Abstract
Vibrio cholerae, the causative agent of major epidemics of diarrheal disease in Bangladesh, South America, Southeastern Asia, and Africa, was isolated from clinical samples and from aquatic environments during and between epidemics over the past 20 years. To determine the evolutionary relationships and molecular diversity of these strains, in order to understand sources, origin, and epidemiology, a novel DNA fingerprinting technique, amplified fragment length polymorphism (AFLP), was employed. Two sets of restriction enzyme-primer combinations were tested for fingerprinting of V. cholerae serogroup O1, O139, and non-O1, O139 isolates. Amplification of HindIII- and TaqI-digested genomic DNA produced 30 to 50 bands for each strain. However, this combination, although capable of separating environmental isolates of O1 and non-O1 strains, was unable to distinguish between O1 and O139 clinical strains. This result confirmed that clinical O1 and O139 strains are genetically closely related. On the other hand, AFLP analyses of restriction enzyme ApaI- and TaqI-digested genomic DNA yielded 20 to 30 bands for each strain, but were able to separate O1 from O139 strains. Of the 74 strains examined with the latter combination, 26 serogroup O1 strains showed identical banding patterns and were represented by the O1 El Tor strain of the seventh pandemic. A second group, represented by O139 Bengal, included 12 strains of O139 clinical isolates, with 7 from Thailand, 3 from Bangladesh, and 2 from India. Interestingly, an O1 clinical isolate from Africa also grouped with the O139 clinical isolates. Eight clinical O1 isolates from Mexico grouped separately from the O1 El Tor of the seventh pandemic, suggesting an independent origin of these isolates. Identical fingerprints were observed between an O1 environmental isolate from a river in Chile and an O1 clinical strain from Kenya, both isolated more than 10 years apart. Both strains were distinct from the O1 seventh pandemic strain. Two O139 clinical isolates from Africa clustered with environmental non-O1 isolates, independent of other O139 strains included in the study. These results suggest that although a single clone of pathogenic V. cholerae appears responsible for many cases of cholera in Asia, Africa, and Latin America during the seventh pandemic, other cases of clinical cholera were caused by toxigenic V. cholerae strains that appear to have been derived locally from environmental O1 or non-O1 strains.
Vibrio cholerae, the etiologic agent for the diarrheal disease cholera, continues to be an important cause of morbidity and mortality in many areas of Asia, Africa, and Latin America. The World Health Organization describes cholera as a tragedy because this theoretically “most preventable disease” is one of the top causes of human morbidity and mortality in the world. The incidence of cholera is estimated to exceed five million cases each year (18). Cholera is an ancient disease in the midst of a modern resurgence. During the last decade, remarkable changes have been reported regarding the incidence and characteristics of V. cholerae, including the “entry” of V. cholerae O1 into Latin America in 1991 (4) and the emergence and rapid spread of O139 serotypes of V. cholerae in Southeast Asia in 1992 (6). A significant turning point in our understanding of the toxigenicity of V. cholerae may prove to be the discovery of a lysogenic filamentous bacteriophage encoding the cholera toxin, CTXφ, which uses the toxin-coregulated pilus as a receptor and is transferable to nontoxigenic V. cholerae (8, 23). Most recently, Trucksis et al. (19) reported that V. cholerae contains two unique circular chromosomes, with a copy of the V. cholerae CTXφ element coding for ctxAB on each of the replicons. Thus, despite more than a century of study, this aquatic species still presents surprises and challenges.
The genetic diversity and molecular epidemiology of V. cholerae serogroups O1 and O139 have been studied extensively in recent years due to the availability of various molecular techniques. These techniques include the analysis of restriction fragment length polymorphisms (RFLPs) in different genes. By using gene probes to study RFLPs in the cholera toxin genes and their flanking DNA sequences, it was observed that clinical isolates from the U.S. Gulf Coast region are different from other seventh pandemic isolates (13). RFLPs in conserved rRNA genes have been used to differentiate V. cholerae strains into ribotypes. Analysis of clinical isolates from the Latin America epidemic that occurred in 1991 showed that these strains were related to seventh pandemic isolates from other parts of the world, suggesting the Latin American cholera epidemic was an extension of the seventh pandemic (9, 21, 22).
The results of pulsed-field gel electrophoresis (PFGE) analysis of chromosomal DNA indicate that several V. cholerae strains belonging to different serovars and biotypes have distinct restriction patterns (5). Four different PFGE patterns were detected among isolates that were presumed to be identical, based on the DNA sequence of the cholera toxin B unit and multilocus enzyme electrophoresis markers (16). Genotypic evolution in V. cholerae O139 Bengal was suggested based on changes in PFGE patterns among O139 isolates obtained from Bangladesh between 1993 and 1996 (1).
A study of the DNA sequences of the asd genes from 45 isolates of V. cholerae indicated that there were no differences between sixth and seventh pandemic isolates; however, variation was found between the two forms and among the non-O1 isolates. The O139 isolates had asd sequences identical to those of seventh pandemic isolates, suggesting they are seventh pandemic derived (14). These results also suggest that the sixth pandemic, seventh pandemic, and U.S. Gulf clones evolved independently from different lineages of environmental, nontoxigenic, non-O1 V. cholerae isolates.
PCR was also used to amplify the enterobacterial repetitive intergenic consensus (ERIC) sequences of Vibrio cholerae O1, O139, and non-O1 strains. However, these earlier studies failed to separate toxigenic V. cholerae O1 strains from the O139 serogroup (17).
In spite of the rapid advancements in the molecular epidemiology of V. cholerae due to the development of techniques mentioned above, the level of resolution between individual strains is still limited. Many of the techniques are also time-consuming and labor intensive and therefore are not suited to rapid epidemiological identification. Amplified fragment length polymorphism (AFLP) is a high-resolution genomic fingerprinting method. The successful application of this method to identification and classification of bacteria has recently been reported, and it has been demonstrated to have both a greater capacity for genome coverage and better reproducibility than other genotyping technologies (10, 12, 20). Compared with PFGE, this method is rapid, cost-effective, and useful for fingerprinting a large number of strains simultaneously. In the study reported here, we applied the AFLP fingerprinting method to examination of molecular evolution and diversity in V. cholerae.
MATERIALS AND METHODS
V. cholerae isolates and DNA preparation.
The V. cholerae strains used in this study were collected during the past 20 years from Asia, Africa, and Latin America and included strains from both clinical and environmental samples collected during and between epidemic periods. Detailed information on the location and date of isolation is presented in Fig. 1 and 2 and Tables 2 and 3. Isolates were stored in liquid nitrogen until used for this study. Frozen stocks were subcultured on Luria-Bertani (LB) agar (Difco) plates for reisolation. A single colony of each strain was inoculated into LB broth and grown overnight at 37°C. Chromosomal DNA was extracted by using a CTAB (cetylethylammonium bromide) protocol previously described (3). The purity and quality of the DNA were determined by UV absorption at wavelengths of 260 and 280 nm by Beckman DU 640 spectrophotometry (Beckman Instruments, Inc., Fullerton, Calif.).
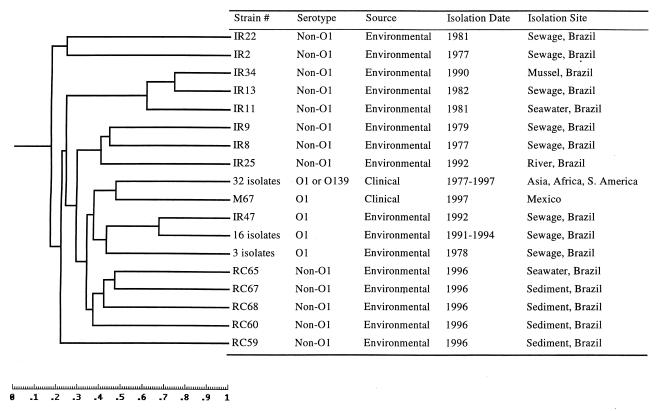
FIG. 1.
Representative gel image of AFLP analysis of V. cholerae from clinical and environmental sources with the HindIII-TaqI combination. Lanes: 1 and 2, environmental non-O1 (ir11 and ir2); 3 to 5 and 7 and 8, clinical O1 and O139 strains (from left to right, m45, m44, m42, m30, and m27); 6 and 9 to 15, environmental non-O1 strains (from left to right, ir34, rc68, rc67, rc65, rc64, rc61, rc60, and rc59).
FIG. 2.
Similarity analysis of AFLP fingerprints of V. cholerae isolates with the restriction enzyme HindIII-TaqI combination. Strains were collected from clinical and environmental samples during the past 20 years. The dendrogram was created by computing the similarity values according to the position of the bands.
TABLE 2.
Bacterial strains included in group Aa
| Strainb | Serotype | Isolation date | Isolation site |
|---|---|---|---|
| m63 | O1 | 1997 | Africa |
| m10 | O139 | NAc | Bangladesh |
| m13 | O139 | NA | Bangladesh |
| m14 | O139 | NA | Bangladesh |
| m11 | O139 | 1984 | India |
| m12 | O139 | 1985 | India |
| m26 | O139 | 1983 | Thailand |
| m27 | O139 | 1993 | Thailand |
| m32 | O139 | 1993 | Thailand |
| m28 | O139 | 1993 | Thailand |
| m29 | O139 | 1993 | Thailand |
| m30 | O139 | 1993 | Thailand |
TABLE 3.
Bacterial strains (serotype O1) included in group Ba
| Strainb | Isolation date | Isolation site |
|---|---|---|
| m42 | Before 1991 | Bangladesh |
| m40 | Before 1991 | Bangladesh |
| m25 | 1991 | Indonesia |
| m17 | 1991 | Iran |
| m1 | NAc | Tanzania |
| m2 | NA | West Africa |
| m23 | 1980 | West Africa |
| m9 | NA | Kenya |
| m15 | 1985 | Kenya |
| m72 | 1991 | Mexico (1st case) |
| m8 | 1991 | Mexico |
| m37 | 1992 | Mexico |
| m34 | 1993 | Mexico |
| m73 | 1993 | Mexico |
| m69 | 1995 | Mexico |
| m75 | 1997 | Mexico (1st case) |
| m74 | 1997 | Peru |
| m18 | NA | Peru |
| m20 | NA | Peru |
| m24 | 1991 | Peru |
| m19 | NA | Brazil |
| m16 | 1991 | Chile |
| m21 | 1991 | Chile |
| m31 | NA | Chile |
| m53 | 1947 | ATCC 11623 |
| m54 | 1961 | ATCC 14035 |
See the dendrogram in Fig. 3.
All strains listed here are clinical isolates, except for m74 (environmental).
NA, not available.
AFLP fingerprinting.
All procedures were performed as described by Janssen et al. (11), with slight modification. In brief, 1 μg of DNA was digested with restriction enzyme TaqI at 65°C for 1 h and then the second restriction enzyme (ApaI or HindIII) was added and incubated at 30 or 37°C, according to the optimal temperature for enzyme activity, for 1 to 3 h. Following digestion, adapters (Table 1) were added to a final concentration of 0.4 μM for TaqI adapters and 0.04 μM for ApaI and HindIII adapters. Ligation reactions were performed at 16°C overnight. Ligated template DNA was purified by ethanol precipitation and resuspended in TE0.1 buffer (10 mM Tris, 0.1 mM EDTA [pH 8.0]), stored at −20°C, and used for PCR amplification within 48 h.
TABLE 1.
Adapters and primers used in the AFLP analyses
| Adaptor or primer | Oligonucleotide sequence |
|---|---|
| ApaI adaptor | 5′-TCGTAGACTGCGTACAGGCC-3′ |
| 3′-CATCTGACGCATGT-5′ | |
| A01 primer | 5′-GACTGCGTACAGGCCCA-3′ |
| HindIII adaptor | 5′-CTCGTAGACTGCGTACC-3′ |
| 3′-CTGACGCATGGTCGA-5′ | |
| H01 primer | 5′-GACTGCGTACCAGCTTA-3′ |
| TaqI adaptor | 5′-GACGATGAGTCCTGAC-3′ |
| 3′-TACTCAGGACTGGC-5′ | |
| T01 primer | 5′-CGATGAGTCCTGACCGAA-3′ |
The PCR was performed with an MJ Research thermal cycler by employing the program described by Janssen et al. (11): i.e., cycle 1 of 94°C for 1 min, 65°C for 0.5 min, and 72°C for 1 min; followed by 11 cycles with the annealing temperature reduced 0.7°C at each cycle; and cycles 13 to 24 of 94°C for 1 min, 56°C for 0.5 min, and 72°C for 1 min. Prior to PCR, primer T01 (Table 1) was end labeled with [γ-32P]ATP. Two microliters of template DNA was used for amplification in a total reaction volume of 25 μl. A reference strain is included in each experiment as a PCR control and a universal standard for gel analysis as described below. Amplification products were separated on 5% denaturing polyacrylamide gels and exposed to X-ray film (Kodak, Inc.).
Analysis of fingerprint patterns.
Autoradiographs were digitized by an HP scanner fitted with a transparency adapter (Hewlett-Packard, Inc.). Digital images were straightened and unwarped by using Molecular Analyst/Fingerprinting software (Bio-Rad Laboratories) according to the manufacturer's instructions. Fingerprints were normalized and sized by alignment to both a reference strain included in each gel at multiple places and a radiolabeled molecular weight ladder (RST Ready-label 100-bp DNA ladder; GibcoBRL, Gaithersburg, Md.). Digital images were combined by assigning one reference track as a global standard and by alignment of all other strains with this standard. Bands were automatically selected and visually corrected for addition or subtraction of bands. Similarity values were computed based on shared and unshared bands, by using Molecular Analyst/fingerprinting software (Bio-Rad Laboratories), and were graphically represented in a dendrogram.
RESULTS
AFLP analysis with TaqI and HindIII restriction enzymes.
Restriction enzymes TaqI and HindIII were used to generate AFLP template DNA from 66 environmental and clinical isolates of V. cholerae. More than 50 bands were observed after PCR amplification with the T01-H01 primer set for each strain. While dramatically different banding patterns were evident among V. cholerae non-O1 strains and some O1 strains, no genetic differences were detected between serogroup O1 and O139 strains with this restriction enzyme combination (Fig. 1). Similarity analysis of the fingerprints suggested that this enzyme combination was unsuitable for distinguishing genetic differences between V. cholerae serogroups O1 and O139 (Fig. 2). However, a clinical O1 isolate from Mexico in 1997 (m67) was separated from the clinical O1 and O139 group, suggesting that O1 and O139 strains were genetically more closely related than the relationship within some of the O1 strains. Brazilian O1 isolates from sewage (ir47: 16 isolates between 1991 and 1994 and 3 isolates from 1978) were more closely related to O1 clinical isolates than to non-O1 environmental isolates according to this assay (Fig. 2).
AFLP analysis with TaqI and ApaI restriction enzymes.
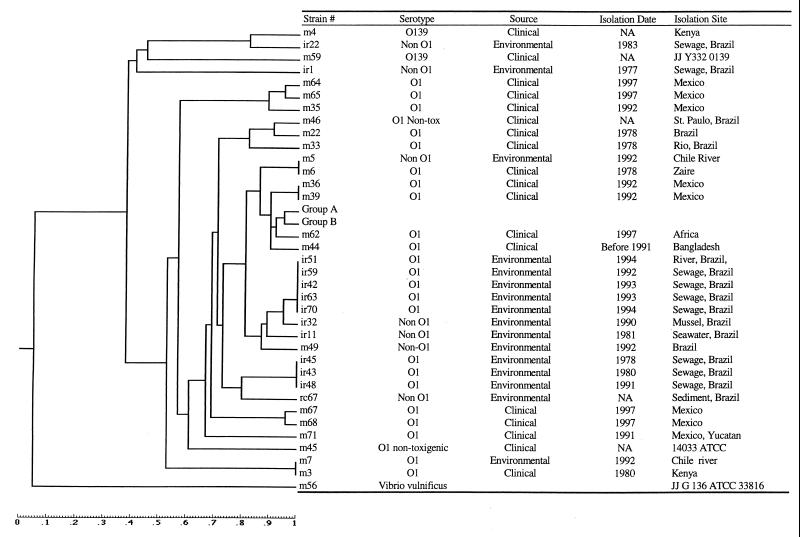
Restriction enzymes TaqI and ApaI were used to generate AFLP template DNA from 73 V. cholerae strains and a Vibrio vulnificus strain. AFLP amplification of template DNA by using the T01-A01 primer set generated between 20 and 30 bands for most of the strains tested. Analyses of fingerprints of 73 V. cholerae isolates from Asia, Africa, and Latin America, isolated between 1977 and 1997, indicated that 26 strains belonging to serogroup O1 from all three continents had identical fingerprinting patterns (Fig. 3 [group B] and Table 2). All but one isolate in this group were from clinical sources. Twelve O139 clinical strains from Asia (Table 1 [group A]) were separated from the O1 group, but were most closely related to the O1 clinical group (Table 3 [group B]). Interestingly, a clinical O1 strain recently (1997) isolated from Africa (m63) also showed a fingerprinting pattern identical to that of the O139 group. Eight clinical O1 isolates from Mexico (m64, m65, m35, m36, m39, m67, m68, and m71) were separated from the seventh pandemic El Tor strains, including some of the Mexico clinical isolates (Fig. 3). The Mexico O1 clinical isolates appear to have an independent origin. Identical fingerprints were observed for an O1 clinical strain from Africa and a non-O1 environmental isolate from a river in Chile, both isolated more than 10 years apart and both distinct from O1 seventh pandemic El Tor strains. A deep branching lineage was also observed for two clinical O139 isolates from Africa that appeared more closely related to non-O1 environmental isolates than the other clinical strains tested.
FIG. 3.
Similarity analysis of AFLP fingerprints of V. cholerae isolates with the restriction enzyme ApaI-TaqI combination. Strains were collected from clinical and environmental samples during the past 20 years. The dendrogram was created by computing the similarity values according to the position of the bands.
Environmental isolates from Brazil clustered together, with the exception of two non-O1 isolates (ir22 and ir1) obtained during 1983 and 1977. Strains collected in the early 1990s (ir51, ir59, ir42, ir63, ir70, ir32, and m49) from both sewage and seawater were more closely related to clinical groups A and B than to the other O1 clinical isolates.
DISCUSSION
AFLP fingerprinting of DNA was first described in 1995 as a technique to detect genomic restriction fragments by PCR amplification and as being useful for DNAs of any origin or complexity (20). Fingerprints are produced without prior sequence knowledge, by using a limited set of generic primers. The AFLP technique is also highly reproducible because stringent reaction conditions are used for primer annealing. Since 1995, this technique has been widely used and has been demonstrated to have superb resolution power for bacterial classification, molecular typing, and molecular epidemiology (7, 10–12, 15). In a previous study, intraspecific diversity among V. vulnificus isolates was examined by seven methods, including API20E, BIOLOG, total protein profiles, serotyping, enzyme-linked immunosorbent assay, ribotyping, and AFLP. AFLP was found to be the best method for differentiating between strains belonging to the same serovar (2). The study reported here is the first application of AFLP for analysis of the molecular diversity and epidemiology of V. cholerae.
The genetic resolution of the AFLP method largely relies on the selection of the appropriate restriction enzymes. Janssen and colleagues (11) suggested that the restriction enzyme HindIII, combined with TaqI, gave an adequate number of suitably sized restriction fragments for bacteria with a G+C ratio of 40 to 50 mol%, and EcoRI-MseI and ApaI-TaqI were most suited for bacterial genomes with low and high G+C contents, respectively. Huys and colleagues (10) emphasized the importance of evenly distributed bands along the length of the gel lane, because a good band distribution was critical for optimal normalization and a high level of discrimination in cluster analyses. Compared with ribotyping, Huys et al. (10) suggested that a larger number of DNA polymorphisms yields a more accurate comparative analysis and facilitates strain-to-strain discrimination. Our experience with the application of AFLP to the study of the genetic diversity of V. cholerae indicated the choice of restriction enzymes should be based on the target bacterial species and the desired degree of discrimination between strains. The choice of restriction enzymes and primer sequences should derive from the results of preliminary research with target bacteria. A larger number of DNA polymorphisms did not necessarily favor more accurate discrimination. For example, the restriction enzyme HindIII-TaqI combination produced a larger number of evenly distributed bands along the length of the gel lane, but was not helpful in discriminating between V. cholerae serogroups O1 and O139. In contrast, ApaI-TaqI yielded fewer bands, but had greater resolving power. The HindIII-TaqI combination is better suited for analysis of genetic diversity among environmental isolates, where greater diversity is expected. The restriction enzyme ApaI-TaqI combination appears to be more helpful for “fine-tuning” relatedness among strains.
The results of AFLP analyses of clinical isolates of V. cholerae support previous results showing that a single clone or clones from Asia and of the same origin were responsible for the spread of the seventh cholera pandemic over three continents. A fingerprint pattern identical to that of the seventh pandemic strains was also detected in an environmental strain isolated in Peru (group B) during the recent epidemic there, suggesting that the aquatic environment does serve as a reservoir for transmission and spread of cholera. The results of this study also confirm that serogroups O1 and O139 are closely related, as has been hypothesized by other investigators. However, an interesting finding in this study was that a very recent isolate of clinical O1 from Africa revealed a pattern that grouped it with O139 strains, rather than O1 strains. The African O1 strain may be a transition state in the evolution from O1 to O139. On the other hand, if the O139 strain evolved from an O1 El Tor strain via horizontal gene transfer, was that gene transfer reversible, or was the transferred gene, or portion of the gene, lost in its progenies? Albert and colleagues (1) concluded that rapid genotypic evolution had occurred in V. cholerae O139 Bengal, based on the results of examination of O139 strains collected between 1993 and 1996.
Use of gene probes to study RFLPs in cholera toxin genes and their flanking DNA sequences has provided evidence that U.S. Gulf clinical isolates are different from other seventh pandemic isolates and evolved independently from other pandemic strains. AFLP analysis of the strains in our culture collection suggests that a significant number of cases of cholera were caused by pathogenic V. cholerae strains that had evolved independently and that this occurred more frequently than expected. The most obvious evidence was strains found among isolates from Mexico. That is, a significant number of clinical isolates from Mexico differed from the seventh pandemic type strains and clustered in different groupings in the dendrogram (Fig. 3). In addition, variations were observed among O1 isolates from Brazil, Kenya, Zaire, and Bangladesh. These results raise several questions concerning the source and reservoir of V. cholerae between epidemics. Throughout the history of cholera epidemics and pandemics, the disease was believed to originate in one area of the world and spread to other continents via human contact and/or transport of contaminated water, food, etc. However, if pathogenic V. cholerae can evolve independently from nontoxigenic strains in the environment, coastal waters are likely to be the source for new epidemic strains. The genetic similarity between environmental non-O1 strains and clinical O1, O139 strains is evident. From data presented in this study, it is concluded that cholera epidemics potentially may arise from multiple independent sources. Although the original clone of the seventh cholera pandemic from Asia was found to be similar to the strains isolated across Africa and Latin America, other cases of cholera are likely the result of pathogenic V. cholerae strains that evolved independently in the local coastal environment. Future research will focus on the abundance and diversity of V. cholerae in the coastal environment and the potential for cholera toxin gene transfer via transduction.
ACKNOWLEDGMENTS
This research was supported by the U.S. Environmental Protection Agency (grant R824995-01-0) and was also partially supported by the National Institutes of Health (grant 1RO1A13912901) and NASA (grant NAG2-1195).
REFERENCES
- 1.Albert M J, Bhuiyan N A, Talukder K A, Faruque A S G, Nahar S, Faruque S M, Ansaruzzaman M, Rahman M. Phenotypic and genotypic changes in Vibrio cholerae O139 Bengal. J Clin Microbiol. 1997;35:2588–2592. doi: 10.1128/jcm.35.10.2588-2592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias C, Verdonck L, Swings J, Aznar R, Garay E. A polyphasic approach to study the intraspecific diversity amongst Vibrio vulnificus isolates. Syst Appl Microbiol. 1997;20:622–633. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Centers for Disease Control. Cholera—Peru, 1991. Morbid Mortal Weekly Rep. 1991;40:108–110. [Google Scholar]
- 5.Choudhury S R, Bhadra R K, Das J. Genome size and restriction fragment length polymorphism analysis of Vibrio cholerae strains belonging to different serovars and biotypes. FEMS Microbiol Lett. 1994;115:329–334. doi: 10.1111/j.1574-6968.1994.tb06659.x. [DOI] [PubMed] [Google Scholar]
- 6.Dalsgaard A, Forslund A, Tam N V, Vinh D X, Cam P D. Cholera in Vietnam: changes in genotypes and emergence of class I integrons containing aminoglycoside resistance gene cassettes in Vibrio cholerae O1 strains isolated from 1979 to 1996. J Clin Microbiol. 1999;37:734–741. doi: 10.1128/jcm.37.3.734-741.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkshoorn L, Aucken H, Gerner-Smidt P, Janssen P, Kaufmann M E, Garaizar J, Ursing J, Pitt T L. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J Clin Microbiol. 1996;34:1519–1525. doi: 10.1128/jcm.34.6.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque S M, Albert M J, Mekalanos J J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque S M, Albert M J. Genetic relation between Vibrio cholerae O1 strains in Ecuador and Bangladesh. Lancet. 1992;339:740–741. doi: 10.1016/0140-6736(92)90636-h. [DOI] [PubMed] [Google Scholar]
- 10.Huys G, Coopman R, Janssen P, Kersters K. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int J Syst Bacteriol. 1996;46:572–580. doi: 10.1099/00207713-46-2-572. [DOI] [PubMed] [Google Scholar]
- 11.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 12.Janssen P, Dijkshoorn L. High resolution DNA fingerprinting of Acinetobacter outbreak strains. FEMS Microbiol Lett. 1996;142:191–194. doi: 10.1111/j.1574-6968.1996.tb08429.x. [DOI] [PubMed] [Google Scholar]
- 13.Kaper J B, Bradford H B, Roberts N C, Falkow S. Molecular epidemiology of Vibrio cholerae in the U.S. Gulf Coast. J Clin Microbiol. 1982;16:129–134. doi: 10.1128/jcm.16.1.129-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karaolis D K R, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popovic T, Fields P I, Olsvik O, Wells J G, Evins G M, Cameron D N, Farmer III J J, Bopp C A, Wachsmuth K, Sack R B, Albert M J, Nair G B, Shimada T, Feeley J C. Molecular subtyping of toxigenic Vibrio cholerae O139 causing epidemic cholera in India and Bangladesh, 1992–1993. J Infect Dis. 1995;171:122–127. doi: 10.1093/infdis/171.1.122. [DOI] [PubMed] [Google Scholar]
- 17.Rivera I G, Chowdhury M A R, Huq A, Jacobs D, Martins M T, Colwell R R. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl Environ Microbiol. 1995;61:2898–2904. doi: 10.1128/aem.61.8.2898-2904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tauxe R, Seminario L, Tapia R, Libel M. The Latin American epidemic. In: Wachsmuth I K, Blake P A, Olsvik Ø, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C.: American Society for Microbiology; 1994. pp. 321–344. [Google Scholar]
- 19.Trucksis M, Michalski J, Deng Y K, Kaper J B. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wachsmuth I K, Bopp C A, Fields P I, Carrillo C. Difference between toxigenic Vibrio cholerae O1 from South America and US Gulf Coast. Lancet. 1991;337:1097–1098. doi: 10.1016/0140-6736(91)91744-f. [DOI] [PubMed] [Google Scholar]
- 22.Wachsmuth I K, Evins G M, Fields P I, Olsvik O, Popovic T, Bopp D A, Wells J G, Carrillo C, Blake P. The molecular epidemiology of cholera in Latin America. J Infect Dis. 1993;167:621–626. doi: 10.1093/infdis/167.3.621. [DOI] [PubMed] [Google Scholar]
- 23.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]