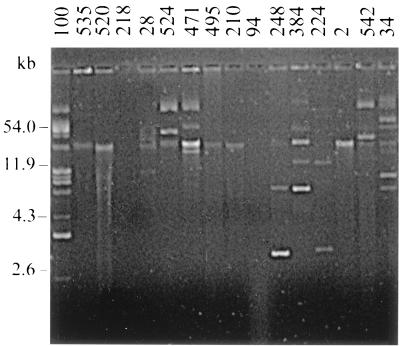
A total of 18 Enterobacteriaceae and 16 Aeromonas strains exhibiting one to six resistance markers were selected for plasmid content and transfer analysis. All strains except three contained one or more plasmids. Enterobacteria carried 0 to 7 plasmid bands, ranging in size from >100 to 1.9 kb (Fig. 4; Table 3), and Aeromonas strains harbored 0 to 11 plasmid bands, of >100 to 2.4 kb (Fig. 5; Table 4). Some singly resistant strains contained multiple plasmid bands (e.g., Ec233 and Ac100) (Ec = E. coli; Ac = A. caviae), whether some multiresistant strains contained a single plasmid band (e.g., Ec173 and Ac2). Resistance determinants were considered to be chromosomally located in the absence of plasmids or when no transformation with plasmid DNA was obtained. These resistances were thought to be plasmid mediated when transformation experiments gave positive results, providing that the plasmid profiles of the transformant and the donor strain were consistent (presence of a common plasmid band) (data not shown). Plasmids were demonstrated to be conjugative or not, depending on the results of mating experiments between the transformants and the adequate recipient. For enterobacteria (Table 3), among the 51 acquired resistance determinants tested (including 44 high-level and 7 low-level resistances), 15 (all high-level resistances) were present on plasmid DNA, 8 of which were transferred to E. coli K-12 Nalr. Except for nalidixic acid and fosfomycin resistance determinants, which were always situated on the chromosome, markers were found either on plasmids or on the chromosome. The distribution of the plasmid locations for high-level resistance determinants was as follows ticarcillin, 83%; sulfamethoxazole, 50%; chloramphenicol, 50%; tetracycline, 30%; and trimethoprim, 14%. Only 10 of the 18 strains tested (56%) carried R plasmids, encoding one (8 strains), two (1 strain) or five (1 strain) resistances; only 4 of the 10 R plasmids (40%) were conjugative, including that in the strain that carried the five markers. For Aeromonas, none of the 45 acquired resistance determinants tested (including 32 high-level and 13 low-level resistances) could be transferred by transformation, using either A. caviae CIP 7616 or E. coli K-12 as the recipient strain. Furthermore, no transformant could be obtained with plasmid DNAs of the four enterobacteria that carried conjugative plasmids (E. coli 233, E. coli 163, Enterobacter cloacae 436, and E. coli 173) and A. caviae CIP 7616 as the recipient.
FIG. 4.
Agarose gel electrophoresis of plasmid DNAs extracted from 18 representative enterobacterial strains. The sizes indicated on the left are those of plasmids pUC19 (2.6 kb), pBR322 (4.3 kb), pKK3535 (11.9 kb), and RP4 (54.0 kb).
FIG. 5.
Agarose gel electrophoresis of plasmid DNAs extracted from 16 representative Aeromonas strains. The sizes indicated on the left are those of plasmids pUC19 (2.6 kb), pBR322 (4.3 kb), pKK3535 (11.9 kb), and RP4 (54.0 kb).
DISCUSSION
Urban effluents are known to contain high levels of antibiotics and antibiotic-resistant bacteria belonging to the human and animal commensal flora, mainly Enterobacteriaceae (9, 22, 27). In order to study the impact of an urban effluent on the antimicrobial resistance of the microbial riverine flora, the acquired antibiotic resistance rates of enterobacteria and Aeromonas spp. (typical freshwater bacteria) upstream and downstream from a polluted discharge have been determined.
In our opinion, such studies should differentiate intrinsic and acquired resistances, and antibiotic resistances should be analyzed by chemical family. Indeed, various antibiotics are differentially effective against different groups of bacteria. For example, the benzyl- and isoxazolylpenicillins are mainly effective against gram-positive bacteria, and nalidixic acid is mainly effective against gram-negative bacteria (36). This intrinsic resistance is a function of the genetic inheritance of each species (17). Moreover, several molecules belonging to the same chemical family are usually affected by a single mechanism of resistance (cross-resistance). For example, both ampicillin and cephalothin are inactivated by chromosomal beta-lactamases produced by many enterobacterial (17) and Aeromonas (40) species. Most studies on bacterial antibiotic resistances in sewage (3, 18), freshwater (3, 9, 27, 33, 34), and seawater (8, 38) did not take into account these elements, leading to unexpected conclusions. For example, Jones et al. (27) found a higher incidence of resistance in the bacteria isolated from remote upland tarns than in those isolated from a polluted lake or a sewage, with the highest values being observed for pseudomonads, which are naturally multiresistant organisms. Similarly, McKeon et al. (32) have reported that 100% of A. hydrophila strains isolated from rural groundwater supplies were resistant to at least two antibiotics, but among the tested antibiotics were ampicillin and cephalothin, to which A. hydrophila is naturally resistant (40).
In this study, two types of acquired resistances were found: single nalidixic acid resistance and other resistance profiles. Strains resistant only to nalidixic acid represented more than half of the resistant strains of Aeromonas, but a very low percentage of resistant enterobacteria, and in any case at a level similar to that for other antibiotic resistances. Surprisingly, the highest rates of single nalidixic acid resistance in Aeromonas were encountered upstream from the wastewater discharge and at the farthest site downstream. This observation suggests that the source of the quinolone resistance was located upstream from the discharge and that it was not related to wastewaters. Previous studies have demonstrated that quinolone resistance was less than 25% among environmental isolates (2, 11, 21, 28, 32) and less than 5% for clinical Aeromonas isolates (29). Quinolones (with co-trimoxazole) have even been recommended as the first choice for treatment of infections caused by Aeromonas (29). These synthetic antibiotics are naturally absent in freshwaters. Quinolone resistances are exclusively due to chromosomal mutations (31) and are thus highly stable and not transferable, as confirmed in this study. Quinolone-resistant strains of Aeromonas probably have been selected by heavy discharges of these compounds into the river. In Spain, the agricultural and veterinary use of antibiotics has been roughly estimated at two-thirds of the amount consumed by humans (7). Several quinolones (oxolinic acid, flumequine, and enrofloxacin, etc.) are commonly used as therapeutic agents for animals (22). The river flows through agricultural catchments and may be contaminated by runoff waters (22). Quinolones are excreted mostly as unchanged substances, and they are among the most persistent antibiotics in the environment (half-lives of 150 days for flumequine and of 150 to 1,000 days for oxolinic acid) (22). An extensive use of veterinary quinolones in the farms located near the river Arga, upstream from Pamplona, is thus questionable.
Downstream from the polluted discharge, there was a similar increase of rates of acquired resistance to antibiotics other than quinolones for both bacterial groups. These rates remained high until Belascoain (16 km downstream from the discharge). For enterobacteria, the levels at Puente la Reina (30 km downstream from the discharge) were similar to those at the most polluted sites. In contrast, for Aeromonas they decreased to a level similar to that upstream from the wastewater effluent. An increase of resistances was also observed in strains isolated from rivers receiving urban discharge (9) or hospital and pharmaceutical plant wastewaters (21). Tetracycline resistance rates similar to or higher than those found in this study have been reported (2, 3, 21, 28, 34). Beta-lactam resistance rates found in the literature are difficult to analyze, since many coliforms are intrinsically resistant to these drugs. However, Al-Jebouri (3) and Al-Ghazali et al. (2) found that 45 and 90% of E. coli strains were resistant to ampicillin, respectively, which are higher rates than those found here. Beta-lactams, co-trimoxazole, and tetracyclines are widely used in human and veterinary practices. In contrast, chloramphenicol resistances are rare in most studies (3, 9, 28, 34), possibly as the result of the restricted use of this drug. Acquired resistances to these antibiotics are usually encoded by plasmids and/or transposable elements (36).
In order to investigate the hypothesis of resistance transfer between allochthonous and autochthonous riverine flora, genetic analysis was undertaken for representative strains. Most of the tested strains carried several plasmids, including some with a high molecular weight. However, no correlation was found between the number of antibiotic resistance markers and the number of plasmid bands, as previously reported (15), suggesting that most of these plasmids either encoded characters other than antibiotic resistances or were cryptic (13). In enterobacteria, resistances which are known to be exclusively (quinolones) or mainly (fosfomycin) due to mutations were found to be chromosomally located. Only one-third of the high-level resistance determinants was carried by plasmids, including most ticarcillin resistances and half of sulfonamide and chloramphenicol resistances. Very few of the tetracycline and trimethoprim resistances were found to be plasmid mediated, although chromosomal mutations are rarely involved, at least in the former case (36). Most of the R plasmids coded for single resistances and were not conjugative, except for one strain in which multiple resistance markers were carried by a single conjugative plasmid of ca. 46.8 kb. Thus, most of the high-level acquired resistances in enterobacteria were probably encoded by transposable elements or plasmids integrated into the chromosome. Low-level resistances to sulfonamides, trimethoprim, and chloramphenicol have been related to mutations leading to a decreased permeability of the outer membrane (36). In Aeromonas, although numerous plasmids were present in most strains, all resistances appeared to be governed by chromosomal genes. Indeed, half of them were to nalidixic acid and fosfomycin or were at a low level. In addition, strains of Aeromonas that are highly resistant to beta-lactams (including ticarcillin) are primarily derepressed mutants overproducing their chromosomal beta-lactamases (23, 40). However, high-level tetracycline resistances have been found to be encoded in Aeromonas by class A or D genes as part of transposons located on plasmids (1, 16, 33), and plasmid transfer between enterobacteria and Aeromonas has been occasionally demonstrated (1, 15, 24). Whatever the source of antibiotic resistance determinants in Aeromonas was, a discharge of antibiotics in the urban effluent might have selected preexisting resistant strains; the decreasing frequency of Aeromonas strains resistant to antibiotics other than quinolones along the study zone might be explained by the dilution with susceptible strains (20).
In conclusion, in this study, the urban discharge resulted in the increase of resistant strains of riverine autochthonous and allochthonous bacteria. However, our data are consistent with limited genetic exchanges between enterobacteria and Aeromonas. Quinolone-resistant bacteria, particularly Aeromonas strains, were more frequent upstream than downstream from the discharge, suggesting an origin other than the urban wastewater effluent for these resistances. The survey of antibiotic resistances in the microbial flora of freshwaters allows detection of hidden uses which contribute to the increase of bacterial resistances and thus limit the efficiency of these drugs in the treatment of human and animal infections.
ACKNOWLEDGMENTS
We thank C. Lizarraga and the Depuradora de Arazuri (Mancomunidad de Aguas de la Comarca de Pamplona) for determining chemical and biological parameters of the Arga River, the Arazuri Task Force for technical assistance during sampling, and M. H. Canron for help during genetic experiments.
This work was supported by a Ph.D. grant to M.G.-U. from the Navarra Regional Council.
REFERENCES
- 1.Adams C A, Austin B, Meaden P G, McIntosh D. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl Environ Microbiol. 1998;64:4194–4201. doi: 10.1128/aem.64.11.4194-4201.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Ghazali M R, Jazrawi S F, Al-Doori Z A. Antibiotic resistance among pollution indicator bacteria isolated from Al-Khair river, Baghdad. Water Res. 1988;22:641–644. [Google Scholar]
- 3.Al-Jebouri M M. A note on antibiotic resistance in the bacterial flora of raw sewage and sewage-polluted river Tigris in Mosul, Iraq. J Appl Bacteriol. 1985;58:401–405. doi: 10.1111/j.1365-2672.1985.tb01479.x. [DOI] [PubMed] [Google Scholar]
- 4.Andersen S R, Sandaa R A. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl Environ Microbiol. 1994;60:908–912. doi: 10.1128/aem.60.3.908-912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin B, Altwegg M, Gosling P J, Joseph S. The genus Aeromonas. Chichester, West Sussex, United Kingdom: Wiley and Sons Ltd.; 1996. [Google Scholar]
- 6.Balows A, Hausler W J, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society of Microbiology; 1991. [Google Scholar]
- 7.Baquero F the Task Force of the General Direction for Health Planning of the Spanish Ministry of Health. Antibiotic resistance in Spain: what can be done? Clin Infect Dis. 1996;23:819–823. doi: 10.1093/clinids/23.4.819. [DOI] [PubMed] [Google Scholar]
- 8.Baya A M, Brayton P R, Brown V L, Grimes D J, Russek-Cohen E, Colwell R R. Coincident plasmids and antimicrobial resistance in marine bacteria isolated from polluted and unpolluted Atlantic Ocean samples. Appl Environ Microbiol. 1986;51:1285–1292. doi: 10.1128/aem.51.6.1285-1292.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacherjee J W, Pathak S P, Gaur A. Antibiotic resistance and metal tolerance of coliform bacteria isolated from Gomati River water at Lucknow city. J Gen Appl Microbiol. 1988;34:391–399. [Google Scholar]
- 10.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 11.Boon P I. Antibiotic resistance of aquatic bacteria and its implications for limnological research. Aust J Mar Freshwater Res. 1992;43:847–859. [Google Scholar]
- 12.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noder H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 13.Brown R L, Sanderson K, Kirov S M. Plasmids and Aeromonas virulence. FEMS Immunol Med Microbiol. 1997;17:217–223. doi: 10.1111/j.1574-695X.1997.tb01015.x. [DOI] [PubMed] [Google Scholar]
- 14.Casas Alvero C. Antibiotic resistance of heterotrophic bacterial flora of two lakes. Syst Appl Microbiol. 1987;9:169–172. [Google Scholar]
- 15.Chang B J, Bolton S M. Plasmids and resistance to antimicrobial agents in Aeromonas sobria and Aeromonas hydrophila clinical isolates. Antimicrob Agents Chemother. 1987;31:1281–1282. doi: 10.1128/aac.31.8.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chopra I, Hawkey P M, Hinton M. Tetracyclines, molecules and clinical aspects. J Antimicrob Chemother. 1992;29:245–277. doi: 10.1093/jac/29.3.245. [DOI] [PubMed] [Google Scholar]
- 17.Comité de l'Antibiogramme de la Société Française de Microbiologie. Communiqué du Comité de l'Antibiogramme de la Société Française de Microbiologie. Bull Soc Fr Microbiol. 1998;13:243–258. [Google Scholar]
- 18.Fernandez Astorga A, Fernandez de Aranguiz A, Umaran A, Cisterna R. Comparison of antibiotic resistance and plasmid bands between two identical sets of raw sewage enterobacterial strains. Acta Hydrochim Hydrobiol. 1990;18:345–350. [Google Scholar]
- 19.Freney J, Husson M O, Gavini F, Madier S, Martra A, Izard D, Leclerc H, Fleurette J. Susceptibilities to antibiotics and antiseptics of new species of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1988;32:873–876. doi: 10.1128/aac.32.6.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalo M P, Arribas R M, Latorre E, Baquero F, Martinez J L. Sewage dilution and loss of antibiotic resistance and virulence determinants in E. coli. FEMS Microbiol Lett. 1989;59:93–96. doi: 10.1016/0378-1097(89)90465-5. [DOI] [PubMed] [Google Scholar]
- 21.Guardabassi L, Petersen A, Olsen J E, Dalsgaard A. Antibiotic resistance in Acinetobacter spp. isolated from sewers receiving waste effluent from a hospital and a pharmaceutical plant. Appl Environ Microbiol. 1998;64:3499–3502. doi: 10.1128/aem.64.9.3499-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halling-Sørensen B, Nors Nielsen S, Lanzky P F, Ingerslev F, Holten Lützhøft H C, Jørgensen S E. Ocurrence, fate and effects of pharmaceutical substances in the environment—a review. Chemosphere. 1998;36:357–393. doi: 10.1016/s0045-6535(97)00354-8. [DOI] [PubMed] [Google Scholar]
- 23.Hayes M V, Thomson C J, Amyes S G B. The “hidden” carbapenemase of Aeromonas hydrophila. J Antimicrob Chemother. 1996;37:33–44. doi: 10.1093/jac/37.1.33. [DOI] [PubMed] [Google Scholar]
- 24.Hedges R W, Smith P, Brazil G. Resistance plasmids of Aeromonads. J Gen Microbiol. 1985;131:2091–2095. [Google Scholar]
- 25.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T, editors. Bergey's manual of determinative bacteriology. 9th ed. Baltimore, Md: Williams and Wilkins, Co.; 1994. [Google Scholar]
- 26.Jacob A E, Grinter N J. Plasmid RP4 as a vector replicon in genetic engineering. Nature. 1975;255:504–506. doi: 10.1038/255504a0. [DOI] [PubMed] [Google Scholar]
- 27.Jones J G, Gardener S, Simon B M, Pickup R W. Antibiotic resistant bacteria in Windermere and two remote upland tarns in the English Lake District. J Appl Bacteriol. 1986;60:443–453. doi: 10.1111/j.1365-2672.1986.tb05090.x. [DOI] [PubMed] [Google Scholar]
- 28.Kelch W J, Lee J S. Antibiotic resistance patterns of gram-negative bacteria isolated from environmental sources. Appl Environ Microbiol. 1978;36:450–456. doi: 10.1128/aem.36.3.450-456.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko W C, Yu K W, Liu C Y, Huang C T, Leu H S, Chuang Y C. Increasing antibiotic resistance in clinical isolates of Aeromonas strains in Taiwan. Antimicrob Agents Chemother. 1996;40:1260–1262. doi: 10.1128/aac.40.5.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kralikova K, Krcmery V, Krcmery V., Jr Antibiotic resistance and transferable resistance in Enterobacteriaceae in municipal waste waters. Recomb DNA Technol Bull. 1986;9:59–64. [PubMed] [Google Scholar]
- 31.Liu L F, editor. DNA topoisomerases: biochemistry and molecular biology. Advances in pharmacology. 29A. New York, N.Y: Academic Press; 1994. [Google Scholar]
- 32.McKeon D M, Calabrese J P, Bissonnette G K. Antibiotic resistant gram-negative bacteria in rural groundwater supplies. Water Res. 1995;29:1902–1908. [Google Scholar]
- 33.Ogan M T, Nwiika D E. Studies on the ecology of aquatic bacteria of the Lower Niger Delta: multiple antibiotic resistance among the standard plate count organisms. J Appl Bacteriol. 1993;74:595–602. [PubMed] [Google Scholar]
- 34.Pathak S P, Bhattacherjee J W, Ray P K. Seasonal variation in survival and antibiotic resistance among various bacterial populations in a tropical river. J Gen Appl Microbiol. 1993;39:47–56. [Google Scholar]
- 35.Popoff M, Véron M. A taxonomic study of the Aeromonas hydrophila-Aeromonas punctata group. J Gen Microbiol. 1976;94:11–22. doi: 10.1099/00221287-94-1-11. [DOI] [PubMed] [Google Scholar]
- 36.Rice L B, Bonomo R A. In: Genetic and biochemical mechanisms of bacterial resistance to antimicrobial agents. Lorian V, editor. Baltimore, Md: Antibiotics in laboratory medicine Williams and Wilkins Co.; 1996. pp. 453–501. [Google Scholar]
- 37.Richardson C J L, Robinson J O, Wagener L B, Burke V. In-vitro susceptibility of Aeromonas spp. to antimicrobial agents. J Antimicrob Chemother. 1982;9:267–274. doi: 10.1093/jac/9.4.267. [DOI] [PubMed] [Google Scholar]
- 38.Sabry S A, Ghozlan H A, Abou-Zeid D M. Metal tolerance and antibiotic resistance patterns of a bacterial population isolated from sea water. J Appl Microbiol. 1997;82:245–252. doi: 10.1111/j.1365-2672.1997.tb02858.x. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Walsh T R, Payne D J, MacGowan A P, Bennett P M. A clinical isolate of Aeromonas sobria with three chromosomally mediated inductible beta-lactamases: a cephalosporine, a penicillinase and a third enzyme, displaying carbapenemase activity. J Antimicrob Chemother. 1995;35:271–279. doi: 10.1093/jac/35.2.271. [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]