Abstract
Modifications of DNA nucleobases are present in all forms of life. The purpose of these modifications in eukaryotic cells, however, is not always clear. Although the role of 5-methylcytosine (m5C) in epigenetic regulation and the maintenance of stability in plant genomes is becoming better understood, knowledge pertaining to the origin and function of oxidized nucleobases is still scarce. The formation of 5-hydroxymetylcytosine (hm5C) in plant genomes is especially debatable. DNA modifications, functioning as regulatory factors or serving as DNA injury markers, may have an effect on DNA structure and the interaction of genomic DNA with proteins. Thus, these modifications can influence plant development and adaptation to environmental stress. Here, for the first time, the changes in DNA global levels of m5C, hm5C, and 8-oxo-7,8-dihydroguanine (8-oxoG) measured by ELISA have been documented in recalcitrant embryonic axes subjected to desiccation and accelerated aging. We demonstrated that tissue desiccation induces a similar trend in changes in the global level of hm5C and 8-oxoG, which may suggest that they both originate from the activity of reactive oxygen species (ROS). Our study supports the premise that m5C can serve as a marker of plant tissue viability whereas oxidized nucleobases, although indicating a cellular redox state, cannot.
Keywords: Acer pseudoplatanus, biomarker, desiccation, DNA oxidative damage, ELISA, embryonic axes, hm5C, m5C, 8-oxoG, recalcitrant seeds, sycamore, viability
1. Introduction
DNA is a highly dynamic molecule that is continuously subjected to events that may result in alterations in the nucleobases and the DNA backbone structure. The four canonical bases that make up genomic DNA, adenine (A), thymine (T), guanine (G), and cytosine (C), are subjected to a wide variety of chemical modifications. While some of these alterations reflect regulated modifications of DNA, others represent DNA damage resulting from superimposed events, such as aging. Research conducted over the past decade has resulted in the identification of the enzymes and non-enzymatic agents responsible for induction and processing of DNA modifications [1,2].
One of the well-known DNA modifications is the methylation of cytosine which was initially observed and reported more than a century ago [3]. The presence of 5-methylcytosine (m5C) in DNA was demonstrated by Hotchkiss [4] in 1948, and seven decades of subsequent studies have clearly established that the nucleotide sequence is not the only information contained in DNA. Notably, it is the most common DNA modification in eukaryotes and often referred to as the “fifth base”. The m5C modification represents a conserved epigenetic mark that plays a role in many essential biological processes, including heterochromatin formation, cell proliferation, genomic imprinting, regulation of endogenous gene expression, as well as defense against transposon and silencing transgenes [5]. The total level of cytosine methylation of DNA ranges between 5–25%, depending on the plant species [6,7,8,9,10,11,12].
DNA methylation is catalyzed by DNA methyltransferases, which utilize the methyl group from S-adenosyl-L-methionine. In plants, m5C is found in the context of CG, CHG, and CHH sequences (where H represents A, C, or T). The de novo induction and maintenance of methylation are associated with different pathways. The first, which occurs in all sequence contexts, is driven by RNA (RNA-directed DNA methylation, RdDM) and catalyzed by DOMAINS REARRANGED METHYLASE 2 (DRM2). Post-replication maintenance of the methylation event within CHH and CHG sequences is regulated by DRM2, CMT2 (CHROMOMETHYLASE 2), and CMT3 (CHROMOMETHYLASE 3). Lastly, CG dinucleotides are methylated by MET1 (METHYLOTRASNFERASE 1), which recognizes hemimethylated DNA [13].
Notably, 5-methylcytosine can be passively removed from DNA when DNA methyltransferase activity is absent, or when the source for the methyl group donor is deficient. Active demethylation occurs by base excision (base excision repair, BER), which is carried out by enzymes involved in DNA repair. In contrast to the single enzyme involved in m5C synthesis, the removal of m5C requires the activity of several enzymes and is initiated by DNA demethylases. While m5C removal in mammals is accomplished by DNA demethylation involving m5C oxidation and/or deamination, plants can directly remove m5C from any sequence context [13]. Arabidopsis thaliana has four active m5C DNA demethylases: ROS1 (REPRESSOR OF SILENCING 1), DME (TRANSCRIPTIONAL ACTIVATOR DEMETER), DML2, and 3 (DEMETER-LIKE PROTEIN) [6]. Currently, DNA methylation is one of the best-characterized epigenetic processes, and m5C patterning when properly established and maintained, is essential for the normal functioning of living cells [7].
In addition to the normal, regulated modifications of DNA, like m5C, that are responsible for epigenetic control, DNA modifications can also be induced by environmental and endogenous factors resulting in DNA damage. In particular, reactive oxygen species (ROS), among the multiple agents that damage DNA, deserve special attention due to their omnipresence and their ability to impact DNA structure and function [1,14,15,16,17,18]. ROS have a high potential to cause strand breaks (SB) in DNA that severely impact genomic integrity [19]. In most cases, superoxide anion radicals (O2−) are the primary type of ROS produced and are readily converted to H2O2 and other peroxides. H2O2 is the most stable type of ROS and can freely migrate across cell membranes. Peroxides are involved in the metal-catalyzed Haber–Weiss reaction in the presence of reduced transition metals (Fe2+; Fenton reaction) [20] and Cu+, which yield hydroxyl radicals (OH), the strongest type of cellular oxidant [1,14,21,22,23]. Additionally, metal-independent ·OH formation has also been established, which occurs by a mechanism that relies on polyhalogenated quinone-mediated H2O2 homolytic decomposition [24]. The OH radical reacts immediately with every molecular species in the vicinity and its activity is only restricted by its diffusion rate. This radical can form adducts with all four bases, and in the process, extracts a hydrogen atom from 2′-deoxyribose, leading to strand scission, base release, and/or oxidation. Notably, guanine (G) is the most sensitive of the nucleobases to oxidation as it has the lowest redox potential. As a result, 8-oxo-7,8-dihydroguanine (8-oxoG) represents the most abundant oxidative DNA base lesion, and can arise through the activity of ·OH, hydroperoxide radicals (OOH), or rare singlet oxygen (1O2) [9,10,16,25]. Peroxynitrite anions (ONOO−), a reactive nitrogen species (RNS), can also form 8-oxoG, although at a slower rate than by other methods due to an unfavorable reaction barrier [25]. The most recent reports indicate, however, that carbonate radicals (CO3−) rather than ·OH radicals are the major product of the Fenton reaction, and that the carbonate radical specifically generates C8 or C5-oxidized G but does not cause direct DNA strand breaks [16,21,22,26,27,28]. These new findings deserve further study, as they generate new questions and ideas about the plausible regulatory function of 8-oxoG. Despite the fact that 8-oxoG is efficiently removed from DNA by BER and NER (nucleotide excision repair), it can still be detected and used as an indicator of DNA exposure to ROS [18,29].
The compound 5-hydroxymethylcytosine (hm5C) is a new epigenetic mark that has received tremendous attention in epigenetic research [5]. It was first identified in bacteriophages by Wyatt and Cohen (1952) [30], and in vertebrates in 1972 [31], but its function was poorly understood until 2009 when it was shown to comprise 0.6 and 0.2% of the nucleotides in Purkinje and granule cells [32,33]. Those studies revitalized the interest in hm5C and it is now often referred to as the “sixth base” [5,6]. It is currently believed that this DNA modification plays a critical role in many biological processes in eukaryotes [5,6].
While active DNA demethylation in plants can be achieved through direct cleavage of DNA by DNA glycosylases, followed by the replacement of m5C with C utilizing BER machinery, studies in mammals have demonstrated that m5C can be sequentially oxidized to hm5C, 5-formylcytosine (fo5C), and 5-carboxycytosine (ca5C) by members of the TET/JBP (ten-eleven translocation/J-binding proteins) family of iron-(II)/α-ketoglutarate-dependent dioxygenases [7,34,35]. The m5C oxidation products are subsequently recognized and cleaved by thymine-DNA glycosylase (TDG), thus, restoring unmethylated C via the BER pathway [7]. The consecutive oxidation of m5C in mammalian genomes raises a question pertaining to the potential presence of consecutive oxidation products in plant genomes [8]. In this regard, hm5C has been detected and reported in several plant genomes, with levels estimated to range between 0.068 and 0.075% in Arabidopsis, soybean, and rice DNA [36,37,38,39,40]. The origin of this modification is still in question, however, because TET/JBP proteins and UHRF2 (ubiquitin-like PHD and Ring Finger Domains 2), the proteins associated with hm5C, have not been identified in plants [6,34,38,41]. Therefore, it is assumed that hm5C may not be a product of enzymatic activity. In contrast, Tang et al. [7] demonstrated the presence of fo5C and ca5C in genomic DNA of various plant tissues and suggested that in addition to direct DNA cleavage by glycosylase, active DNA demethylation in plants may be achieved through an alternative pathway similar to mammals. Finally, the formation of hm5C and fo5C has been proposed to arise from H-atom extraction from the methyl group of m5C by ·OH [42]. As a result, a clear distinction between hm5C as a plant epigenetic mark or marker of DNA damage is still unresolved.
Therefore, in order to address this issue in the present study, we examined the changes in m5C, hm5C, and 8-oxoG in embryonic axes of sycamore maple (Acer pseudoplatanus L.) under desiccation stress conditions and after accelerated aging. We previously used sycamore seeds as a model in research on recalcitrant (desiccation-sensitive) seeds [10,43]. Here, we report for the first time on the changes in the level of three DNA modifications (m5C, hm5C, and 8-oxoG) while simultaneously tracking the level of ROS and antioxidants (AOX). We discuss the nature of those modifications and the implications of their-genome wide imbalances, as well as why only global methylation status is a good biomarker of tissue viability.
2. Materials and Methods
2.1. Plant Material and Treatments
Samaras from sycamore (Acer pseudoplatanus L.) were collected from trees in provenances located in Olsztyn (53.7553° N, 20.4561° E), in northeastern Poland. The collected samaras were dried at room temperature and 35% RH (relative humidity) and stored at 3 °C in tightly sealed, plastic containers. Embryonic axes were excised from the seeds no later than eight weeks after collection. The desiccation of embryonic axes was conducted as previously described [9,10,43]. The duration of the desiccation protocol ranged from 1 to 18 h. and was calculated on a fresh weight basis as previously described [44] (Table 1). In the accelerated aging experiment (storage at high temperature and high MC), fresh embryonic axes at a MC of 50% were kept in 2 mL tubes at 45 °C for three days. Unless otherwise indicated, all chemicals and materials were purchased from Merck (Darmstadt, Germany).
Table 1.
Moisture content (MC) and water content (WC) of Acer pseudoplatanus L. embryonic axes after desiccation between 1 and 18 h.
| Desiccation (h) | MC, % (WC, g H2O g−1 Dry Weight) |
|---|---|
| 0 | 50.2 ± 0.08 |
| (1.01 ± 0.03) | |
| 1 | 19.3 ± 0.07 |
| (0.24 ± 0.01) | |
| 4 | 11.9 ± 0.08 |
| (0.13 ± 0.01) | |
| 6 | 9.2 ± 0.4 |
| (0.10 ± 0.01) | |
| 18 | 5.7 ± 1.2 |
| (0.06 ± 0.01) |
2.2. Viability Assessment
A tetrazolium chloride (TTC) assay was used to assess the viability of axes subjected to accelerated ageing as previously described [43,44,45]. Four biological replicates comprising ten embryonic axes in each replicate were stained in 1% 2,3,5-triphenyltetrazolium chloride (TTC) solution. Embryonic axes were considered dead when remaining green, while those stained pink to red were classified as metabolically active and alive.
An in vitro regrowth assay was conducted as previously described [43] using five biological replicates comprising 10–15 embryonic axes in each replicate. After eight weeks, regrowth of embryonic axes was assessed and only counted as regenerated if the embryonic axis had developed either a shoot or a shoot with a root.
2.3. ROS Detection
ROS levels were quantified in aged embryonic axes using the fluorogenic dye, 2′,7′- dichlorodihydrofluorescein diacetate (H2DCFDA; Invitrogen, Waltham, MA, USA) as previously described [43]. Embryonic axes were incubated in 1 mL of 10 µM H2DCFDA in darkness for 15 min. Fluorescence was measured at an excitation wavelength of 492 nm and an emission wavelength of 525 nm using an Infinite M200 PRO plate reader (Tecan, Männedorf, Switzerland). Four replicates comprising five embryonic axes in each replicate were analyzed. Results are expressed as relative fluorescence units per gram of dry weight (RFU g−1 DW).
2.4. Measurement of Total and Non-Protein Antioxidant Capacity
Total antioxidant capacity (TAC) was measured using a commercially available Total Antioxidant Capacity Assay kit (Merck (Darmstadt, Germany). Five embryonic axes were used as replicates in each experiment and each experiment was replicated four times. Explants were homogenized in liquid nitrogen (LN) and samples were extracted in 0.8 mL of ice cold 1X Phosphate Buffered Saline (PBS) to maintain the recorded antioxidant values within the range of the standards provided in the commercial kit. Absorbance of the samples (10 µL) was assessed at λ 570 nm according to the manufacturer’s instructions. Antioxidant capacity was calculated based on trolox equivalent antioxidant capacity, that is, the amount of trolox producing the same effect as the sample studied. Calculations were made based on standard curves obtained for a trolox solution. Values were then normalized to tissue dry weight. For non-protein antioxidant capacity (NPAC), samples were diluted 1:1 with Protein Mask provided by the manufacturer.
2.5. DNA Isolation and Measurement of DNA Modifications
Total genomic DNA was extracted from control embryonic axes, embryos subjected to desiccation, and embryos subjected to accelerated ageing after the axes were homogenized in LN using a NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. Five embryonic axes were used as replicates in each experiment and each experiment was replicated four times. DNA concentration and quality (A260/A280 = 1.8–1.9) were measured with a NanoQuant Plate M200 PRO (Tecan, Männedorf, Switzerland). DNA modifications were quantified colorimetrically using MethylFlash Global DNA Methylation (5-mC), Hydroxymethylation (5-hmC) ELISA Easy Kits, and EpiQuik 8-OHdG DNA Damage Quantification Direct Kit (Epigentec, Farmingdale, NY, USA) according to the manufacturer’s instructions. Two technical replicates of each biological replicate were measured. A total of 100 ng of DNA was used for the m5C and hm5C measurements, while 200 ng was used for the 8-oxoG measurements. The relative quantity of the different entities was calculated according to the formula provided in the commercial kits using the provided standards for the preparation of standard curves.
2.6. Statistical Analysis
Graphical visualization and statistical analysis of the obtained data was conducted with R software (R Core Team 2020; https://www.r-project.org; accessed on 1 April 2022). The effect of desiccation on viability (TTC staining assay, regrowth) was evaluated separately using a general linear model (GLM) with a binomial distribution. The impact of desiccation and accelerated aging on the level of m5C, hm5C, and the assessment of the total amount of antioxidants, were evaluated using a linear model. An assessment of normality of the data was conducted using the Shapiro–Wilk test, and the Levene’s test was used to test homogeneity of variance. For non-normally distributed data or data with non-homogenous variation, such as the level of 8-oxoG, ROS, and the amount of non-protein antioxidants, a non-parametric Kruskal–Wallis test was used. A one-way ANOVA was used to test significant differences between mean values of data that were normally distributed. Pairwise comparisons between treatments were performed using a Duncan’s multiple range test at p ≤ 0.05. The correlation between in vitro regrowth and viability measured by TTC, and the levels of hm5C, m5C, 8-oxoG, ROS, the assessment of the total amount of antioxidants and the amount of non-protein antioxidants after different desiccation time and accelerated aging, were analyzed using a Spearman correlation coefficient analysis. An R-package, ‘corrplot‘, was used for the analysis and construction of the correlation between the measured parameters. Principal component analysis (PCA) was applied to z-scores of the data. R-packages ‘ggplot2’, ‘factoextra’ and ‘FactoMineR’, were used for the principal component analysis (PCA), while ‘ggplot2’ was used for the graphical visualization of the data.
3. Results
3.1. Explant Viability: TTC Assay and Regrowth
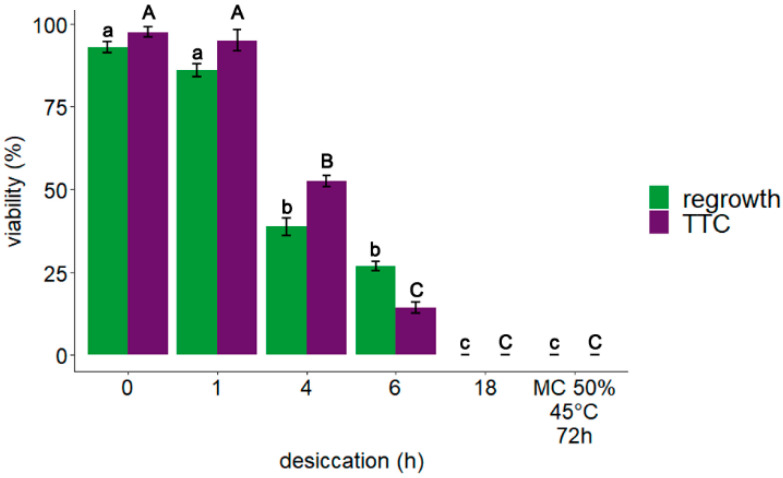
The impact of desiccation on the viability of embryonic axes of A. pseudoplatanus was assessed by the level of metabolic activity of the tissues determined by TTC staining assay and as regrowth of seedlings after 8 weeks of in vitro culture. Data obtained from a previous study [43], along with data for metabolic activity of embryonic axes subjected to accelerated ageing, are also presented (Figure 1). Altogether results indicated that a desiccation period of four and six hours significantly decreased the viability of embryonic axes while explants desiccated for 18 h, or subjected to accelerated aging, were dead.
Figure 1.
Effect of desiccation on Acer pseudoplatanus L. embryonic axes regrowth (n = 5) and respiratory activity measured by TTC assay (n = 4) [43]. The presented graph is supplemented with data obtained after storage of embryonic axes in accelerated aging conditions (72 h, 45 °C, MC of 50%). Statistical analyses were conducted on separate data sets. Capital and small letters correspond to analysis of TTC staining and tissue regrowth, respectively. Values labelled with different letters are significantly different at p ≤ 0.05, according to the Duncan test. Data represents mean ± SE.
3.2. Measurement of Reactive Oxygen Species
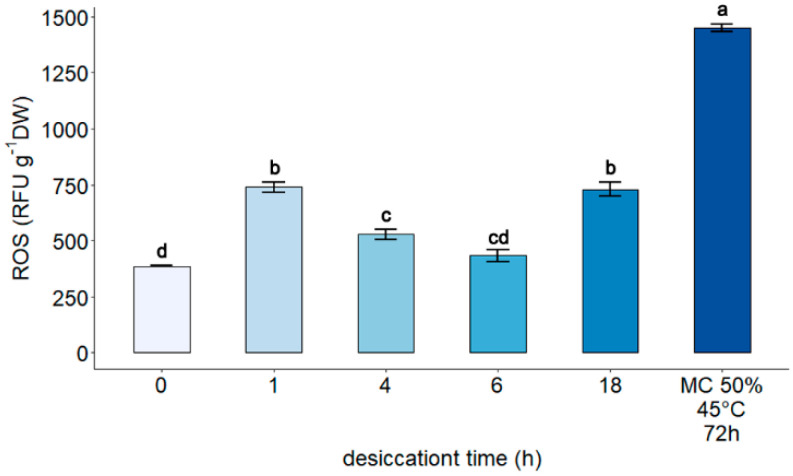
The impact of gradual desiccation and accelerated aging on ROS was determined using the H2DCFDA assay. Previously presented data, along with data for embryonic axes subjected to accelerated aging, are also presented. The highest level of ROS was detected in explants subjected to accelerated aging for 3 days at 45 °C (1440 RFU g−1 DW), (Figure 2).
Figure 2.
Influence of gradual desiccation on ROS level in embryonic axes of Acer pseudoplatanus L. [43]. The presented graph is supplemented with data obtained after storage of embryonic axes in accelerated aging conditions (72 h, 45 °C, MC of 50%). Values labelled with different letters are significantly different at p ≤ 0.05, according to the Duncan test. Data represents mean ± SE, n = 4.
3.3. Measurement of Total Antioxidant Capacity and Non-Protein Antioxidant Capacity
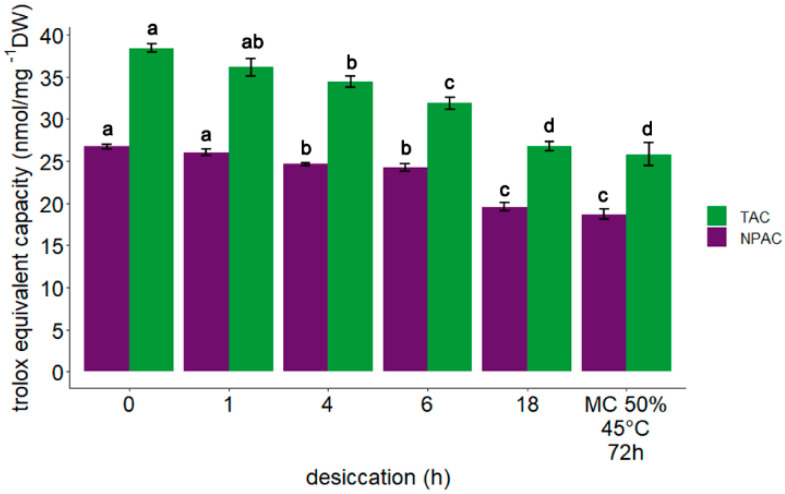
The highest total antioxidant capacity (TAC) (38.4 nmol/µL g−1 DW) was observed in control embryonic axes. Desiccation of embryonic axes for 1 h resulted in only a slight decrease in the level of TAC (36.1 nmol/µL g−1 DW). Further desiccation for 4 h, 6 h, and 18 h caused a significant progressive decline in TAC to 34.4, 31.9, and 26.7 nmol/µL g−1 DW, respectively (Figure 3). The lowest level of TAC (25.8 nmol/µL g−1 DW) was detected in embryonic axes subjected to accelerated aging.
Figure 3.
Influence of gradual desiccation and accelerated aging (72 h, 45 °C, MC of 50%) on total antioxidants capacity (TAC) and non-protein antioxidant capacity (NPAC) in embryonic axes of Acer pseudoplatanus L. Values labelled with different letters are significantly different at p ≤ 0.05, according to the Duncan test. Data represents mean ± SE, n = 4.
Non-treated (control) embryonic axes of A. pseudoplatanus exhibited the highest non-protein antioxidant capacity (NPAC) (26.7 nmol/µL g−1 DW). A similar level was observed after 1 h desiccation, however, after desiccation for 4 h and 6 h, a decrease in NPAC was detected (24.4 and 24.2 nmol/µL g−1 DW, respectively) (Figure 3). The NPAC after desiccation for 18 h and accelerated ageing was comparable (19.6 nmol/µL g−1 DW and 18.7 6 nmol/µL g−1 DW, respectively).
3.4. Assessment of the Genomic Level of m5C, 8-oxoG, and hm5C in the DNA of Embryonic Axes of Acer pseudoplatanus
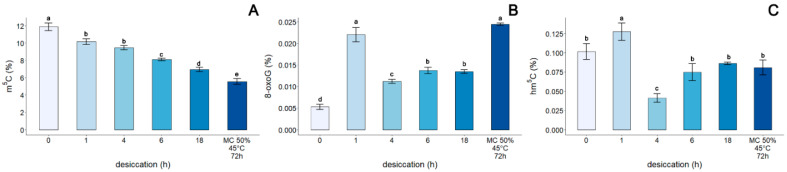
Non-treated (control) embryonic axes of A. pseudoplatanus exhibited the highest level of m5C (11.9%) (Figure 4A). As desiccation time increased, a significant decrease in the level of m5C in DNA was observed to 10.2%, 9.48%, 8.11%, and 6.96% after desiccation for 1, 4, 6, 18 h, respectively. Accelerated aging had the strongest impact on the level of global m5C, reducing it to half the level (5.56%) that it was in control embryonic axes.
Figure 4.
Influence of gradual desiccation and accelerated aging conditions (72 h, 45 °C, MC of 50%) on the relative global level of (A) 5-methylcytosine; (B) 8-oxo-7,8-dihydroguanine; (C) 5-hydroxymethycytosine. Values labelled with different letters are significantly different at p ≤ 0.05, according to the Duncan test. Data represents mean ± SE, n = 3.
The lowest percentage of 8-oxoG in DNA (0.0053%) was detected in non-treated, control embryonic axes. Desiccation of embryonic axes for 1 h caused a significant increase in the detected amount of 8-oxoG, increasing the percentage to 0.022% (Figure 4B). Further desiccation resulted in a decline in 8-oxoG to 0.0112%. No difference was observed between the percentage of 8-oxoG in embryonic axes desiccated for 6 and 18 h (0.0137% and 0.0135%, respectively). In contrast, a significant 4.6-fold increase in the level of 8-oxoG, relative to the control, was observed in embryonic axes after 3 days of accelerated aging at 45 °C.
A global hm5C level of 0.102% was detected in control embryonic axes (Figure 4C). Desiccation for 1 h induced a significant increase in the detected level of hm5C to 0.128%. After desiccation of embryonic axes for 4 h, however, a significant decrease in the level of this modification to 0.041% was observed. Notably, further desiccation for 6 h and 18 h, as well as accelerated ageing conditions, did not significantly affect the level of hm5C, which remained in the range of 0.075–0.081%.
3.5. Correlation and Principal Component Analyses
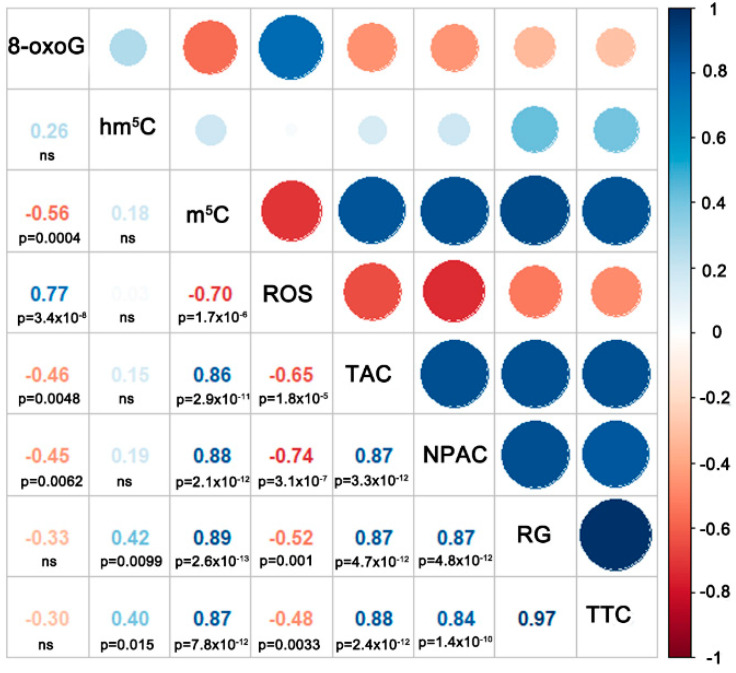
Spearman correlation coefficient analysis of the detected levels of m5C, hm5C, 8-oxoG, ROS, TAC, and NPAC indicated that the level of 8-oxoG was significantly negatively correlated with the level of m5C, TAC, and NPAC (Figure 5). The level of 8-oxoG was also significantly positively correlated with the detected level of ROS. The level of m5C exhibited a highly significant negative correlation with the level of ROS, and a highly significant strong positive correlation with TAC and NPAC. No significant correlation was observed between the level of hm5C and any of the other measured parameters.
Figure 5.
The Spearman correlation coefficient between the means of oxidative stress measurements (ROS), viability measurements (TTC, regrowth RG), global level of m5C, hm5C, 8-oxoG, total antioxidant capacity (TAC), non-protein antioxidant capacity (NPAC) measured in Acer pseudoplatanus L. embryonic axes directly after desiccation or accelerated aging (72 h, 45 °C, MC of 50%). The size of the circles represents the level of correlation (r), bigger circles indicate that a given trait correlates at higher level. Blue color indicates positive correlations and red indicates negative correlations.
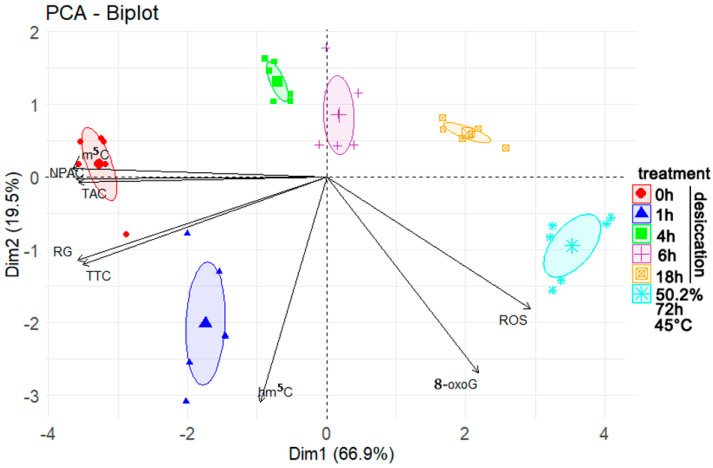
The PCA results (Figure 6) revealed a negative relationship between the viability of embryonic axes (as measured by either metabolic activity or regrowth), the level of m5C, TAC, and NPAC, with the level of 8-oxoG and ROS. This negative relationship was reflected in the opposite coordinates of these two groups and the high loading of these variables in principal component 1 (Prin 1), (Supplementary Table S1). The level of m5C, TAC, NPAC, metabolic activity of the tissues (TTC), and regrowth of seedlings in in vitro culture (RG) had the biggest influence on principal component 1 (Prin 1). In contrast, hm5C and 8-oxoG had the biggest influence on principal component 2. This influence was reflected in the high presence of these variables in Prin1 and Prin2, respectively (Supplementary Table S2). The first two principal components accounted for 66.9 and 19.5% of the observed variance, respectively. The embryonic axes clustered into six groups based on Prin1 and Prin2, clustering in groups corresponding to their treatment. The results indicated that the groups differed mostly based on their level of m5C, TAC, and NPAC.
Figure 6.
Results of principal component analysis (PCA) applied on the correlations of regrowth (RG), metabolic activity (TTC), ROS level, relative amount (%) of m5C, hm5C, 8-oxoG, total antioxidant capacity (TAC), non-protein antioxidant capacity (NPAC) in embryonic axes directly after desiccation or accelerated aging (72 h, 45 °C, MC of 50%). Ellipse confidence level = 0.95.
4. Discussion
Seeds of Acer pseudoplatanus L. (sycamore) are desiccation sensitive, a feature that complicates their long-term storage since the seeds do not tolerate desiccation below 30% of MC. Owing to their recalcitrance, sycamore seeds can only be stored under standard conditions for 2–3 years at −3 °C. Thus, sycamore seeds have become a model system for desiccation studies of temperate zone recalcitrant seeds [10,43,46,47,48,49,50]. Notably, the predominant categorization system utilized to classify seeds is based on their tolerance to water withdrawal [51]. Desiccation studies have received considerable attention by seed researchers since desiccation tolerance is a determining factor for successful seed preservation under both conventional and cryogenic conditions [12,52,53,54].
The current study was conducted to provide information on the changes in nucleobase modifications that occur in seeds when they are exposed to the stress of desiccation. The objective was to determine the changes that occur in the global level of three nucleobase modifications in embryonic axes in response to desiccation, to better understand the relationship between these modified nucleobases. Notably, we were able to document for the first time changes in the global level of hm5C under conditions that have not been previously examined. To determine if the observed changes are characteristic only for tissues with decreasing viability due to desiccation, we also measured changes in these three nucleotide modifications in embryonic axes subjected to accelerated aging at high moisture content and temperature. This methodology was used to enable us to simulate, over a short period of time, natural aging processes that occur in plant tissues over an extended time period.
We previously used 2D-TLC to monitor changes in global DNA methylation in seeds, which allowed us to measure the level of m5C in DNA [10,11,44], even in problematic seed samples that are rich in lipids and proteins. Although this method has several notable advantages, it does not allow for quick, high-throughput analyses, preferably performed in a 96-well format, that provides the ability to assess multiple biological samples simultaneously in a statistically sound manner. Therefore, we decided to use an enzyme-linked immunosorbent assay (ELISA) in the present study. This method has been successfully used by others [55,56,57], although it has never been used in the context of recalcitrant seed desiccation studies.
Desiccation induces oxidative stress and a redox imbalance. Cells, however, possess a system of enzymatic proteins (dismutase, peroxidases, catalase), non-enzymatic proteins (peroxiredoxins, thioredoxins, glutaredoxins, and metallothionein), and low molecular weight antioxidants (glutathione ascorbate, carotenoids) [1] to prevent the formation and accumulation of ROS and to maintain the redox equilibrium. In the present study, the total, as well as non-protein small molecule antioxidant capacity, decreased with time of desiccation and in samples subjected to accelerated aging. We tracked ROS levels in a previous study using the fluorescent probe H2DCFDA, which is oxidized by multiple oxidative agents [43], thus, providing information on the oxidative stress level present in plant tissues. In the present study, we measured both the oxidative and antioxidant status of cells at each stage of desiccation and after accelerated aging, along with concomitant changes in the level and type of DNA modifications. As a result, we demonstrated a correlation between the redox state of tissues and the 8-oxoG and m5C level, although no clear correlation was observed for hm5C. Further we explain, nevertheless, why we consider hm5C as a result of oxidation.
Chemical modifications to DNA add an additional layer of complexity to cellular processes due to their ability to function in a regulatory network that modulates chromatin structure and genome function. Cytosine methylation is an element of the epigenome, a system of non-sequence based, potentially heritable traits. The composition of the epigenome within a given cell is a function of both genetic determinants and external factors [6,7,58]. In this regard, desiccation stress has been reported to induce changes in the pattern and level of DNA methylation in various plant tissues, including recalcitrant and orthodox (desiccation resistant) seeds [9,10,44]. Our current results confirm previous observations. Rapid, severe desiccation of isolated recalcitrant embryonic axes resulted in a decrease in the percentage of total m5C in genomic DNA. This decline was highly correlated with a decline in viability (Figure 5). Notably, the reduction in m5C was induced by both desiccation and accelerated aging, therefore treatment differing in tissue moisture content and temperature. We previously demonstrated that the decrease in viability of Quercus robur L. recalcitrant seeds during storage (natural aging) also resulted in a decline in global m5C levels [11]. Additionally, it was also reported that epigenetic changes occurred upon accelerated aging conditions in orthodox seeds of Secale cereale L. and Mentha aquatica L., even before a major loss of viability was observed [59,60]. Therefore, collectively the data indicate that the decline in DNA methylation that occurs in plant embryos that also undergo a reduction in viability is a universal process resulting from being subjected to different stresses (water withdrawal vs. high temperature and high MC) [11,61]. As a result, reductions in m5C levels can be considered as a viability marker, which was proved by high significant correlation (Figure 5) and PCA analysis (Figure 6).
Pro-oxidant species are relatively abundant within cells and are continuously generated by endogenous metabolism and in response to exposure to external stress factors [29]. ROS-mediated DNA oxidation represents a threat to the stability of genetic material and has been estimated to occur at a rate that is only 50% lower than that occurring for protein oxidation [29,62]. While ROS can oxidize DNA either by altering the four bases or deoxyribose, the most stable and frequent DNA oxidation product in DNA is 8-oxoG [29], and as a result, this alteration is considered as a marker for oxidative damage to DNA. In the present study, the changes in 8-oxoG were the result of oxidative stress induced by desiccation and accelerated aging. We previously analyzed desiccation-driven DNA damage and repair after rehydration of seeds using an enzyme-modified alkaline comet assay with formamidopyrimidine [fapy]-DNA glycosylase (Fpg) enzyme, which recognizes oxidized forms of guanine [44]. By using another method in the present study, we confirmed that elevated levels of oxidative stress induced by desiccation negatively impact DNA integrity. A high percentage of oxidized G was also detected in embryonic axes that lost their viability due to accelerated ageing. Notably, 8-oxoG was also reported to be the most common and abundant DNA lesion in mammalian aged cells [63]. Accumulation of ROS resulting in 8-oxoG increase in DNA and lower seed viability was also reported [64,65,66]. Moreover, transcriptional changes during seed ageing were accompanied by a progressive shift towards a more oxidizing cellular environment and nucleic acid degradation, which occurred prior to any loss of seed viability [67]. However, in current research, in contrast to the m5C epigenetic mark, high levels of oxidative damage can clearly be detected in viable tissue affected by desiccation-driven oxidative stress, as well as in dead tissue. Therefore, an increase in 8-oxoG does not appear to be specific to the physiological state of the tissue, which suggests that one needs to track DNA repair efficiency over time to distinguish viable and non-viable tissues [43]. Therefore, the evaluation of 8-oxoG as a single time point measurement performed immediately after any treatment may not represent an accurate assessment of viability. This premise agrees with previous research [68] showing that biomarkers such as ROS are useful in describing the nature of stress and the reason for cellular damage but may not be helpful as direct markers of the actual viability state of a tissue.
Nevertheless, oxidative damage to G presents a severe threat to genome and epigenome stability. It represents a pre-mutagenic lesion as its syn arrangement pairs with adenine leading to a G:C-T:A transversion upon replication [29,69]. In vitro experiments have demonstrated that the presence of 8-oxoG destabilizes DNA fragments possessing different methylation patterns. The dynamic and thermodynamic effects are not additive in fully methylated oxidized CpG, indicating that the introduced modifications interact with each other. The close proximity of m5C and 8-oxoG within a major groove, together with steric and ion-dipole interactions between 8-oxoG and the sugar-phosphate moiety of the 8-oxoG nucleotide, may be expected to disturb the conformational dynamics of methylated and oxidized CpG sites [69]. Finally, 8-oxoG is known to interfere with the action of many DNA-binding proteins. Thus, the conformational and dynamic effects of spurious DNA oxidation in the regulatory CpG dinucleotides can have a far-reaching biological impact [69]. The presence of 8-oxoG was shown to bias the recognition of methylated CpG dinucleotides by ROS1, with an approximately fourfold preference for the oxidized DNA strand [69]. Additionally, 8-oxoG was also shown to affect cytosine methylation by interfering with the binding of DNA methyltransferases [29]. Consequently, a negative correlation between m5C content and 8-oxoG was observed and was also evident in our present study (Figure 5). Indeed, cells that are deficient in the antioxidant enzyme CuZn-SOD exhibit an accumulation of 8-oxoG and massive levels of DNA hypomethylation [29,70,71]. Therefore, it seems that oxidative DNA damage has a substantial impact on epigenetic markers by blocking their synthesis and enhancing their removal.
Interestingly, it was recently suggested that 8-oxoG can play a role in the regulation of genomic functions through mechanisms responsible for its formation and processing [29]. Epigenetics studies focus on heritable changes in genome function that are not due to alterations in the DNA sequences. In this regard, m5C is a common epigenetic mark that provides a new level of epigenetic information, while 8-oxoG also represents a stable change in DNA structure similar to m5C and may provide an additional regulatory influence on the primary sequence that is dictated by the redox environment surrounding the genome [29]. In this regard, 8-oxoG is produced by fluxes of H2O2 from the cytosol to the nucleus, thus reflecting the intracellular redox balance. It can also be produced, however, by a local reaction involving the formation of ·OH from H2O2 by iron-containing complexes associated with DNA structure in close proximity to chromatin. H2O2 is also sourced through the activity of nuclear oxidases and FAD-dependent lysine-specific histone demethylases (LDL1, LDL2, LDL3 FLD) [1,72] that participate in the regulation of gene expression [1,29]. It is still not known if these two pathways (cytosolic and nucleolar) cooperate and if 8-oxoG encodes transmissible information capable to durably alter cellular behavior [29].
The last modification we tracked was hm5C, which is also recognized as a stable epigenetic mark [35]. In general, oxidized forms of m5C in plants are very limited and there is no consensus currently if the oxidation is a result of ROS or enzymatic activity [5,7,8,34,41]. The relatively low levels of m5C oxidation products suggest that they most likely arise due to the activity of ROS in cells, which is supported by the lack of any functional Tet-family dioxygenase enzymes (writer and primary eraser of hm5C) and UHFR2 (reader of hm5C) [5,8,35] being identified. Moricova et al., 2013 [55], however, detected hm5C in protoplasts of Cucumis sativus L. and Brassica oleracea L. but stated that oxidative damage was not the main source of hm5C. Only a few studies have investigated the role of hm5C in plants under stress conditions [73]. In the present study, the changes in global hm5C induced in the genome of A. pseudoplatanus by progressive steps of desiccation mirrored the changes observed for guanine oxidative damage (Figure 4). An initial increase in hm5C was observed after 1 h of desiccation when the change in MC was the highest, followed by a drop after 4 h, in association with corresponding DNA repair in viable tissues [43], followed by a plateau in tissues with low viability or that were dead (after 6 and 18 h of desiccation, respectively). Although the initial hm5C increase is smaller in comparison to 8-oxoG, the similarity between these changes in global DNA modification during subsequent desiccation steps is striking. However, when the data obtained from embryonic axes subjected to accelerated ageing were analyzed, concomitant increases in ROS and 8-oxoG were clearly evident, while no such increase in hm5C was observed in embryonic axes subjected to accelerated aging. This is contrary to what one would expect for oxidative DNA damage resulting from increased levels of ROS (Figure 2), however it is in concordance with previous studies, as it has been reported that there is no correlation between the age of adult mice and the level of hm5C in Purkinje and granule cells [32], while a decrease in hm5C was recently reported in mouse cells and tissues subjected to physiological and accelerate aging [74]. Therefore, it could be hypothesized that the level of hm5C can be affected by the accumulation of its oxidized forms (fo5C and ca5C), which have been detected in plant genomes [7]. Environmental stresses, such as drought and salinity, can alter the level of fo5C and ca5C in plant genomes, suggesting that these modifications play a role in plant response to environmental stress [7]. A second potential scenario is that a decrease of m5C, resulting from demethylation or 8-oxoG proximity (discussed above), may also reduce the amount of hm5C since it is derived from m5C. Indeed, a decrease in m5C was observed in aging plant tissues concomitant with a decrease in CMT3 and MET1 expression, and increased expression of ROS1 [75]. This suggests that regions of DNA in plants may undergo demethylation during plant aging due to a decrease in DNA methylation and the activation of DNA demethylation processes [75,76]. Therefore, the lack of correlation between ROS and hm5C (Figure 5) may result from aging-related m5C decline. Moreover, hm5C does not appear to be a direct marker of tissue viability (Figure 5), although it may exert an impact on genome stability and gene expression. For example, hm5C in plants may be involved in passive demethylation during cell division. The A. thaliana VIM1 protein has been reported to recognize hm5C in vitro, and it has been suggested that hm5C at a CpG site may trigger VIM-mediated passive loss of cytosine methylation during Arabidopsis DNA replication [36]. In addition, plants possess a DNA glycosylase that can directly cleave m5C, and both DME and ROS1 can also react with hm5C [77], also participating in the demethylation process by removal of oxidized m5C.
5. Conclusions
Our data revealed and confirmed the presence of modified DNA bases in the sycamore genome. Our study supports previous reports that desiccation induces oxidative stress and leads to a decline in the pool of antioxidants and increase in ROS. For the first time, however, we demonstrated that these changes occur along with both a decrease in global DNA methylation and concomitant mirroring changes in the level of oxidized m5C and G in embryonic axes of sycamore subjected to desiccation stress, suggesting that generated ROS react with both G and m5C and that hm5C may serve as both an epigenetic mark and an oxidative stress marker. However, only m5C can be considered as a viability marker, as we did not detect any significant correlation between 8-oxoG or hm5C and tissue viability.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11111748/s1, Table S1: Table of coordinates of variables used in principal component analysis; Table S2: Table of loadings matrix of variables used in principal component analysis.
Author Contributions
B.P.P.-M.: conceptualization, methodology, investigation, data curation, validation, formal analysis, resources, writing—original draft preparation, project administration, funding acquisition, M.L.: methodology, writing—review and editing, M.M.: investigation, data curation, formal analysis, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by National Science Center, Poland, grant number UMO-2016/21/D/NZ9/02489 awarded to B.P.P-M.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gorini F., Scala G., Cooke M.S., Majello B., Amente S. Towards a Comprehensive View of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine: Highlighting the Intertwined Roles of DNA Damage and Epigenetics in Genomic Instability. DNA Repair. 2021;97:103027. doi: 10.1016/j.dnarep.2020.103027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bilyard M.K., Becker S., Balasubramanian S. Natural, Modified DNA Bases. Curr. Opin Chem. Biol. 2020;57:1–7. doi: 10.1016/j.cbpa.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Ruppel W.G. Zur Chemie der Tuberkelbacillen. Hoppe-Seyler’S Z. Für Physiol. Chem. 1899;26:218–232. doi: 10.1515/bchm2.1899.26.3-4.218. [DOI] [Google Scholar]
- 4.Hotchkiss R.D. The Quantitative Separation Of Purines, Pyrimidines, and Nucleosides by Paper Chromatography. J. Biol. Chem. 1948;175:315–332. doi: 10.1016/S0021-9258(18)57261-6. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Song S., Wu Y.-S., Li Y.-L., Chen T., Huang Z., Liu S., Dunwell T.L., Pfeifer G.P., Dunwell J.M., et al. Genome-Wide Mapping of 5-Hydroxymethylcytosine in Three Rice Cultivars Reveals Its Preferential Localization in Transcriptionally Silent Transposable Element Genes. J. Exp. Bot. 2015;66:6651–6663. doi: 10.1093/jxb/erv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi D.-Q., Ali I., Tang J., Yang W.-C. New Insights into 5hmC DNA Modification: Generation, Distribution and Function. Front. Genet. 2017;8:100. doi: 10.3389/fgene.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y., Xiong J., Jiang H.-P., Zheng S.-J., Feng Y.-Q., Yuan B.-F. Determination of Oxidation Products of 5-Methylcytosine in Plants by Chemical Derivatization Coupled with Liquid Chromatography/Tandem Mass Spectrometry Analysis. Anal. Chem. 2014;86:7764–7772. doi: 10.1021/ac5016886. [DOI] [PubMed] [Google Scholar]
- 8.Chen T., Li E. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 9.Plitta B.P., Michalak M., Bujarska-Borkowska B., Barciszewska M.Z., Barciszewski J., Chmielarz P. Effect of Desiccation on the Dynamics of Genome-Wide DNA Methylation in Orthodox Seeds of Acer platanoides L. Plant Physiol. Biochem. 2014;85:71–77. doi: 10.1016/j.plaphy.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Plitta-Michalak B.P., Naskręt-Barciszewska M.Z., Kotlarski S., Tomaszewski D., Tylkowski T., Barciszewski J., Chmielarz P., Michalak M. Changes in Genomic 5-Methylcytosine Level Mirror the Response of Orthodox (Acer platanoides L.) and Recalcitrant (Acer pseudoplatanus L.) Seeds to Severe Desiccation. Tree Physiol. 2018;38:617–629. doi: 10.1093/treephys/tpx134. [DOI] [PubMed] [Google Scholar]
- 11.Michalak M., Plitta-Michalak B., Nasktęt-Barciszewska M., Barciszewski J., Bujarska-Borkowska B., Chmielarz P. Global 5-Methylcytosine Alterations in DNA during Ageing of Quercus robur Seeds. Ann. Bot-London. 2015;116 doi: 10.1093/aob/mcv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plitta B.P., Michalak M., Naskręt-Barciszewska M.Z., Barciszewski J., Chmielarz P. DNA Methylation of Quercus robur L. Plumules Following Cryo-Pretreatment and Cryopreservation. Plant Cell Tiss. Organ Cult. 2014;117:31–37. doi: 10.1007/s11240-013-0417-9. [DOI] [Google Scholar]
- 13.Zhang H., Lang Z., Zhu J.-K. Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- 14.Smolikova G., Leonova T., Vashurina N., Frolov A., Medvedev S. Desiccation Tolerance as the Basis of Long-Term Seed Viability. Int. J. Mol. Sci. 2021;22:101. doi: 10.3390/ijms22010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurek K., Plitta-Michalak B., Ratajczak E. Reactive Oxygen Species as Potential Drivers of the Seed Aging Process. Plants. 2019;8:174. doi: 10.3390/plants8060174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma P., Jha A.B., Dubey R.S., Pessarakli M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012;2012:1–26. doi: 10.1155/2012/217037. [DOI] [Google Scholar]
- 17.Adetunji A.E., Adetunji T.L., Varghese B., Sershen, Pammenter N.W. Oxidative Stress, Ageing and Methods of Seed Invigoration: An Overview and Perspectives. Agronomy. 2021;11:2369. doi: 10.3390/agronomy11122369. [DOI] [Google Scholar]
- 18.Lee T.-H., Kang T.-H. DNA Oxidation and Excision Repair Pathways. Int. J. Mol. Sci. 2019;20:6092. doi: 10.3390/ijms20236092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karanjawala Z.E., Murphy N., Hinton D.R., Hsieh C.-L., Lieber M.R. Oxygen Metabolism Causes Chromosome Breaks and Is Associated with the Neuronal Apoptosis Observed in DNA Double-Strand Break Repair Mutants. Curr. Biol. 2002;12:397–402. doi: 10.1016/S0960-9822(02)00684-X. [DOI] [PubMed] [Google Scholar]
- 20.Fenton H.J.H. Oxidation of Tartaric Acid in Presence of Iron. J. Chem. Soc. Trans. 1894;65:899–910. doi: 10.1039/CT8946500899. [DOI] [Google Scholar]
- 21.Fleming A.M., Burrows C. On the Irrelevancy of Hydroxyl Radical to DNA Damage from Oxidative Stress and Implications for Epigenetics. Chem. Soc. Rev. 2020;49:6524–6528. doi: 10.1039/D0CS00579G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleming A.M., Burrows C.J. Iron Fenton Oxidation of 2′-Deoxyguanosine in Physiological Bicarbonate Buffer Yields Products Consistent with the Reactive Oxygen Species Carbonate Radical Anion Not the Hydroxyl Radical. Chem. Commun. 2020;56:9779–9782. doi: 10.1039/D0CC04138F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatgilialoglu C., Ferreri C., Krokidis M.G., Masi A., Terzidis M.A. On the Relevance of Hydroxyl Radical to Purine DNA Damage. Free Radic. Res. 2021;55:384–404. doi: 10.1080/10715762.2021.1876855. [DOI] [PubMed] [Google Scholar]
- 24.Yin R., Zhang D., Song Y., Zhu B.-Z., Wang H. Potent DNA Damage by Polyhalogenated Quinones and H2O2 via a Metal-Independent and Intercalation-Enhanced Oxidation Mechanism. Sci. Rep. 2013;3:1269. doi: 10.1038/srep01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jena N.R., Mishra P.C. Formation of 8-Nitroguanine and 8-Oxoguanine due to Reactions of Peroxynitrite with Guanine. J. Comput. Chem. 2007;28:1321–1335. doi: 10.1002/jcc.20607. [DOI] [PubMed] [Google Scholar]
- 26.Halliwell B., Adhikary A., Dingfelder M., Dizdaroglu M. Hydroxyl Radical Is a Significant Player in Oxidative DNA Damage in Vivo. Chem. Soc. Rev. 2021;50:8355–8360. doi: 10.1039/D1CS00044F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Illés E., Mizrahi A., Marks V., Meyerstein D. Carbonate-Radical-Anions, and Not Hydroxyl Radicals, Are the Products of the Fenton Reaction in Neutral Solutions Containing Bicarbonate. Free Radical Biol. Med. 2019;131:1–6. doi: 10.1016/j.freeradbiomed.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Illés E., Patra S.G., Marks V., Mizrahi A., Meyerstein D. The FeII(Citrate) Fenton Reaction under Physiological Conditions. J. Inorg. Biochem. 2020;206:111018. doi: 10.1016/j.jinorgbio.2020.111018. [DOI] [PubMed] [Google Scholar]
- 29.Giorgio M., Dellino G.I., Gambino V., Roda N., Pelicci P.G. On the Epigenetic Role of Guanosine Oxidation. Redox Biol. 2020;29:101398. doi: 10.1016/j.redox.2019.101398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyatt G.R., Cohen S.S. A New Pyrimidine Base from Bacteriophage Nucleic Acids. Nature. 1952;170:1072–1073. doi: 10.1038/1701072a0. [DOI] [PubMed] [Google Scholar]
- 31.Penn N.W., Suwalski R., O’Riley C., Bojanowski K., Yura R. The Presence of 5-Hydroxymethylcytosine in Animal Deoxyribonucleic Acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriaucionis S., Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakovlev I.A., Gackowski D., Abakir A., Viejo M., Ruzov A., Olinski R., Starczak M., Fossdal C.G., Krutovsky K. Mass Spectrometry Reveals the Presence of Specific Set of Epigenetic DNA Modifications in the Norway Spruce Genome. Sci. Rep. 2019;9:19314. doi: 10.1038/s41598-019-55826-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rausch C., Hastert F.D., Cardoso M.C. DNA Modification Readers and Writers and Their Interplay. J. Mol. Biol. 2020;432:1731–1746. doi: 10.1016/j.jmb.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 36.Yao Q., Song C.-X., He C., Kumaran D., Dunn J.J. Heterologous Expression and Purification of Arabidopsis Thaliana VIM1 Protein: In Vitro Evidence for Its Inability to Recognize Hydroxymethylcytosine, a Rare Base in Arabidopsis DNA. Protein Expr. Purif. 2012;83:104–111. doi: 10.1016/j.pep.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Terragni J., Bitinaite J., Zheng Y., Pradhan S. Correction to Biochemical Characterization of Recombinant β-Glucosyltransferase and Analysis of Global 5-Hydroxymethylcytosine in Unique Genomes. Biochemistry. 2012;51:6261. doi: 10.1021/bi3008643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S., Dunwell T.L., Pfeifer G.P., Dunwell J.M., Ullah I., Wang Y. Detection of Oxidation Products of 5-Methyl-2′-Deoxycytidine in Arabidopsis DNA. PLoS ONE. 2013;8:e84620. doi: 10.1371/journal.pone.0084620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W., Huang F., Qin Q., Zhao X., Li Z., Fu B. Comparative Analysis of DNA Methylation Changes in Two Rice Genotypes under Salt Stress and Subsequent Recovery. Biochem. Biophys. Res. Commun. 2015;465:790–796. doi: 10.1016/j.bbrc.2015.08.089. [DOI] [PubMed] [Google Scholar]
- 40.Mahmood A.M., Dunwell J.M. Evidence for Novel Epigenetic Marks within Plants. AIMS Genet. 2019;06:070–087. doi: 10.3934/genet.2019.4.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erdmann R.M., Souza A.L., Clish C.B., Gehring M. 5-Hydroxymethylcytosine Is Not Present in Appreciable Quantities in Arabidopsis DNA. G3-Genes Genomes Genet. 2015;5:1–8. doi: 10.1534/g3.114.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madugundu G.S., Cadet J., Wagner J.R. Hydroxyl-Radical-Induced Oxidation of 5-Methylcytosine in Isolated and Cellular DNA. Nucleic Acids Res. 2014;42:7450–7460. doi: 10.1093/nar/gku334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plitta-Michalak B.P., Ramos A.A., Pupel P., Michalak M. Oxidative Damage and DNA Repair in Desiccated Recalcitrant Embryonic Axes of Acer pseudoplatanus L. BMC Plant Biol. 2022;22:40. doi: 10.1186/s12870-021-03419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michalak M., Barciszewska M.Z., Barciszewski J., Plitta B.P., Chmielarz P. Global Changes in DNA Methylation in Seeds and Seedlings of Pyrus Communis after Seed Desiccation and Storage. PLoS ONE. 2013;8:e70693. doi: 10.1371/journal.pone.0070693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.International Rules for Seed Testing 2021-ISTA Online-International Seed Testing Association. [(accessed on 1 March 2022)]. Available online: https://www.seedtest.org/en/international-rules-for-seed-testing-_content---1--1083.html.
- 46.Tylkowski T. Short-Term Storage of after Ripened Seeds of Acer platanoides L. and Acer pseudoplatanus L. Arbor. Kórnickie. 1989;34:135–141. [Google Scholar]
- 47.Tylkowski T. Przedsiewne Traktowanie Nasion Drzew, Krzewów, Pnączy i Krzewinek. Centrum Informacyjne Lasów Państwowych; Warszawa, Poland: 2016. [Google Scholar]
- 48.Hong T.D., Ellis R.H. A Comparison of Maturation Drying, Germination, and Desiccation Tolerance between Developing Seeds of Acer pseudoplatanus L. and Acer platanoides L. New Phytol. 1990;116:589–596. doi: 10.1111/j.1469-8137.1990.tb00543.x. [DOI] [Google Scholar]
- 49.Daws M.I., Cleland H., Chmielarz P., Gorian F., Leprince O., Mullins C.E., Thanos C.A., Vandvik V., Pritchard H.W., Daws M.I., et al. Variable Desiccation Tolerance in Acer Pseudoplatanus Seeds in Relation to Developmental Conditions: A Case of Phenotypic Recalcitrance? Funct. Plant Biol. 2006;33:59–66. doi: 10.1071/FP04206. [DOI] [PubMed] [Google Scholar]
- 50.Berjak P., Pammenter N.W. Implications of the Lack of Desiccation Tolerance in Recalcitrant Seeds. Front. Plant. Sci. 2013;4:478. doi: 10.3389/fpls.2013.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts E.H. Predicting the Storage Life of Seeds. Seed Sci. Technol. 1973;1:499–514. [Google Scholar]
- 52.Plitta-Michalak B., Michalak M., Kotlarski S., Chmielarz P. Cryogenic Storage of Seeds. Sylwan. 2013;157:723–729. [Google Scholar]
- 53.Pence V.C., Ballesteros D., Walters C., Reed B.M., Philpott M., Dixon K.W., Pritchard H.W., Culley T.M., Vanhove A.-C. Cryobiotechnologies: Tools for Expanding Long-Term Ex Situ Conservation to All Plant Species. Biol. Conserv. 2020;250:108736. doi: 10.1016/j.biocon.2020.108736. [DOI] [Google Scholar]
- 54.Walters C., Pence V.C. The Unique Role of Seed Banking and Cryobiotechnologies in Plant Conservation. Plants People Planet. 2021;3:83–91. doi: 10.1002/ppp3.10121. [DOI] [Google Scholar]
- 55.Moricová P., Ondřej V., Navrátilová B., Luhová L. Changes of DNA Methylation and Hydroxymethylation in Plant Protoplast Cultures. Acta Biochim. Pol. 2013;60:33–36. doi: 10.18388/abp.2013_1947. [DOI] [PubMed] [Google Scholar]
- 56.Visscher A.M., Latorre Frances A., Yeo M., Yan J., Colville L., Gomez Barreiro P., Pritchard H.W. Comparative Analyses of Extreme Dry Seed Thermotolerance in Five Cactaceae Species. Environ. Exp. Bot. 2021;188:104514. doi: 10.1016/j.envexpbot.2021.104514. [DOI] [Google Scholar]
- 57.Golubov A., Kovalchuk I. Analysis of DNA Hydroxymethylation Using Colorimetric Assay. In: Kovalchuk I., editor. Plant Epigenetics: Methods and Protocols. 2nd ed. Volume 1456. Humana Press Inc.; Totowa, Japan: 2017. pp. 89–97. [DOI] [PubMed] [Google Scholar]
- 58.Bernstein B.E., Meissner A., Lander E.S. The Mammalian Epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 59.Mira S., Pirredda M., Martín-Sánchez M., Marchessi J., Martín C. DNA methylation and integrity in aged seeds and regenerated plants. Seed Sci. Res. 2020;30:92–100. doi: 10.1017/S0960258520000021. [DOI] [Google Scholar]
- 60.Pirredda M., González-Benito M.E., Martín C., Mira S. Genetic and Epigenetic Stability in Rye Seeds under Different Storage Conditions: Ageing and Oxygen Effect. Plants. 2020;9:393. doi: 10.3390/plants9030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plitta-Michalak B.P., Naskręt-Barciszewska M.Z., Barciszewski J., Chmielarz P., Michalak M. Epigenetic Integrity of Orthodox Seeds Stored under Conventional and Cryogenic Conditions. Forests. 2021;12:288. doi: 10.3390/f12030288. [DOI] [Google Scholar]
- 62.Domej W., Oettl K., Renner W. Oxidative Stress and Free Radicals in COPD–Implications and Relevance for Treatment. Int. J. Chron. Obstruct. Pulmon. Dis. 2014;9:1207–1224. doi: 10.2147/COPD.S51226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dianov G.L., Souza-Pinto N., Nyaga S.G., Thybo T., Stevnsner T., Bohr V.A. Base Excision Repair in Nuclear and Mitochondrial DNA. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- 64.Kiran K.R., Deepika V.B., Swathy P.S., Prasad K., Kabekkodu S.P., Murali T.S., Satyamoorthy K., Muthusamy A. ROS-dependent DNA damage and repair during germination of NaCl primed seeds. J. Photochem. Photobiol. B Biol. 2020;213:112050. doi: 10.1016/j.jphotobiol.2020.112050. [DOI] [PubMed] [Google Scholar]
- 65.Balestrazzi A., Confalonieri M., Macovei A., Carbonera D. Seed imbibition in Medicago truncatula Gaertn.: Expression profiles of DNA repair genes in relation to PEG-mediated stress. J. Plant Physiol. 2011;168:706–713. doi: 10.1016/j.jplph.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Chandra J., Parkhey S., Keshavkant S. Ageing-regulated changes in genetic integrity of two recalcitrant seeded species having contrasting longevity. Trees. 2018;32:109–123. doi: 10.1007/s00468-017-1615-6. [DOI] [Google Scholar]
- 67.Chen H., Osuna D., Colville L., Lorenzo O., Graeber K., Küster H., Leubner-Metzger G., Kranner I. Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell death. PLoS ONE. 2013;8:e78471. doi: 10.1371/journal.pone.0078471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sershen, Varghese B., Naidoo C., Pammenter N.W. The Use of Plant Stress Biomarkers in Assessing the Effects of Desiccation in Zygotic Embryos from Recalcitrant Seeds: Challenges and Considerations. Plant Biol. 2016;18:433–444. doi: 10.1111/plb.12428. [DOI] [PubMed] [Google Scholar]
- 69.Gruber D.R., Toner J.J., Miears H.L., Shernyukov A.V., Kiryutin A.S., Lomzov A.A., Endutkin A.V., Grin I.R., Petrova D.V., Kupryushkin M.S., et al. Oxidative Damage to Epigenetically Methylated Sites Affects DNA Stability, Dynamics and Enzymatic Demethylation. Nucleic Acids Res. 2018;46:10827–10839. doi: 10.1093/nar/gky893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guz J., Foksinski M., Siomek A., Gackowski D., Rozalski R., Dziaman T., Szpila A., Olinski R. The Relationship between 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine Level and Extent of Cytosine Methylation in Leukocytes DNA of Healthy Subjects and in Patients with Colon Adenomas and Carcinomas. Mutat. Res. 2008;640:170–173. doi: 10.1016/j.mrfmmm.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 71.Bhusari S.S., Dobosy J.R., Fu V., Almassi N., Oberley T., Jarrard D.F. Superoxide Dismutase 1 Knockdown Induces Oxidative Stress and DNA Methylation Loss in the Prostate. Epigenetics. 2010;5:402–409. doi: 10.4161/epi.5.5.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martignago D., Bernardini B., Polticelli F., Salvi D., Cona A., Angelini R., Tavladoraki P. The Four FAD-Dependent Histone Demethylases of Arabidopsis Are Differently Involved in the Control of Flowering Time. Front. Plant Sci. 2019;10:669. doi: 10.3389/fpls.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Castander-Olarieta A., Pereira C., Sales E., Meijon M., Arrillaga I., Jesus Canal M., Goicoa T., Dolores Ugarte M., Moncalean P., Montalban I.A. Induction of Radiata Pine Somatic Embryogenesis at High Temperatures Provokes a Long-Term Decrease in DNA Methylation/Hydroxymethylation and Differential Expression of Stress-Related Genes. Plants-Basel. 2020;9:1762. doi: 10.3390/plants9121762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zarakowska E., Czerwinska J., Tupalska A., Yousefzadeh M.J., Gregg S.Q., Croix C.M.S., Niedernhofer L.J., Foksinski M., Gackowski D., Szpila A., et al. Oxidation Products of 5-Methylcytosine Are Decreased in Senescent Cells and Tissues of Progeroid Mice. J. Gerontol. A Biol. Sci. 2018;73:1003–1009. doi: 10.1093/gerona/gly012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogneva Z.V., Dubrovina A.S., Kiselev K.V. Age-Associated Alterations in DNA Methylation and Expression of Methyltransferase and Demethylase Genes in Arabidopsis thaliana. Biol. Plant. 2016;60:628–634. doi: 10.1007/s10535-016-0638-y. [DOI] [Google Scholar]
- 76.Wang W.-S., Pan Y.-J., Zhao X.-Q., Dwivedi D., Zhu L.-H., Ali J., Fu B.-Y., Li Z.-K. Drought-Induced Site-Specific DNA Methylation and Its Association with Drought Tolerance in Rice (Oryza sativa L.) J. Exp. Bot. 2011;62:1951–1960. doi: 10.1093/jxb/erq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jang H., Shin H., Eichman B.F., Huh J.H. Excision of 5-Hydroxymethylcytosine by DEMETER Family DNA Glycosylases. Biochem. Biophys. Res. Commun. 2014;446:1067–1072. doi: 10.1016/j.bbrc.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.