Abstract
Vibrio cholerae is indigenous to the aquatic environment, and serotype non-O1 strains are readily isolated from coastal waters. However, in comparison with intensive studies of the O1 group, relatively little effort has been made to analyze the population structure and molecular evolution of non-O1 V. cholerae. In this study, high-resolution genomic DNA fingerprinting, amplified fragment length polymorphism (AFLP), was used to characterize the temporal and spatial genetic diversity of 67 V. cholerae strains isolated from Chesapeake Bay during April through July 1998, at four different sampling sites. Isolation of V. cholerae during the winter months (January through March) was unsuccessful, as observed in earlier studies (J. H. L. Kaper, R. R. Colwell, and S. W. Joseph, Appl. Environ. Microbiol. 37:91–103, 1979). AFLP fingerprints subjected to similarity analysis yielded a grouping of isolates into three large clusters, reflecting time of the year when the strains were isolated. April and May isolates were closely related, while July isolates were genetically diverse and did not cluster with the isolates obtained earlier in the year. The results suggest that the population structure of V. cholerae undergoes a shift in genotype that is linked to changes in environmental conditions. From January to July, the water temperature increased from 3°C to 27.5°C, bacterial direct counts increased nearly an order of magnitude, and the chlorophyll a concentration tripled (or even quadrupled at some sites). No correlation was observed between genetic similarity among isolates and geographical source of isolation, since isolates found at a single sampling site were genetically diverse and genetically identical isolates were found at several of the sampling sites. Thus, V. cholerae populations may be transported by surface currents throughout the entire Bay, or, more likely, similar environmental conditions may be selected for a specific genotype. The dynamic nature of the population structure of this bacterial species in Chesapeake Bay provides new insight into the ecology and molecular evolution of V. cholerae in the natural environment.
Vibrio cholerae is indigenous to the aquatic environment (42). However, contaminated water supplies in some parts of the world have caused selected, pathogenic clones of the species to become dominant in epidemics. Thus, cholera continues to be an important cause of morbidity and mortality in many areas of Asia, Africa, and Latin America. Historically, the genetic diversity of Vibrio cholerae was characterized by serotyping, with the result that ca. 200 serogroups can be distinguished on the basis of epitopic variation in the cell surface lipopolysaccharide (LPS) (43). Until recently, all recorded pandemic and epidemic cases of cholera were associated with strains carrying the type O1 antigen. The remaining non-O1 strains were considered of little epidemic importance. However, an epidemic which began in India late in 1992 and spread to several neighboring countries was caused by an O139 strain (1, 34, 37). The V. cholerae O139 strain proved to be genetically similar to V. cholerae O1 and is hypothesized as having evolved from strains of the early seventh pandemic (22, 31) by a mechanism involving insertion of exogenous DNA encoding the O139 LPS (6, 7).
The recent discovery of a lysogenic filamentous phage, CTXΦ, that encodes the toxin genes shines new light upon the evolution of pathogenicity in V. cholerae (41). The receptor for CTXΦ, the toxin-coregulated pilus, is encoded by an operon that is part of the transmissible pathogenicity island (PAI). Karaolis et al. (29) reported finding the cholera PAI in two clinical non-O1, non-O139 cholera toxin (CT)-positive strains and suggested that PAI can be transferred among V. cholerae strains.
Intrigued by the emergence of the O139 strain, investigators have focused on the genetic diversity and population structure of V. cholerae non-O1, O139 strains (4, 19). Yamai et al. (43) examined 1,898 strains of V. cholerae non-O1, O139 collected worldwide and found approximately 2% of the strains produced CT. Dalsgaard and colleagues (19) found CT-producing strains were prevalent in serogroup O141, with 10 of 16 strains testing positive for CT, including 7 strains recovered from stool and water samples in the United States. A clone of serogroup O37 that demonstrated epidemic potential in the 1960s was shown to be genetically closely related to the pandemic O1 and O139 clones, suggesting the new cholera clones arose by modification of a lineage that is already epidemic or is closely related to such clones (4).
In recent years, outbreaks of cholera-like disease caused by non-O1, non-O139 strains have been reported on several occasions (5, 20, 39). An unusual upsurge in the incidence of cholera-like disease in Calcutta, India, between February and May 1996 was attributed to the non-O1, non-O139 serogroups (39). Results of PCR assays indicated that none of the non-O1, non-O139 strains were positive for the toxin-encoding genes, suggesting that these serogroups of V. cholerae cause diarrhea by a mechanism quite different from that of toxigenic V. cholerae O1 and O139 (39).
V. cholerae non-O1, O139 strains are readily isolated from the coastal environment (17, 42). The geographic distribution of V. cholerae in the Chesapeake Bay has been correlated with salinity (28). V. cholerae strains were isolated at stations ranging in salinity from 4 to 14 ppt, but were not detected when salinity dropped below 4 ppt or above 14 ppt. In spite of repetitive efforts, O1 is not readily isolated from environmental samples, even in cholera-endemic areas (16). A study of the cell surface characteristics of V. cholerae O1 and non-O1 revealed that environmental non-O1 strains possess exposed phospholipids in their outer membrane and are resistant to lytic agents (10). V. cholerae O1 is susceptible to lysis; therefore, entering a viable but nonculturable (VBNC) state serves well as a survival strategy (16). Furthermore, environmental non-O1 strains appear to be capable of converting to the O1 serogroup under the influence of selected environmental conditions (16). Although the majority of environmental V. cholerae isolates do not contain toxin genes (42), a study of the molecular diversity of naturally occurring V. cholerae strains should offer insight into the ecology, evolution, and epidemiology of V. cholerae as a species. To date, relatively little effort has been made to study the population structure and molecular evolution of this bacterial species in its native habitat, the aquatic environment.
Amplified fragment length polymorphism (AFLP) is a high-resolution DNA fingerprinting technique that has been effectively used to distinguish closely related bacterial strains (2, 25, 26, 32). In a previous study (27), we evaluated this technique for use in examining the genetic diversity of V. cholerae and found it to be highly sensitive and reproducible. In the study reported here, the technique was used to analyze the temporal and spatial diversity of V. cholerae in the Chesapeake Bay.
MATERIALS AND METHODS
Isolation of V. cholerae from Chesapeake Bay.
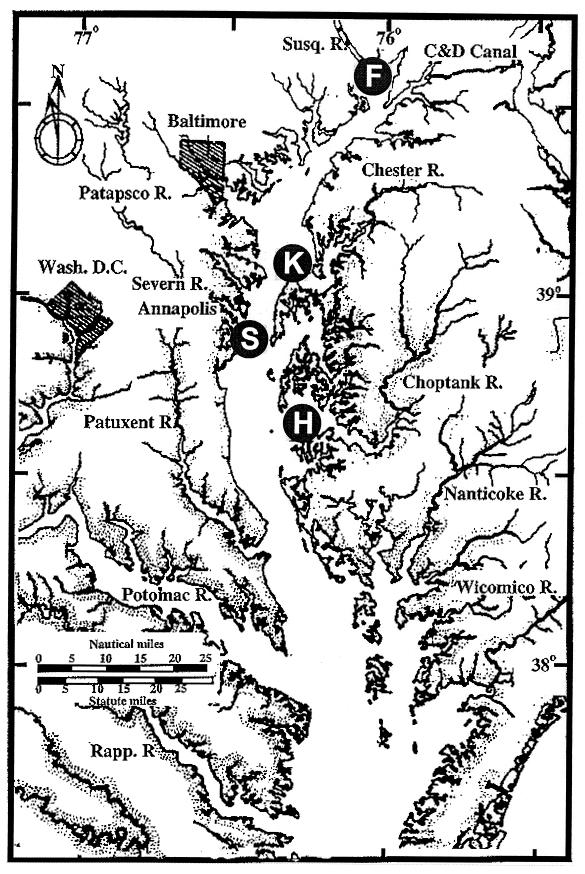
Water and plankton samples (phytoplankton and zooplankton) were collected at four stations in the mid- to upper Chesapeake Bay over a 7-month period from January to July 1998 (Fig. 1). Station F (39°34′N, 76°01′W) was located on the Susquehanna River flats near the mouth of the Susquehanna River. Station S (38°53′N, 76°32′W) was located at the Smithsonian Environmental Research Center (SERC) south of Annapolis on the Western Shore of the Bay. Station K (38°52′N, 76°20′W) was at Kent Island in the middle of the Bay. Station H (38°35′N, 76°08′W) was off the dock at the University of Maryland Horn Point Laboratory, Cambridge, Md., on the Eastern Shore. Samples were taken at the second or third week of each month, except during June, when two samples were taken (the first one during the second week, and the second during the fourth week). Ca. 200- to 300-ml water samples were collected aseptically, concentrated on 0.2-μm-pore-diameter filters, and enriched in alkaline-peptone-water (APW [1% peptone, 1% NaCl; pH 8.4 to 8.6]) for isolation of V. cholerae (28). Plankton samples were collected by pumping 100 to 600 liters of water through 64- and 20-μm-mesh-size plankton nets. The concentrated plankton samples were also enriched for V. cholerae. Enrichments were incubated at 25 to 30°C for 9 to 12 h. Bacterial colonies were isolated from the enrichment cultures by using thiosulfate-citrate-bile-salts (TCBS) agar. The colonies on TCBS were confirmed to be V. cholerae by both biochemical testing and PCR amplification with V. cholerae rRNA-intergenic spacer-specific primers, as described previously (14). Isolates were also examined for the presence of the cholera toxin gene (ctx) by PCR amplification with ctxA-specific primers, as described previously (33).
FIG. 1.
Chesapeake Bay sampling stations included in this study. F, Susquehanna River flats; S, SERC; K, Kent Island; H, Horn Point laboratory.
Determination of environmental parameters.
Both water temperature and salinity were measured on site. For total bacterial direct counts, 20 to 50 ml of water was fixed with a final concentration of 2% formalin and stained with DAPI (4′,6′ diamindino-2-phenylindole), as previously described (35). Ca. 1- to 3-ml fixed samples were filtered onto a 0.2-μm-pore-diameter black polycarbonate filter (Millipore, Inc.). Bacterial direct counts were performed with an Olympus epifluorescence microscope (Olympus, Inc.). Samples for chlorophyll a determination were collected onto Whatman GF/F filters and stored at −80°C until analyzed. Chlorophyll a was extracted from the filter with methanol, and concentrations were determined fluorometrically (23).
AFLP analysis of V. cholerae genomic DNA.
Genomic DNA from each isolate (n = 67) was extracted from an overnight culture by using the CTAB (cetylethylammonium bromide) protocol, as previously described (3). The purity and quality of the DNA were determined by UV absorption with a UV spectrophotometer (Beckman Instruments, Inc., Fullerton, Calif.). One microgram of genomic DNA was used for AFLP analysis. The procedures used for template DNA preparation, PCR amplification, and gel electrophoresis are described elsewhere (28). Restriction enzymes HindIII and TaqI were used in combination to generate template DNAs for AFLP analysis. The primer and adapter sequences used for AFLP are shown in Table 1.
TABLE 1.
Primer and adapter sequences used for AFLP analysis of V. cholerae from the Chesapeake Bay
| Adaptor or primer | Oligonucleotide sequence |
|---|---|
| HindIII adaptor | 5′-CTCGTAGACTGCGTACC-3′ |
| 3′-CTGACGCATGGTCGA-5′ | |
| H01 primer | 5′-GACTGCGTACCAGCTTA-3′ |
| TaqI adaptor | 5′-GACGATGAGTCCTGAC-3′ |
| 3′-TACTCAGGACTGGC-5′ | |
| T02 primer | 5′-CGATGAGTCCTGACCGAAC-3′ |
Data analysis.
Digitized AFLP fingerprints were analyzed by using the Molecular Analyst/Fingerprinting software (Bio-Rad Laboratories, Richmond, Calif.) following the manufacturer's instructions. Images were straightened, unwarped, and normalized by alignment to a reference strain included in each gel and/or to an RTS-Ready Label molecular weight marker (Bio-Rad Laboratories). Bands were selected by the computer program, with visual assistance for correction or addition of bands. Dendrograms were created by computing similarity values according to the position of the bands.
RESULTS
Environmental parameters.
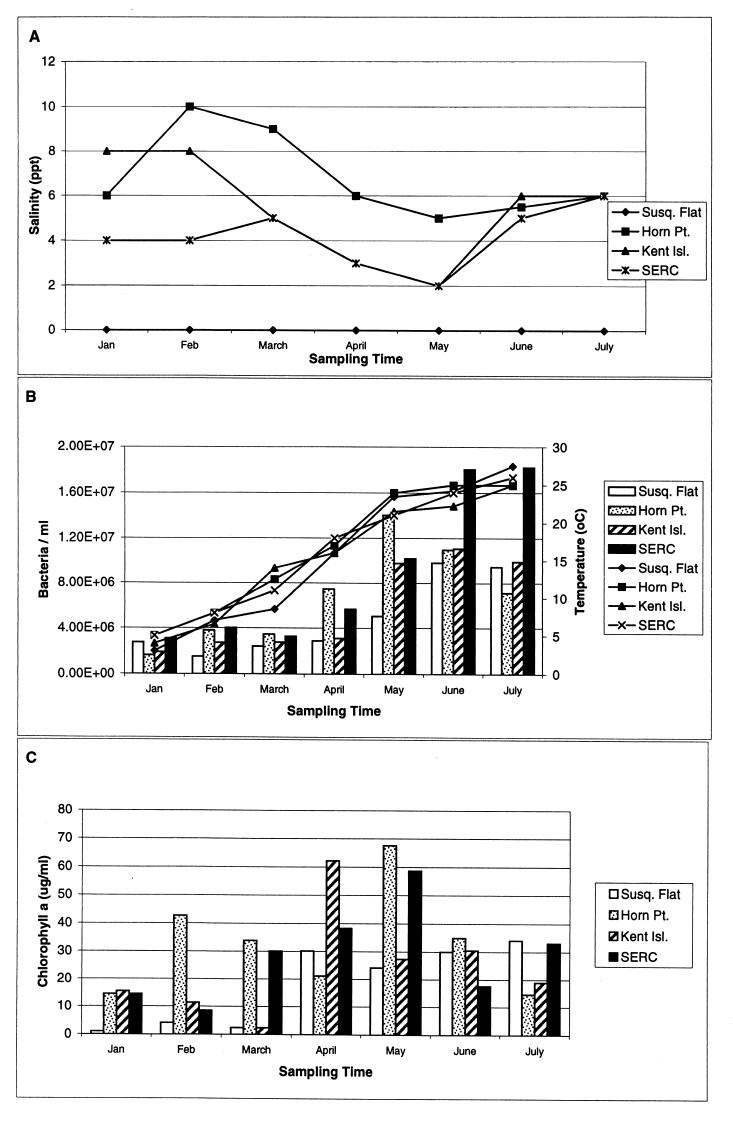
Seasonal fluctuation of salinity in the upper and mid-Chesapeake Bay is shown in Fig. 2A. The salinity in the Susquehanna River flats was essentially freshwater throughout the sampling period. At Kent Island and SERC, salinity fluctuated between 2 and 8 ppt, while the highest salinity recorded in this study, ranging from 5 to 10 ppt, was at Horn Point.
FIG. 2.
Environmental parameters measured at the Susquehanna River flats, Horn Point, Kent Island, and SERC stations between January and July 1998. (A) Salinity. (B) Bacterial abundance and temperature. Bacterial abundance (bar graph) was determined by epifluorescence microscopy and is presented as the number of bacteria per milliliter of seawater. Temperature was determined in situ. (C) Chlorophyll a (measured fluorometrically in micrograms per milliliter of seawater).
Water temperature in the Chesapeake Bay changed dramatically over the study period, increasing from a low of 3°C in January to a maximum of 27.5°C in July (Fig. 2B). A dramatic increase in the bacterial population was detected in April at station H (Horn Point), and between March and June, the bacterial abundance at station S (SERC) increased nearly an order of magnitude. Bacterial abundance was found to be strongly correlated with water temperature (R2 = 0.7) at all four stations in the Chesapeake Bay. A bloom of phytoplankton, indicated by a peak in chlorophyll a concentration, was detected at station H as early as February 1998 (Fig. 2C). Fluctuations in the phytoplankton standing stock were observed at all sampling sites.
Isolation and confirmation of V. cholerae from Chesapeake Bay.
V. cholerae was first isolated from both water samples and plankton fractions at three of the four sites in April 1998 (Table 2). Between May and July, all three brackish water sampling sites (Horn Point, Kent Island, and SERC) tested positive for V. cholerae. V. cholerae was detected at the Susquehanna River flats in the sample collected in late June 1998. Since isolation was achieved by enrichment culture, quantitative measurement of the true abundance of V. cholerae in bay water or associated with plankton is not currently available, because viable but nonculturable V. cholerae isolates were not measured. However, the percentage of isolates confirmed positive for V. cholerae among the total isolates on TCBS increased significantly from April to June at all sampling sites, except at the Susquehanna River flats (Fig. 3). Thus, the abundance of V. cholerae increases during the summer months (June and July) in the Chesapeake Bay, as shown in earlier studies (28). The frequency of isolation was correlated with both water temperature (R2 = 0.8) and total bacterial abundance (R2 = 0.5).
TABLE 2.
Numbers of V. cholerae strains isolated by enrichment from Chesapeake Bay between January and July 1998
| Mo | No. of strains
|
|||
|---|---|---|---|---|
| Susquehanna River flats | Horn Point | Kent Island | SERC | |
| January | 0 | 0 | 0 | 0 |
| February | 0 | 0 | 0 | 0 |
| March | 0 | 0 | 0 | 0 |
| April | 7 | 8 | 0 | 1 |
| May | 0 | 10 | 1 | 3 |
| June | 2 | 32 | 33 | 32 |
| July | 0 | 18 | 22 | 20 |
FIG. 3.
Frequency of V. cholerae isolation among total bacterial isolates on TCBS at the Susquehanna River flats, Horn Point, Kent Island, and SERC stations in Chesapeake Bay between January and July 1998. V. cholerae was identified by biochemical testing and confirmed by PCR amplification with V. cholerae-specific primers.
Isolates from TCBS agar were confirmed as V. cholerae by a combination of biochemical testing and PCR amplification, with V. cholerae-specific primers. An interesting observation was that many PCR-positive isolates showed slight variations in the standard biochemical test results. The frequent variations were observed for methyl red, the Voges-Proskauer test, and growth in nutrient broth with 0% NaCl. Occasional variations were observed in the tests for lysine decarboxylase, ornithine decarboxylase, and acid production from d-mannitol. A total of 189 isolates tested PCR positive with V. cholerae-specific primers (gel picture not shown). However, none of the isolates tested PCR positive for the presence of the ctxA gene (data not shown).
AFLP analysis of V. cholerae isolates.
Sixty-seven V. cholerae isolates, recovered from Chesapeake Bay in April through July, were fingerprinted by the AFLP technique. Isolates showing differences in biochemical tests were selected from the June and July samples for analysis to avoid repetitive testing of the same clone. All isolates collected from April through May were subjected to AFLP fingerprinting, since only a few isolates were obtained during this period. AFLP analyses with the adapters and primers listed in Table 1 yielded 40 to 60 bands for each isolate (Fig. 4). A similarity analysis of the fingerprints yielded a grouping of isolates into three large clusters separated by date of isolation (Fig. 5). These results indicated that April and May isolates were genetically similar, and July isolates were distantly related to isolates obtained early in the year.
FIG. 4.
AFLP fingerprints of V. cholerae isolated from Chesapeake Bay by using adapters and primer sequences described in Table 1. Lanes: M, reference marker; 1, H19; 2, H8; 3, H12; 4, H23; 5, H30; 6, H21; 7, H11; 8, H27; 9, S20, 10, H10.
FIG. 5.
Similarity analysis of AFLP fingerprints for 67 V. cholerae isolates from the Susquehanna River flats, Horn Point, Kent Island, and SERC stations in Chesapeake Bay collected between April and July 1998. The dendrogram was created by computing the similarity values according to the position of the bands with Molecular Analyst/Fingerprinting software (Bio-Rad Laboratories). The isolates were either free-living bacteria from the water column or associated with smaller phytoplankton (>20-μm- to <64-μm-mesh fraction) or larger plankton (>64-μm-mesh fraction).
Genetic diversity of isolates did not correlate with sampling location. That is identical banding patterns were observed for two isolates from two different sampling sites (Fig. 5, S14 and H11). Furthermore, no strong correlation was found between the genetic similarity of the isolates and the source of their isolation.
DISCUSSION
The genetic diversity of V. cholerae O1 and O139 has been studied extensively in recent years due to the availability of advanced molecular techniques (8, 9, 11, 12, 15, 30, 36). In contrast, the diversity of environmental isolates has been studied only sparsely in the context of their relationship to V. cholerae O1 and O139 (12, 31, 39). The molecular diversity and evolution of these groups of aquatic bacteria deserve in-depth investigation in order to understand better their ecological role and function in the aquatic environment. The results of the study reported here provide the first systematic investigation of the temporal and spatial genetic diversity of V. cholerae from an estuarine and riverine environment, the Chesapeake Bay and Susquehanna River flats.
V. cholerae is a dynamic and genetically diverse species in the Chesapeake Bay. The presence, abundance, and genotypical changes of the V. cholerae population were influenced or triggered by environmental conditions, such as temperature, salinity, and/or interaction with other microorganisms (plankton) in the water column. Repetitive attempts to isolate V. cholerae by culturing during the winter season (January to March) were unsuccessful, indicating that V. cholerae organisms are present in low numbers during the winter or are present in the VBNC state. Colwell and Huq (18) reviewed the phenomenon of VBNC in V. cholerae, showing that entrance into the VBNC state is a common strategy for this species to survive low temperatures, low nutrient levels, or other unfavorable environmental conditions. Microcosm studies have demonstrated temperature-induced recovery of V. cholerae from the VBNC state (38), and the presence of V. cholerae and Vibrio mimicus in the Chesapeake Bay during the winter has been shown, with gene probes, by Heidelberg and Colwell (unpublished results).
The frequency of isolation of Vibrio cholerae was correlated with an increase in water temperature in Chesapeake Bay, and large numbers were isolated during the summer months. This may reflect a relationship between growth rate or replication rate and temperature, indicated by the strong correlation between temperature and total bacterial abundance in the Bay. In addition, temperature changes also influence the succession of microbial communities. Temperature increases in the water column between April and July contribute significantly to the genetic diversity and shift in population structure of V. cholerae in the Chesapeake Bay. A high level of genetic diversity was observed among strains isolated during the summer months, with isolates obtained in July being the most distantly related to the early April and May strains.
The correlation between salinity and the presence of V. cholerae in aquatic environments has been provided in several previous studies (18). Kaper et al. (28) found V. cholerae organisms only in stations where the salinity was between 4 and 14 ppt along a transect in the Chesapeake Bay. Colwell and Huq (18) found that the greater frequency of isolation was achieved at sites where the salinity was between 0.2 and 2 ppt. The results of this study are in agreement with those of the earlier studies in our laboratory. The riverine station, Susquehanna River flats, showed the lowest frequency of isolation during the 7-month sampling period. Late summer and early fall may be times of greater abundance of V. cholerae in the Susquehanna River, and this relationship is the subject of further study.
A strong association of V. cholerae with plankton has been demonstrated previously (13, 21, 40). An examination of genetic similarity among V. cholerae strains isolated within each size fraction of plankton should elucidate specific interactions between V. cholerae and plankton species. For example, a certain genotype of V. cholerae may be dominant among free-living V. cholerae, and a distinct genotype may be affiliated with smaller plankton (>20-μm- and <64-μm-mesh fraction, dominated by phytoplankton) or larger plankton (>64-μm-mesh fraction, dominated by zooplankton). However, our results do not suggest a clear separation of genetic diversity or frequency of isolation between organisms from these different fractions of the sample. This may be due to the inclusion of nauplii and juveniles of zooplankton in the smaller plankton. A larger sample size from each fraction, coupled with analysis of specific zooplankton and phytoplankton species, may provide a resolution of this issue. No correlation was detected between phytoplankton abundance (as indicated by chlorophyll a concentration) and frequency of V. cholerae isolation, providing support for the zooplankton-V. cholerae relationship. Thus, analyses of the species of plankton present in the samples are critical to the understanding of interactions of V. cholerae with the planktonic assemblage. Identification of plankton species in preserved samples is in progress.
The genetic diversity of V. cholerae was not correlated with geographical location, since identical clones were isolated from different locations across the Bay at the same time of sampling (H11 and S14). We interpret this finding as the bacteria in Chesapeake Bay either being transported by currents or wind-driven water movement or responding similarly to environmental conditions at the various geographical sites, with a single genotype being selected. However, some degree of localization of similar genotypes was found in the small clusters of isolates from the same site (Fig. 5).
The AFLP technique had been tested previously, with a cross-section of bacterial families and species, including our most recent research findings on the genetic fingerprinting of Vibrio cholerae (24, 27, 32). The results of the studies show that AFLP is a powerful technique, able to differentiate closely related strains, including subbiotype diversity (2). The application of the AFLP technique in this study clearly demonstrated a genetic diversity of V. cholerae over space and time in the Chesapeake Bay. The results of this study represent a preliminary report of a larger study of cholera and climate. A longer-term study of the genetic diversity of V. cholerae on a global scale is under way, the results of which will be critical to our understanding of the ecology and evolution of this aquatic species and provide further insight into the epidemiology of V. cholerae.
ACKNOWLEDGMENTS
This research was supported by the U.S. Environmental Protection Agency (grant R824995-01-0) and was also partially supported by the National Institutes of Health (grant 1RO1A13912901) and NASA (grant NAG2-1195).
REFERENCES
- 1.Albert M J, Siddique A K, Islam M S, Faruque A S G, Ansaruzzaman M, Faruque S M, Sack R B. A large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 2.Arias C, Verdonck L, Swings J, Aznar R, Garay E. A polyphasic approach to study the intraspecific diversity amongst Vibrio vulnificus isolates. Syst Appl Microbiol. 1997;20:622–633. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Beltran P, Delgado G, Navarro A, Trujillo F, Selander R K, Cravioto A. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999;37:581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya M K, Dutta D, Bhattacharya S K, Deb A, Mukhopadhyay A K, Nair G B, Shimada T, Takeda Y, Chowdhury A, Mahalanabis D. Association of a disease approximating cholera caused by Vibrio cholerae of serogroups other than O1 and O139. Epidemiol Infect. 1998;120:1–5. doi: 10.1017/s0950268897008352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bik E M, Gouw R D, Mooi F R. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calia K E, Waldor M K, Calderwood S B. Use of representational difference analysis to identify genomic differences between pathogenic strains of Vibrio cholerae. Infect Immun. 1998;66:849–852. doi: 10.1128/iai.66.2.849-852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron D N, Khambaty F M, Wachsmuth I K, Tauxe R V, Barrett T J. Molecular characterization of Vibrio cholerae O1 strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1685–1690. doi: 10.1128/jcm.32.7.1685-1690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri K, Bhadra R K, Das J. Cell surface characteristics of environmental and clinical isolates of Vibrio cholerae non-O1. Appl Environ Microbiol. 1992;58:3567–3573. doi: 10.1128/aem.58.11.3567-3573.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, Evins G M, Cook W L, Almeida R, Hargrett-Bean N, Wachsmuth K. Genetic diversity among toxigenic and nontoxigenic Vibrio cholerae O1 isolated from the Western Hemisphere. Epidemiol Infect. 1991;107:225–233. doi: 10.1017/s0950268800048846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhury S R, Bhadra R K, Das J. Genome size and restriction fragment length polymorphism analysis of Vibrio cholerae strains belonging to different serovars and biotypes. FEMS Microbiol Lett. 1994;115:329–334. doi: 10.1111/j.1574-6968.1994.tb06659.x. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury M A R, Huq A, Xu B, Madeira F J B, Colwell R R. Effect of alum on free-living and copepod-associated Vibrio cholerae O1 and O139. Appl Environ Microbiol. 1997;63:3323–3326. doi: 10.1128/aem.63.8.3323-3326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun J, Huq A, Colwell R R. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl Environ Microbiol. 1999;65:2202–2208. doi: 10.1128/aem.65.5.2202-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombo M M, Mastrandrea S, Leite F, Santona A, Uzzau S, Rappeli P, Pisano M, Rubino S, Cappuccinelli P. Tracking of clinical and environmental Vibrio cholerae O1 strains by combined analysis of the presence of the toxin cassette, plasmid content and ERIC PCR. FEMS Immunol Med Microbiol. 1997;19:33–54. doi: 10.1111/j.1574-695X.1997.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 16.Colwell R R, Huq A, Chowdhury M A R, Brayton R R, Xu B. Serogroup conversion of Vibrio cholerae. Can J Microbiol. 1995;41:946–950. doi: 10.1139/m95-131. [DOI] [PubMed] [Google Scholar]
- 17.Colwell R R, Spira W M. The ecology of Vibrio cholerae. In: Barua D, Greenough W B, editors. Cholera. New York, N.Y: Plenum; 1992. pp. 107–127. [Google Scholar]
- 18.Colwell R R, Huq A. Vibrios in the environment: viable but nonculturable Vibrio cholerae. In: Wachsmuth I K, Blake P A, Olsvik Ø, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: American Society for Microbiology; 1994. pp. 117–133. [Google Scholar]
- 19.Dalsgaard A, Forslund A, Mortensen H F, Shimada T. Ribotypes of clinical Vibrio cholerae non-O1 non-O139 strains in relation to O-serotypes. Epidemiol Infect. 1998;121:535–545. doi: 10.1017/s0950268898001654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalsgaard A, Albert M J, Taylor D N, Shimada T, Meza R, Serichantalergs O, Echeverria P. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumontet S, Krovacek K, Baloda S B, Grottoli R, Pasquale V, Vanucci S. Ecological relationship between Aeromonas and Vibrio spp. and planktonic copepods in the coastal marine environment in Southern Italy. Comp Immunol Microbiol Infect Dis. 1996;19:245–254. doi: 10.1016/0147-9571(96)00012-4. [DOI] [PubMed] [Google Scholar]
- 22.Dumontier S, Berche P. Vibrio cholerae O22 might be a putative source of exogenous DNA resulting in the emergence of the new strain of Vibrio cholerae O139. FEMS Microbiol Lett. 1998;164:91–98. doi: 10.1111/j.1574-6968.1998.tb13072.x. [DOI] [PubMed] [Google Scholar]
- 23.Holm-Hansen O, Reimann B. Determination of microbial biomass in ocean profiles. Limnol Oceanogr. 1978;14:740–747. [Google Scholar]
- 24.Huys G, Coopman R, Janssen P, Kersters K. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int J Syst Bacteriol. 1996;46:572–580. doi: 10.1099/00207713-46-2-572. [DOI] [PubMed] [Google Scholar]
- 25.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 26.Janssen P, Dijkshoorn L. High resolution DNA fingerprinting of Acinetobacter outbreak strains. FEMS Microbiol Lett. 1996;142:191–194. doi: 10.1111/j.1574-6968.1996.tb08429.x. [DOI] [PubMed] [Google Scholar]
- 27.Jiang S C, Matte M, Mattee G, Huq A, Colwell R R. Genetic diversity of clinical and environmental isolates of Vibrio cholerae determined by amplified fragment length polymorphism (AFLP) Appl Environ Microbiol. 1999;66:148–153. doi: 10.1128/aem.66.1.148-153.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaper J, Lockman H, Colwell R R, Joseph S W. Ecology, serology, and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl Environ Microbiol. 1979;37:91–103. doi: 10.1128/aem.37.1.91-103.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaolis D K, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karaolis D K R, Lan R, Reeves P R. Molecular evolution of the seventh-pandemic clone of Vibrio cholerae and its relationship to other pandemic and epidemic V. cholerae isolates. J Bacteriol. 1994;176:6199–6206. doi: 10.1128/jb.176.20.6199-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karaolis D K R, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mekalanos J J, Swartz D J, Pearson G D N. Cholera toxin gene: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 34.Nair G B, Ramamurthy T, Bhattacharya S K, Mukhopadhyay A K, Garg S, Bhattacharya M K, Takeda T, Shima T, Takeda Y, Deb B C. Spread of Vibrio cholerae O139 Bengal in India. J Infect Dis. 1994;169:1029–1034. doi: 10.1093/infdis/169.5.1029. [DOI] [PubMed] [Google Scholar]
- 35.Paul J H, Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl Environ Microbiol. 1982;43:1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovic T, Bopp C, Olsvik Ø, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. Emergence of a novel strain of Vibrio cholerae with epidemic potential in Southern and Eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 38.Ravel J, Knight I T, Monahan C E, Hill R T, Colwell R R. Temperature-induced recovery of Vibrio cholerae from the viable but nonculturable state: growth or resuscitation? Microbiology. 1995;141:377–383. doi: 10.1099/13500872-141-2-377. [DOI] [PubMed] [Google Scholar]
- 39.Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay A K, Basu A, Mitra R, Basu I, Bhattacharya S K, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair G B. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamplin M L, Gauzens A L, Huq A, Sack D A, Colwell R R. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990;56:1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 42.Xu H S, Robert N, Singleton F L, Attwell R W, Grimes D J, Colwell R R. Survival and viability of non-culturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol. 1982;8:213–223. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 43.Yamai S, Okitsu T, Shimada T, Yatsube Y. Distribution of serogroups of Vibrio cholerae non-O1 non-O139 with specific reference to their ability to produce cholera toxin, and addition of novel serogroups. J Jpn Assoc Infect Dis. 1997;71:1037–1045. doi: 10.11150/kansenshogakuzasshi1970.71.1037. [DOI] [PubMed] [Google Scholar]