Abstract
Aptamers have been the subject of more than 144 000 papers to date. However, there has been a growing concern that discrepancies in the reporting of aptamer research limit the reliability of these reagents for research and other applications. These observations noting inconsistencies in the use of our RNA antilysozyme aptamer served as an impetus for our systematic review of the reporting of aptamer sequences in the literature. Our detailed examination of the literature citing the RNA antilysozyme aptamer revealed that 93% of the 61 publications reviewed reported unexplained altered sequences with 96% of those using DNA variants. The 10 most cited aptamers were examined using a standardized methodology in order to categorize the extent to which the sequences themselves and altered sequences were adequately described in the literature. Our review of 780 aptamer publications spanned decades, multiple journals, and research groups and revealed that 41% of the papers reported unexplained sequence alterations or omitted sequences. We identified 10 common categories of sequence alterations including deletions, substitutions, and additions, among others. Overall, our findings can be used as a starting point for building better practices in author submissions and publication standards, elevating the rigor and reproducibility of aptamer research.
The irreproducibility of research has become increasingly evident in several fields in recent years1 and poses a serious threat to the efficacy and validity of the research and the general public’s perception of science. The field of aptamer research may be particularly susceptible to this reproducibility crisis.2−5 In part, the ability to develop affinity reagents quickly and simply based on molecular biology manipulations alone provides a ready entry point for a variety of researchers with varying backgrounds. The development of aptamers for use stands in stark contrast to the development of monoclonal antibodies via hybridoma and other technologies, where there is a much larger and longer trail of publications and standards, both academic and commercial, and where there is much larger use, allowing greater cross-validation. The reliability of aptamers has at times been drawn into question.3,6
As an early example of the issues relating to the reproducibility and reliability of aptamer research, we first noted the misinterpretation of an 80-mer RNA antilysozyme aptamer (clone 1).2,7 As we now report in greater detail, subsequent works not only altered the aptamer sequence (using DNA as opposed to the original RNA aptamer) but also truncated the aptamer.4,8
We hypothesized that the sequence alteration of the antilysozyme aptamer may not have been an isolated event. To better elucidate the extent of in silico aptamer sequence alterations, we sampled a large swath of aptamer literature to gauge the fidelity of aptamer sequence reporting. We examined aptamers against the most frequently used aptamer targets: thrombin, adenosine triphosphate (ATP), vascular endothelial growth factor (VEGF), platelet derived growth factor (PDGF) BB, cocaine, theophylline, lysozyme, nucleolin, immunoglobulin E (IgE), and ochratoxin A (OTA; top 10 targets listed by application-based publication frequency).9 We identified original aptamer sequences and followed their subsequent descriptions in cited literature, characterizing apparent sequence alterations as adequately explained, omitted, or unexplained. The unexplained aptamer sequence findings were then organized into phylogenic trees that aided in our identification of common sources and types of apparent mutation.
In our review of 780 publications, we provide evidence of widespread unexplained aptamer sequence alteration over time. Aptamers appear to “mutate” in the literature.
Informatics Methodology
Examination of the Reported Sequences for the RNA Anti-Lysozyme Aptamer, Clone 17
An antilysozyme aptamer selected in 2001 had previously been reported to “mutate” in the literature over time, and we therefore used this aptamer as a starting point for establishing a broader methodology.2,4 As part of this methodology, we identified publications by using Google Scholar and identifying papers that cited the original work using the “cited by” option for this work. From the top literature results, those that were written in English, primary literature (i.e., not reviews, etc.), and used the aptamer in vitro or in vivo were analyzed for reported sequence information. The aptamer sequence(s) was (were) extracted with notes on any alterations to the sequence, using direct quotes from the text.
Analysis of the collected sequences then included the following:
1. Comparison of the reported sequences to the original aptamer sequence (Clone 1)7 and identification of alterations.
2. If alterations were present, further examination of the cited publication led to an assessment of whether the variants were adequately explained or unexplained.
3. In those cases where there were unexplained sequence alterations, they were classified into 10 categories: (i) deletion, (ii) insertion, (iii) substitution, (iv) complete inversion, (v) 5′/3′ addition, (vi) partial inversion, (vii) complementary sequence, (viii) core binding sequence misidentification (i.e., in a figure or table), (ix) sequence omitted, and (x) unknown (i.e., reported an entirely different sequence).
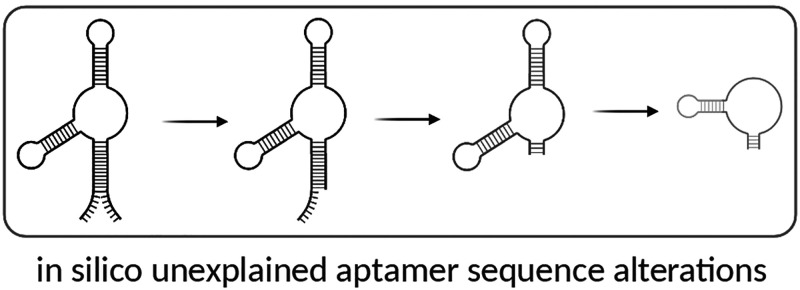
Using phylogenetic trees as a model, we organized the aptamers reported in the literature similarly, using sequence homology to visualize sequence fidelity in our sample. We loosely dubbed this organization an aptamer “phylogenetic” analysis. The nodes in the artificial phylogenetic trees represent groupings of aptamers according to unexplained sequence alterations. Further unexplained derivations were depicted as branching from a previous sequence alteration (i.e., Figure 1, node 4A was derived from Figure 1, node 3A). Publications that correctly reported the original aptamer sequence, including those with fully explained or described additions (e.g., 5′-terminal polyT linker or primary amine for conjugation), were given the same identifying number as the original aptamer.
Figure 1.
Phylogeny depicting unexplained sequence alterations introduced to the clone 1 anti-lysosome RNA aptamer7 collected 01/01/2020. (A) The nodes in the artificial phylogenetic trees represent groupings of aptamers according to unexplained sequence alterations. (B) Reprinted with permission ref (7). Copyright 2001 Elsevier. (C) Reprinted in part with permission from ref (12). Copyright 2004 American Chemical Society. (D) Unexplained sequence alterations were classified into categories.
Examination of Aptamer Sequences Across the Literature
To provide broader insights into the fidelity of aptamer sequence reporting, we reviewed aptamers against the 10 most-used targets9 (Table S1) and again determined a primary (original) aptamer sequence and identified derivatives using the Google Scholar “cited by” option (i.e., works citing the original). From the results generated, publications were further sieved by identifying only those that reported aptamers against the target using the “search within citing articles” function and the search terms “[target]” “aptamer” (Table S1). Without this second search, extraneous papers—for example, those citing the anti-VEGF aptamer in the Introduction—would have been included across multiple reviews or other experimental papers. We then categorized the unexplained sequence alterations according to apparent type and organized the reported sequence derivatives into “phylogenies” according to sequence homology.
The literature search for each aptamer excluded publications that did not use the aptamer sequence experimentally or that were not in the English language; however, no exclusions were made based on the type of journal or date of publication. Publications were searched from some of the original selection experiments in 199010,11 through March 2020.
The sample size for each aptamer examined varied between 9 and 171 publications, although we aimed to acquire at least 50 papers for each phylogeny.
Results and Discussion
A Study of the RNA Anti-Lysozyme Aptamer, Clone 17
Lysozyme is one of the most utilized targets in the field of aptamer research and is among the top 10 in terms of citations.9 This popularity was driven in large measure by the publication of an RNA antilysozyme aptamer by Cox et al.7 We previously noted2 that while the original selection had been for an RNA aptamer, subsequent uses of this aptamer had sometimes involved the synthesis of a DNA version that had not been originally verified as binding to lysozyme; in other words, the aptamer had been mutated in silico. This chemical mutation was perhaps understandable, given that the figure in the original paper showed the DNA version of the RNA aptamers without the primer regions (although the text clearly describes the generation of RNA transcripts and Figures 2 and 4 note the use of RNA). This misreading and misattribution of the original work led to the in silico alterations propagated by subsequent groups.
To examine how in silico alterations can be propagated, we reviewed 61 primary research papers that cited the antilysozyme aptamer, clone 1.7 Of those reports that varied from the original RNA aptamer, the papers were reviewed to ascertain if the reported alterations were adequately described, omitted, or unexplained. Sequence alterations found include deletions, insertions, and/or substitutions.
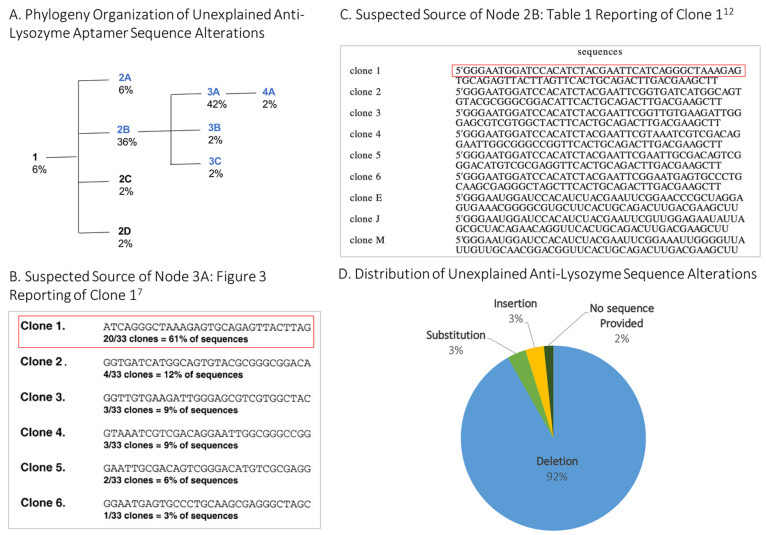
In an effort to illustrate and organize the aptamer sequence data as it “evolved” in the literature, an aptamer phylogeny was created. The phylogeny includes the original/parental 80-mer antilysozyme RNA aptamer (clone 1),7 also called the “root” aptamer, and nodes are organized by sequence homology (i.e., those that more greatly differ from the root aptamer are further away and have a larger node number). As shown in Figure 1A, the phylogeny includes four branch points and nine nodes in all (including the root aptamer) with DNA variants indicated as blue nodes. While we made our best attempts to ascertain what was “reported” versus what was actually “used,” in most cases, we could not distinguish one from the other and thus settled on the liberal use of what was “reported.” Further, in an attempt to ascertain the source of the unexplained sequence alterations, we examined altered sequences and the publications they cited and report our hypotheses here. The sequence alignment of unexplained alterations of the antilysozyme aptamer is shown in Table 1.
Table 1. Unexplained Sequence Alterations Introduced to the Clone 1 Anti-Lysosome RNA Aptamer7a.
| node | # | aptamer sequence reported |
|---|---|---|
| 1 | 4 | GGGAAUGGAUCCACAUCUACGAAUUCAUCAGGGCUAAAGAGUGCAGAGUUACUUAGUUCACUGCAGACUUGACGAAGCUU |
| 2A | 4 | biotin-24polyT-GGGAATGGATCCACATCTACGAATTCATCAGGGCTAAAGAGTGCAGAGTTACTTAGTTCACTGCAGACTTGACGAAGCTT |
| 2B | 20 | GGGAATGGATCCACATCTACGAATTCATCAGGGCTAAAGAGTGCAGAGTTACTTAGTTCACTGCAGACTTGACGAAGCTT |
| 2C | 1 | GGGAAUGGAUCCACAUCUACGAAUUCAUCAGGGCUAAAGAGTGCAGAGUUACUUAGUUCACUGCAGACUUGACGAAGCUU |
| 2D | 1 | GGGAAUGGAUCCACAUCUACGAAUUCAUCAGGGCUAAAGAGUGCAGAGUUACUUAGUUCACUGCAGACUUGACGAAGCUU |
| 3A | 25 | GGGAATGGATCCACATCTACGAATTCATCAGGGCTAAAGAGTGCAGAGTTACTTAGTTCACTGCAGACTTGACGAAGCTT |
| 3B | 1 | HS-(CH2)6-GGGAATGGATCCACATCTACGAATTCATCAGGGCTAAAGAGTGCAGAGTTACTTAGGGGCGCTTCACTGCAGACTTGACGAAGCTT |
| 3C | 1 | GGGAATGGATCCACCAGTGTATCTACGAATTCATCAGGGCTAAAGAGTGAAGAGTTACTTAGTTCACTGCAGACTTGACGAAGCTT |
| 4A | 1 | GGGAATGGATCCACATCTACGAATTCdithiolTTTTTTATCAGGGCTAAAGAGTGCAGAGTTACTTAGTTCACTGCAGACTTGACGAAGCTT |
| 0 | 2 | no sequence in text or supplement |
Unexplained insertions are bold. Unexplained substitutions are bold and underlined. Unexplained deletions are struck out, and justified or explained alterations are in light grey. The number (#) column indicates the number of publications found reporting each sequence in our analysis.
After the root antilysozyme RNA aptamer node (1) in Figure 1, the first branch point leads to four nodes. The first node, 2A, is an unexplained truncated DNA sequence with a 39 nt 3′ deletion, as well as terminal additions (5′ biotinylation and a 24 nt oligo(T) linker). This aptamer was primarily used by a single group.13−16 We hypothesize that the alteration was due to a misinterpretation of Kirby et al. (Table 1),12 shown in Figure 1C, which reported the DNA version of the six original antilysozyme aptamers7 and three novel RNA aptamers with a line break in the sequences. Node 2A includes the first line of this sequence.
The 2B node (Figure 1) is an unexplained DNA sequence with a 24 nt deletion from the 3′ end and a 14 nt deletion from the 5′ end and was found in 36% of papers reviewed. The 2C node is the RNA aptamer sequence, but with a single U to T substitution reported in the paper, an in silico error but likely not in the molecule used, as this is not described in the Methods section.17 Finally, the 2D node is a full-length RNA sequence but with a deletion of four internal nucleotides,18 likely again an error in the description but not in the molecule itself, as the manuscript notes the acquisition of the original aptamer material from the lab where the aptamer originated.
All further unexplained sequence alterations branch from node 2B, the citation of an all-DNA variant (Figure 1A). Organized by sequence homology, the 5′ and 3′ cropped variant at the 2B node leads to a branch point with three nodes: 3A, 3B, and 3C. Node 3A, which makes up 42% of the altered sequences in this phylogeny, is a DNA variant with primer-binding regions deleted (i.e., 5′ 26 nt deletion and 3′ 24 nt deletion). This sequence is equivalent to what was reported in Figure 3 of the 2001 paper,7 although this sequence was not in fact used for experiments. We suspect that the node 3A sequence alteration arose due to a misunderstanding of Figure 3 in the original paper, which reported a DNA version of the 30 nt random sequence region of the selected RNA aptamer, without also including accompanying primer-binding regions.7 Presumably, groups that used the DNA version of the random region obtained it by using this figure.
Variant 3B differs from 3A in that it has an unexplained 3′-terminal addition of GGGCGC, the partial (13 nt) return of the 5′ primer region, and an adequately explained 5′-thiol modification (5′-HS-(CH2)6) for covalent conjugation to gold nanoparticles.19 Variant 3C contains a number of otherwise unexplained alterations: a C to A internal substitution, a 6 nt 5′-terminal addition, a 3 nt 3′-terminal deletion, and a 5′-terminal 3 nt deletion.20
Finally, the 4A sequence (Figure 1) again builds off of the deletion of 5′ and 3′ primer regions described in node 3A and adds a well-described 6 nt polyT linker and thiol modification at the 5′ end for conjugation to silica particles but also includes an unexplained additional 13 nt 3′ deletion.21
In all of the reviewed publications, there were only four papers of the 61 reviewed that reported the correct antilysozyme aptamer, clone 1, sequence and/or adequately explained sequence alterations. Of those that contained unexplained sequence alterations, 96% reported a DNA variant, 92% contained a deletion, 3% contained a substitution, 3% contained an insertion, and 2% failed to include sequence information in the manuscript or supplement.
The question thus becomes not only why so many researchers misinterpreted the primary source of information but also how the derivatives themselves work as reported. The answer is two-fold: first, hen egg-white lysozyme was originally chosen as a target for selection in part because it is extremely basic (pI = 11.35) and thus was very likely to yield high-affinity aptamers. But second, precisely because polyanions will bind oligocations, the DNA aptamer may still bind lysozyme. In an illuminating study, Potty et al.4 examined the binding properties of both the RNA aptamer and DNA of antilysozyme aptamer clone 3 (an entirely different aptamer sequence than the clone 1 sequence that is generally used). These authors found the DNA variant bound nonspecifically to lysozyme through electrostatic interactions.4
This further raises the question of what constitutes specific binding. In general, aptamer studies should contain noncognate binding experiments, in which variants of the aptamers (mutant or scrambled) are shown not to bind to the selection target, and the selected aptamer is shown not to bind to targets other than the selection target. Such studies are rarely carried out.9
Description of Aptamer Sequence Reporting Across the Literature
Given these findings, we broadly sampled the aptamer literature to more fully describe sequence reporting fidelity in the field. On the basis of the extensive set of unexplained aptamer sequence alterations that were found in the literature citing the antilysozyme aptamer,7 we hypothesized that the alteration of original aptamer sequences and/or unexplained sequence alterations could be a pervasive problem. While a full review of the 144 000+ “aptamer” papers on Google Scholar was not feasible, a review of aptamers against the 10 most used targets9 was carried out. This Review systematically sampled a broad swath of the aptamer literature, spanning years, researchers, laboratories, and locations.
Similar to the analysis of the Cox antilysozyme aptamer literature, we first identified an originating aptamer and then examined the literature citing the sequence of this originating aptamer. In cases where multiple clones selected in a paper were used in the literature, all clones were examined as “root” aptamers (23 in all). Unexplained sequence alterations were again categorized, and phylogenies constructed. In all, 780 publications from 23 originating/root aptamers were reviewed using this standardized sampling methodology (Table S2, Figures S1–10). The 23 phylogenies were also grouped based on an originating publication (e.g., three different anti-PDGF aptamer clones from the originating publication are grouped in Figure S4).
Overall, only 59% of the 780 publications reviewed correctly reported the aptamer sequence(s) and/or explained sequence alterations, while 41% contained one or more in silico sequences that were categorized as unexplained.
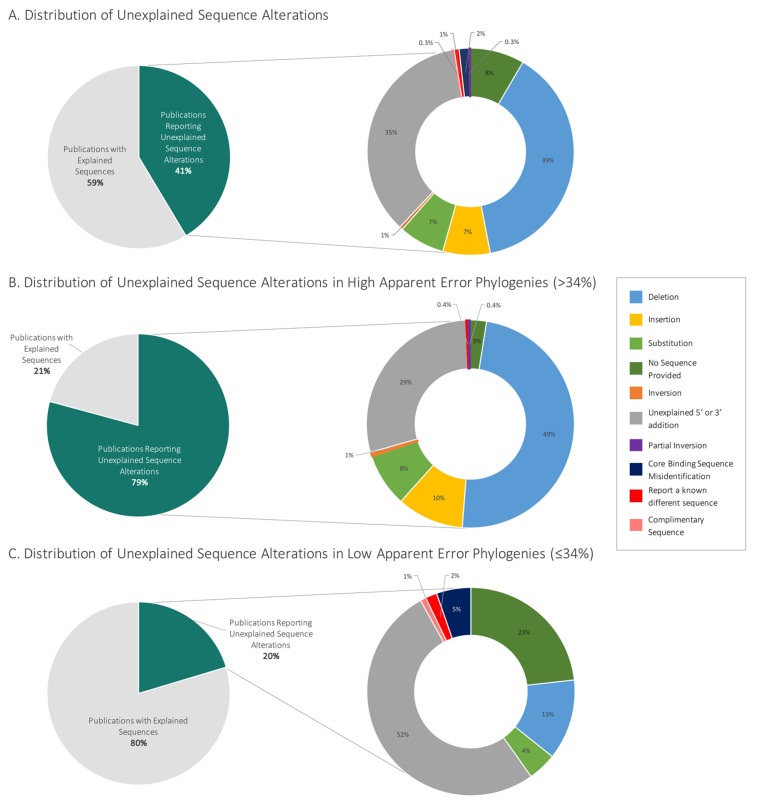
We identified 120 novel sequences that according to our criteria were not adequately explained (Table S3). Some (39%) of these 120 sequences identified contained deletions; 35% contained 5′ or 3′ unexplained additions; 8% did not provide the sequence at all; 7% contained insertions; 7% contained substitutions; and less than 2% contained core binding sequence misidentifications (i.e., boxing the incorrect aptamer sequence, etc.), inversions, the complementary sequence, or an entirely different sequence (Figure 2A).
Figure 2.
Distribution of field-wide unexplained aptamer sequence alterations. (A) The breakdown of all publications examined (n = 780 publications, 23 phylogenies grouped by 11 root aptamer publications). (B) The distributions of phylogenies that contained greater than the median of 34% unexplained internal sequence alteration (five phylogenies: Cox lysozyme, lysozyme (DNA aptamer), ATP, PDGF BB, cocaine). (C) The distribution of phylogenies that contained less than or equal to the median of 34% internal mutations (six phylogenies: nucleolin, IgE, theophylline, VEGF, ochratoxin A, thrombin). The percentage of unexplained sequence alterations within each phylogeny can be found in Table S2.
To further categorize the types of apparent in silico mutations that are perpetuated in the literature, we used a “median split” to divide the phylogenies into two groups: “low”/few apparent mutations and “high”/more apparent mutations.22,23 The median percentage of unexplained sequence alterations within each phylogeny was 34%, and this value was used for the division into groups (Figure 2B and C, Tables S2, S4). There was a larger contribution of deletions (49%), insertions (10%), and substitutions (8%) in the high level of unexplained sequence alteration group relative to the low level of the unexplained sequence alteration group, which had 13% deletions, 4% substitutions, and no insertions (Figure 2B and C). In contrast, the low-level unexplained sequence alteration group contained more unexplained 5′/3′ additions (52%) compared to the high sequence alteration group (29%). This observation generally suggests that deletions and insertions are further propagated in subsequent literature, while 5′/3′ additions that may have a unique, researcher-specific purpose do not. It has previously been noted that many aptamers appear to not utilize flanking sequences during their folding,24 and many 5′/3′ extensions may therefore not have an impact on function.
Since this Perspective describes in silico mutations of aptamers, it is primarily meant as a cautionary tale for authors, rather than as an extensive biophysical or computational characterization of the impact of those in silico mutations. The lack of a comprehensive, updated aptamer database is clearly an impediment to even something as straightforward and potentially useful as analyzing aptamer Kd values. Gathering such data across the literature will require enormous time and analytical commitments, given the extremely diverse way in which aptamer data are already reported (for example, nominal Kd values are typically gathered by a variety of methods and calculated in a variety of ways). However, creating a unified database is something the field as a whole or companies engaged in aptamer research might wish to promote. Overall, sequence data provide the best and currently only reliable means to make comparisons across 30 years of literature.
Conclusions
Our review of 780 publications reveals widespread sequence alterations that are poorly explained or where the aptamer sequence(s) were omitted (41%). This observed systemic mutation of aptamers via the literature buttresses the conclusions of Yan and Levy3 that aptamers can potentially prove unreliable as reagents due to author error. Other reports have raised issues in aptamer research that may similarly impact the propagation of literature mutations relative to originating sequences—the relative ease of creating aptamers based on literature reports alone, the rapid expansion of aptamer research, the rapid increase in application-based publications, and a lack of adherence to publication guidelines.
Echoing previous work,2,3,9 we assert that there is a need both for the collaborative construction of evidence-based publication guidelines and for standardized documentation of aptamer sequence use in the literature (e.g., similar to CiteAb.com). Standardized sequence reporting could be modeled after Nature’s “Life Science Reporting Summary” and Cell’s “STAR Methods,” which create standardized templates for detailed descriptions of experimental design, statistical tests, and materials and reagents. At a minimum, we suggest that three categories of information should be required in aptamer publications: (1) complete sequence, including consistent sequence identifiers; (2) detailed descriptions of binding and experimental conditions; and (3) extensive use of negative and positive controls, for both aptamers and targets. In addition, while aptamer secondary structures are frequently provided based on generally available folding algorithms such as mFOLD,25 authors should generally state that any secondary structures that have been identified computationally are not necessarily accurate in the absence of further experimental characterizations. As Yan and Levy3 suggest, we also support validation practices such as using conditions similar to those found in downstream applications, limiting incubation times and using blocking reagents to minimize nonspecific binding to targets, and performing multiple validations, ideally by different researchers.
In the future, collaborative evidence-based aptamer validation guidelines should be encouraged by journals, including a checklist for the peer review process. Ultimately, we believe that the standardization of aptamer publication guidelines and increased availability of raw data will lead to a more open, nuanced discussion of the data presented and greater success in the translation of aptamer research to the clinic and industry.
Acknowledgments
This research was funded by The University of Texas Freshman Research Initiative, which was supported by Howard Hughes Medical Institute (#52008124) and the College of Natural Sciences. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of The University of Texas at Austin or the Howard Hughes Medical Institute. Apart from Gwendolyn Stovall and Andrew Ellington, the authors began this work as undergraduate researchers in the UT Freshman Research Initiative and would like to thank the FRI’s Aptamer Stream and the 2018 TIDES Science Sprints at the University of Texas at Austin for supporting this project. The authors would also like to thank Jolyn Frnka, Haley Wolf, Shreya Thiagarajan, Eric Lumanog, Christine Mai, Hung Le, Kathy Tran, and Nidhi Pawate for their assistance in the cursory data collection and development of the method.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c04407.
(Table S1) Top 10 targets used and the search terms used during the Google Scholar literature search; (Table S2) percentage of publications with unexplained sequence alterations within each phylogeny; (Table S3) number of unique sequences and number of sequences with unexplained alterations; (Figures S1–S10) extent of publication-induced mutation in the top 10 aptamer targets using the method described here; (Tables S4 and S5) the composite data (PDF)
Author Contributions
A.A.M., S.R.N., A.S.R., C.K., T.R., and T.S. performed initial data collection. A.A.M., S.R.N, and A.S.R. collected all data and analyzed the data. A.A.M., G.M.S., and A.D.E. reviewed and interpreted the results. The methods were drafted by S.R.N. The final manuscript was written and edited by A.A.M., G.M.S., and A.D.E. All authors have approved the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Miyakawa T. No raw data, no science: Another possible source of the reproducibility crisis. Molecular Brain 2020, 13 (1), 1–6. 10.1186/s13041-020-0552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E. J.; Lee J.-W.; Ellington A. D. Applications of Aptamers as Sensors. Annual Review of Analytical Chemistry 2009, 2 (1), 241–264. 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- Yan A. C.; Levy M. Aptamer-Mediated Delivery and Cell-Targeting Aptamers: Room for Improvement. Nucleic Acid Therapeutics 2018, 28 (3), 194–199. 10.1089/nat.2018.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potty A. S. R.; Kourentzi K.; Fang H.; Schuck P.; Willson R. C. Biophysical characterization of DNA and RNA aptamer interactions with hen egg lysozyme. Int. J. Biol. Macromol. 2011, 48 (3), 392–397. 10.1016/j.ijbiomac.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Haßel S. K.; Mayer G. Aptamers as Therapeutic Agents: Has the Initial Euphoria Subsided?. Molecular Diagnosis and Therapy 2019, 23 (3), 301–309. 10.1007/s40291-019-00400-6. [DOI] [PubMed] [Google Scholar]
- Li N.; Ebright J. N.; Stovall G. M.; Chen X.; Nguyen H. H.; Singh A.; Syrett A.; Ellington A. D. Technical and biological issues relevant to cell typing with aptamers. J. Proteome Res. 2009, 8 (5), 2438–2448. 10.1021/pr801048z. [DOI] [PubMed] [Google Scholar]
- Cox J. C.; Ellington A. D. Automated Selection of Anti-Protein Aptamers. Bioorg. Med. Chem. 2001, 9, 2525–2531. 10.1016/S0968-0896(01)00028-1. [DOI] [PubMed] [Google Scholar]
- Padlan C. S.; Malashkevich V. N.; Almo S. C.; Levy M.; Brenowitz M.; Girvin M. E. An RNA aptamer possessing a novel monovalent cation-mediated fold inhibits lysozyme catalysis by inhibiting the binding of long natural substrates. Rna 2014, 20 (4), 447–461. 10.1261/rna.043034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn M. R.; Jimenez R. M.; Chaput J. C. Analysis of aptamer discovery and technology. Nature Reviews Chemistry 2017, 1 (10), 1–16. 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- Ellington A. D.; Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346 (6287), 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249 (4968), 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Kirby R.; Cho E. J.; Gehrke B.; Bayer T.; Park Y. S.; Neikirk D. P.; McDevitt J. T.; Ellington A. D. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal. Chem. 2004, 76 (14), 4066–4075. 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- Mihai I.; Vezeanu A.; Polonschii C.; David S.; Gaspar S.; Bucur B.; Blaszykowski C.; Sheikh S.; Thompson M.; Vasilescu A. Low-fouling SPR detection of lysozyme and its aggregates. Analytical Methods 2014, 6 (19), 7646–7654. 10.1039/C4AY01237B. [DOI] [Google Scholar]
- Mihai I.; Vezeanu A.; Polonschii C.; Albu C.; Radu G. L.; Vasilescu A. Label-free detection of lysozyme in wines using an aptamer based biosensor and SPR detection. Sensors and Actuators, B: Chemical 2015, 206, 198–204. 10.1016/j.snb.2014.09.050. [DOI] [Google Scholar]
- Vasilescu A.; Gaspar S.; Mihai I.; Tache A.; Litescu S. C. Development of a label-free aptasensor for monitoring the self-association of lysozyme. Analyst 2013, 138 (12), 3530–3537. 10.1039/c3an00229b. [DOI] [PubMed] [Google Scholar]
- Vasilescu A.; Purcarea C.; Popa E.; Zamfir M.; Mihai I.; Litescu S.; David S.; Gaspar S.; Gheorghiu M.; Jean-Louis Marty Versatile SPR aptasensor for detection of lysozyme dimer in oligomeric and aggregated mixtures. Biosens. Bioelectron. 2016, 83, 353–360. 10.1016/j.bios.2016.04.080. [DOI] [PubMed] [Google Scholar]
- Cho E. J.; Collett J. R.; Szafranska A. E.; Ellington A. D. Optimization of aptamer microarray technology for multiple protein targets. Anal. Chim. Acta 2006, 564 (1), 82–90. 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- Hybarger G.; Bynum J.; Williams R. F.; Valdes J. J.; Chambers J. P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2006, 384 (1), 191–198. 10.1007/s00216-005-0089-3. [DOI] [PubMed] [Google Scholar]
- Shamsipur M.; Farzin L.; Tabrizi M. A. Ultrasensitive aptamer-based on-off assay for lysozyme using a glassy carbon electrode modified with gold nanoparticles and electrochemically reduced graphene oxide. Microchimica Acta 2016, 183 (10), 2733–2743. 10.1007/s00604-016-1920-6. [DOI] [Google Scholar]
- Zuo L.; Qin G.; Lan Y.; Wei Y.; Dong C. A turn-on phosphorescence aptasensor for ultrasensitive detection of lysozyme in humoral samples. Sensors and Actuators, B: Chemical 2019, 289, 100–105. 10.1016/j.snb.2019.03.088. [DOI] [Google Scholar]
- Bayramoglu G.; Ozalp V. C.; Yilmaz M.; Guler U.; Salih B.; Arica M. Y. Lysozyme specific aptamer immobilized MCM-41 silicate for single-step purification and quartz crystal microbalance (QCM)-based determination of lysozyme from chicken egg white. Microporous Mesoporous Mater. 2015, 207, 95–104. 10.1016/j.micromeso.2015.01.009. [DOI] [Google Scholar]
- Iacobucci D.; Posavac S. S.; Kardes F. R.; Schneider M. J.; Popovich D. L. The median split: Robust, refined, and revived. Journal of Consumer Psychology 2015, 25 (4), 690–704. 10.1016/j.jcps.2015.06.014. [DOI] [Google Scholar]
- DeCoster J.; Gallucci M.; Iselin A.-M. R. Best Practices for Using Median Splits, Artificial Categorization, and their Continuous Alternatives. Journal of Experimental Psychopathology 2011, 2 (2), 197–209. 10.5127/jep.008310. [DOI] [Google Scholar]
- Cowperthwaite M. C.; Ellington A. D. Bioinformatic analysis of the contribution of primer sequences to aptamer structures. Journal of Molecular Evolution 2008, 67 (1), 95–102. 10.1007/s00239-008-9130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31 (11), 3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.