Abstract
Inflammasomes are multiprotein complexes that sense intracellular danger signals and induce pyroptosis. CARD8 and NLRP1 are related inflammasomes that are repressed by the enzymatic activities and protein structures of the dipeptidyl peptidases 8 and 9 (DPP8/9). Potent DPP8/9 inhibitors such as Val-boroPro (VbP) activate both NLRP1 and CARD8, but chemical probes that selectively activate only one have not been identified. Here we report a small molecule called CQ31 that selectively activates CARD8. CQ31 inhibits the M24B aminopeptidases prolidase (PEPD) and Xaa-Pro aminopeptidase 1 (XPNPEP1), leading to the accumulation of proline-containing peptides that inhibit DPP8/9 and thereby activate CARD8. NLRP1 is distinct from CARD8 in that it directly contacts DPP8/9’s active site; these proline-containing peptides, unlike VbP, do not disrupt this repressive interaction and thus do not activate NLRP1. We expect that CQ31 will now become a valuable tool to study CARD8 biology.
At least six human pattern recognition receptors (PRRs) detect danger-associated signals, assemble into multiprotein complexes called inflammasomes and trigger caspase-1 (CASP1)-dependent pyroptosis1-3. CARD8 (caspase activation and recruitment domain-containing 8) and NLRP1 (nucleotide-binding domain leucine-rich repeat pyrin domain-containing 1) are related PRRs that form inflammasomes. Both proteins have function-to-find domains (FIINDs) that undergo post-translational autoproteolysis, generating autoinhibitory N-terminal (NT) and inflammatory C-terminal (CT) polypeptide chains that remain associated and inactive4-6 (Fig. 1a). Several unrelated danger-associated signals induce the proteasome-mediated degradation of the NT fragments of CARD8 and/or NLRP1, thereby releasing free CT fragments that can assemble into inflammasomes. This process has been dubbed ‘functional degradation’7-10.
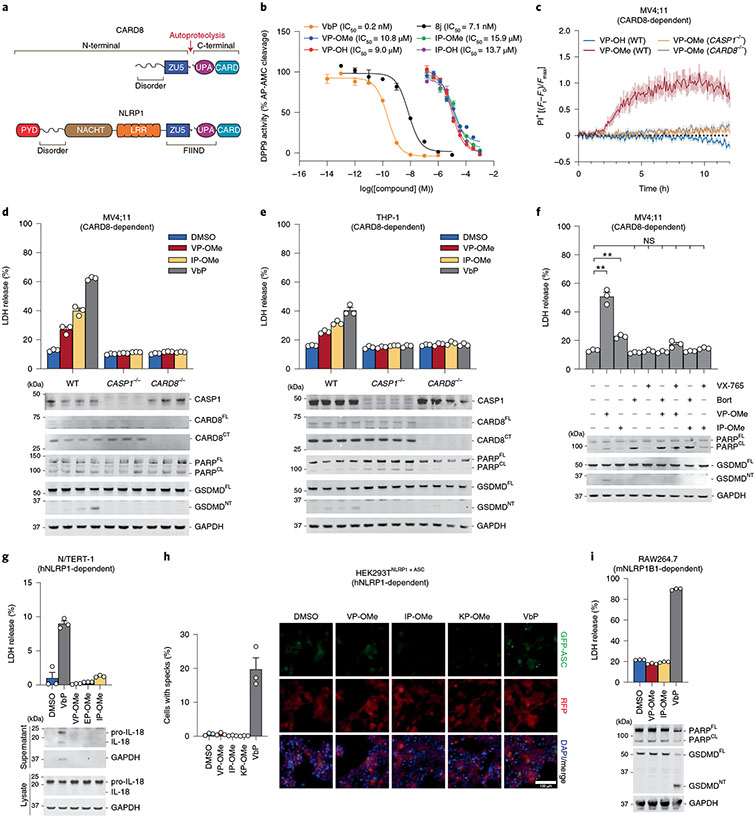
Fig. 1 ∣. Esterified Xaa-Pro dipeptides selectively activate the CARD8 inflammasome.
a, Domain organizations of human NLRP1 and CARD8. Both proteins undergo autoproteolysis between the ZU5 and UPA subdomains of the FIIND. b, Inhibition of recombinant DPP9 activity by the indicated compounds. IC50 values are shown in parentheses. c, The indicated MV4;11 cells were treated with compounds (1 mM) and monitored for PI uptake. Ft, Fluorescence measurement at time t; F0, fluorescence measurement at time 0; Fmax, maximum fluorescence measurement obtained in the experiment. d,e, MV4;11 (d) or THP-1 (e) cells were treated with XP-OMes (1 mM) or VbP (2 μM) for 14 h (d) or 24 h (e) before LDH release and immunoblot analyses. f, MV4;11 cells were treated with VP-OMe (2 mM), IP-OMe (1 mM), bortezomib (bort; 1 μM) and/or VX-765 (50 μM) as indicated for 6 h before LDH release and immunoblot analyses. ***P < 0.001, **P < 0.01 by two-sided Students t-test. NS, not significant. The indicated significant P values are as follows: VP-OMe versus DMSO = 0.0054, IP-OMe versus DMSO = 0.0020. g, N/TERT-1 immortalized keratinocytes were treated with XP-OMes (1 mM) or VbP (10 μM) for 16 h before LDH release and immunoblot analyses. h, HEK 293T cells were transfected with plasmids expressing FLAG-tagged NLRP1 and GFP-tagged ASC and treated with XP-OMes (1 mM) or VbP (10 μM) for 24 h. ASC speck formation was assessed by fluorescence microscopy. Representative images and average cells with specks (%) ± s.e.m. are shown. Scale bar, 100 μm. h, RAW264;7 cells were treated with XP-OMes (1 mM) or VbP (2 μM) for 24 h before LDH release and immunoblot analyses. Data in b (n = 4), c (n = 4) and d-i (n = 3) are presented as means ± s.e.m. of replicates. All data, including immunoblots, are representative of three or more independent experiments.
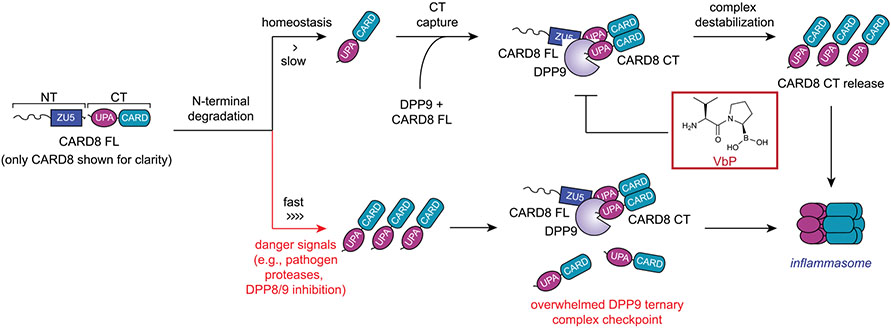
The cytosolic serine dipeptidyl peptidases DPP8 and DPP9 (DPP8/9) directly associate with the FIINDs of CARD8 and NLRP1 and repress inflammasome formation via at least two distinct mechanisms11-16 (Extended Data Fig. 1). First, the enzymatic activities of DPP8/9 restrain the proteasome-mediated degradation of the CARD8 and NLRP1 NT fragments7,13,16,17. The molecular details of the DPP8/9 inhibitor-induced proteasome pathway, which appears to specifically target disordered and misfolded proteins for degradation17, have not been fully established. Second, the complex formed between DPP9 (or DPP8) and the full-length (FL) PRR (either NLRP1 or CARD8) physically sequesters one free CT fragment and thereby prevents inflammasome formation by low levels of the free CT fragment relative to the FL PRR18-20. The CARD8FL–CARD8CT–DPP9 and NLRP1FL–NLRP1CT–DPP9 ternary complexes are similar overall, but only NLRP1 directly interacts with the DPP9 active site19. This contact is mediated by the neo-N terminus of the NLRP1CT fragment.
Several potent small-molecule inhibitors of DPP8/9, including Val-boroPro (VbP), compound 8j21 and 1G244, activate the CARD8 and NLRP1 inflammasomes11,12. These compounds both accelerate NLRP1NT degradation and directly disrupt the NLRP1FL–NLRP1CT–DPP9 ternary complex14,16,18,20, and it is currently thought that both activities contribute to NLRP1 inflammasome formation. These potent DPP8/9 inhibitors also induce CARD8NT degradation7,16, but they do not similarly disrupt the CARD8FL–CARD8CT–DPP9 complex structure in vitro16,19. Intriguingly, however, VbP nevertheless appears to destabilize the CARD8 ternary complex in cells, albeit via an unknown, and possibly indirect, mechanism19.
In addition to their distinct NT domain organizations (Fig. 1a) and interactions with DPP919, CARD8 and NLRP1 also differ in their expression patterns, inflammasome assembly mechanisms and biological outputs. CARD8 is highly expressed and mediates pyroptosis in hematopoietic cells, including T cells, B cells, monocytes and macrophages13,22,23. NLRP1 is highly expressed and mediates pyroptosis in barrier cells and tissues, including keratinocytes and airway epithelial cells14,24-27. In addition, the human NLRP1CT requires the adapter protein ASC (apoptosis-associated speck-like protein containing a CARD) to recruit CASP1, whereas the CARD8CT directly recruits and activates CASP1 without ASC28-30. ASC-dependent inflammasomes, including the human NLRP1 inflammasome, activate a form of CASP1 that cleaves and activates the pore-forming protein gasdermin D (GSDMD) as well as the inflammatory cytokines pro-interleukin-1β (pro-IL-1β) and pro-IL-18, triggering a highly inflammatory form of pyroptosis. By contrast, ASC-independent inflammasomes, including the CARD8 inflammasome, only cleave and activate GSDMD and not the inflammatory cytokines, and thereby trigger a less inflammatory form of lytic cell death28. Consistent with the expression and inflammatory nature of the NLRP1 inflammasome, germline-encoded mutations in NLRP1 cause a number of skin and respiratory tract inflammatory syndromes23-26. No mutations in CARD8 are known that cause human disease.
Potent DPP8/9 inhibitors, and most notably VbP, have served as important tools to characterize CARD8 and NLRP1, but chemical probes have not yet been discovered that selectively modulate only one of these inflammasomes. Because NLRP1 and CARD8 have key differences, we reasoned that it should be possible to identify such selective small-molecule probes, and that these probes would be useful to further characterize the specific biological functions and therapeutic potential of these signaling platforms. In this Article we identify and characterize M24B aminopeptidase inhibitors, which induce the accumulation of proline-containing peptides that weakly inhibit DPP8/9 as selective activators of the CARD8 inflammasome.
Results
Xaa-Pro dipeptides selectively activate CARD8.
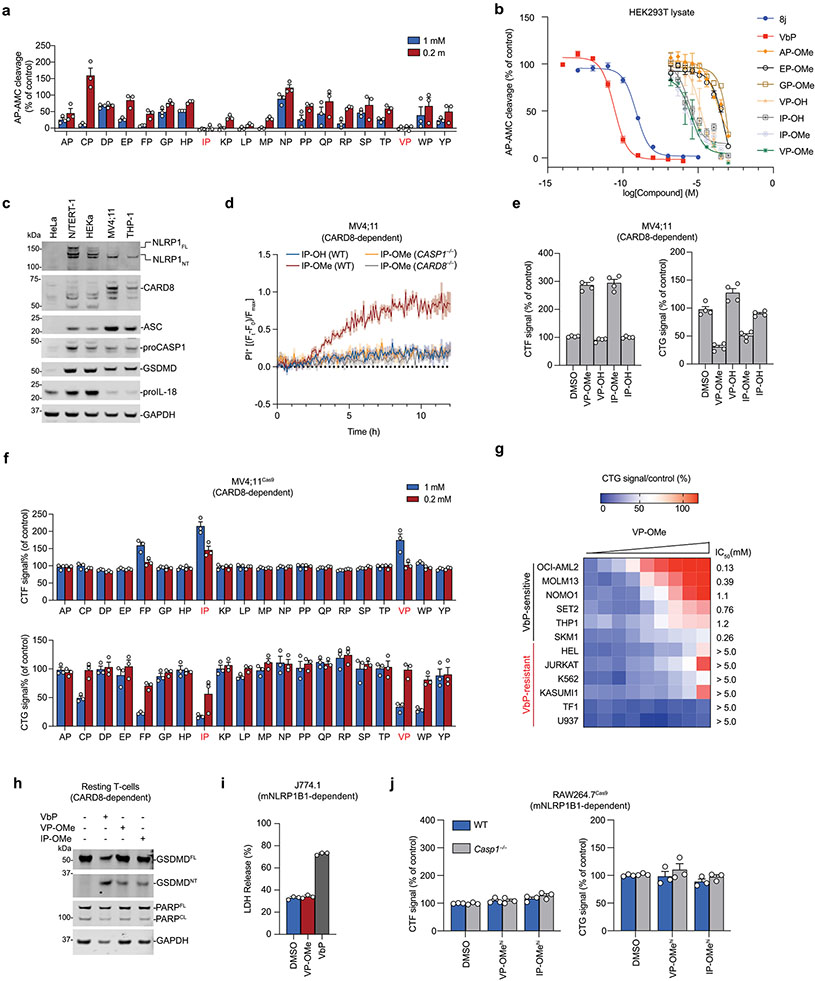
DPP8/9 belong to the S9B serine family of dipeptidyl peptidases, which also includes DPP4 and fibroblast activation protein (FAP). DPP8/9, like DPP4, preferentially cleave after proline residues in the second position of polypeptide substrates, releasing Xaa-Pro dipeptides (where Xaa is any amino acid)31-34. High concentrations (approximately tens to hundreds of micromolar) of several Xaa-Pro dipeptides have been reported to inhibit DPP435-37, and we similarly observed that many Xaa-Pro dipeptides also inhibit recombinant DPP9 (Extended Data Fig. 2a). Ile-Pro and Val-Pro were the two most potent DPP9 inhibitors of the 20 dipeptides, with half-maximum inhibitory concentration (IC50) values of ~10–15 μM in an Ala-Pro-aminomethylcoumarin (AP-AMC)-based DPP9 substrate assay (Fig. 1b and Extended Data Fig. 2b). As such, these dipeptides are several orders of magnitude less potent DPP9 inhibitors than compound 8j and VbP (Fig. 1b and Extended Data Fig. 2b).
We next wanted to evaluate whether these weak DPP8/9 inhibitors could induce pyroptosis. We have previously demonstrated that VbP and 8j induce CARD8-dependent pyroptosis in several human acute myeloid leukemia (AML) cancer cell lines, including MV4;11 and THP-1 cells13 (Extended Data Fig. 2c). We found that high concentrations (1 mM) of the unprotected Ile-Pro and Val-Pro (IP-OH and VP-OH) dipeptides did not induce lytic cell death in MV4;11 cells, as measured by uptake of propidium iodide (PI, which reports on membrane permeability) (Fig. 1c and Extended Data Fig. 2d), Cell-TiterGlo (CTG, which measures intracellular adenosine triphosphate (ATP) levels) and Cyto-ToxFluor (CTF, which detects extracellular protease activity released after membrane rupture) assays (Extended Data Fig. 2e). We reasoned that the charged CT carboxylic acids might limit cell permeability and account for the lack of pyroptosis induction. As esterification often increases the cell permeability of carboxylate-containing molecules38,39, we next synthesized methyl ester versions of the Xaa-Pro dipeptides (XP-OMes). Notably, esterification of Val-Pro and Ile-Pro (VP-OMe and IP-OMe) did not impact DPP9 inhibition (Fig. 1b and Extended Data Fig. 2b). Consistent with our hypothesis regarding permeability, IP-OMe and VP-OMe, and to a lesser extent the methyl ester version of Phe-Pro (FP-OMe), were cytotoxic to MV4;11 cells (Fig. 1c and Extended Data Fig. 2d-f). Moreover, we found that VP-OMe also induced cell death (at concentrations <5 mM) in several additional VbP-sensitive cell lines, including THP-1 cells, but not in VbP-resistant cell lines, indicating a similar mechanism of action (Extended Data Fig. 2g)13.
We next wanted to confirm that VP-OMe and IP-OMe indeed triggered CARD8-dependent pyroptosis in these sensitive cells. As expected, we found that both dipeptides, like VbP, induced the release of intracellular lactate dehydrogenase (LDH) and GSDMD cleavage, two key hallmarks of pyroptosis, in wild-type (WT) MV4;11 and THP-1 cells (Fig. 1d,e). This pyroptotic cell death was completely dependent on both CASP1 and CARD8, as evidenced by the lack of PI uptake, GSDMD cleavage and LDH release in CASP1−/− and CARD8−/− MV4;11 and THP-1 cells (Fig. 1c-e and Extended Data Fig. 2d). Moreover, VbP also activates the CARD8 inflammasome in resting T cells22,23, and we found that both VP-OMe and IP-OMe similarly induced GSDMD cleavage in this primary cell type as well (Extended Data Fig. 2h). Consistent with a ‘functional degradation’ mechanism, pretreatment of MV4;11 cells with either the proteasome inhibitor bortezomib or the CASP1 inhibitor VX-765 completely blocked VP-OMe- and IP-OMe-induced LDH release and GSDMD cleavage (Fig. 1f). It should be noted that bortezomib, but not the dipeptides, caused considerable apoptosis, as evidenced by PARP (poly ADP-ribose polymerase 1) cleavage (Fig. 1d-f). Overall, these data demonstrate that VP-OMe and IP-OMe activate the CARD8 inflammasome.
In addition to activating CARD8, VbP also activates the human and rodent NLRP1 inflammasomes (mice do not have a CARD8 homolog, but have two functional NLRP1 alleles, NLRP1A and NLRP1B). We next wanted to test whether the dipeptide esters similarly activate the NLRP1 inflammasome. Human immortalized N/TERT-1 keratinocytes express the key components of the human NLRP1 inflammasome14, and we therefore treated these cells with VbP, VP-OMe, IP-OMe or EP-OMe (as a negative control). As expected, VbP induced LDH release as well as IL-18 cleavage and release in N/TERT-1 keratinocytes (GSDMD cleavage is difficult to observe in these cells25; Fig. 1g). In contrast, however, the dipeptides did not induce NLRP1 inflammasome activation in these keratinocytes. We next reconstituted the human NLRP1 inflammasome in HEK 293T cells by ectopically expressing human NLRP1 and green fluorescent protein (GFP)-tagged ASC, and found that only VbP, but not VP-OMe, IP-OMe or KP-OMe (a negative control), triggered the formation of ASC specks (Fig. 1h). Similarly, only VbP activated mouse NLRP1B allele 1 (mNLRP1B1) in RAW264.7 and J774.1 macrophages, as determined by LDH release and GSDMD cleavage assays (Fig. 1i and Extended Data Fig. 2i). Notably, we found that even fivefold higher concentrations of VP-OMe and IP-OMe (5 mM) still failed to trigger pyroptosis in RAW264.7 cells (Extended Data Fig. 2j). Thus, VP-OMe and IP-OMe do not activate the human or mouse NLRP1 inflammasomes.
CQ31 inhibits prolidase activity.
The experiments above show that it is possible to selectively activate the CARD8 inflammasome with chemical probes, albeit ones requiring very high concentrations (~1 mM) to trigger this response. We next wanted to identify more drug-like molecules that selectively activate CARD8 at low micromolar concentrations. Intriguingly, it is currently thought that only one mammalian enzyme, the Mn2+-dependent metalloprotease called prolidase (PEPD), can cleave Xaa-Pro dipeptides and release the free amino acids (Fig. 2a)40,41. We therefore hypothesized that PEPD inhibition might cause an accumulation of endogenous Xaa-Pro dipeptides in cells and activate CARD8, essentially mimicking the effects of the exogenously added esterified dipeptides.
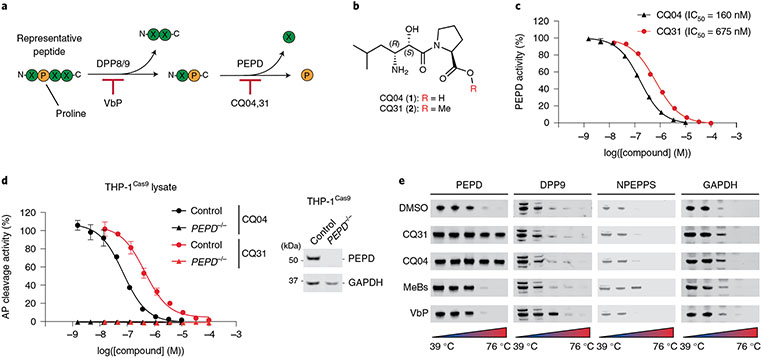
Fig. 2 ∣. CQ04 and CQ31 inhibit PEPD.
a, Schematic of DPP8/9 and PEPD activity. b, Structures of CQ04 and CQ31. c, Inhibition of recombinant PEPD activity. IC50 values are shown in parentheses. d, Evaluation of PEPD activity (AP cleavage) in control and PEPD−/− THP-1 cell lysates treated with the indicated compounds. The release of free alanine in c and d was determined using an alanine assay kit (Sigma). Data in c and d are presented as means ± s.e.m. of three replicates. e, CETSA analyses of the indicated compounds (10 μM) in HEK 293T lysates. All data, including immunoblots and CETSA, are representative of three or more independent experiments.
In the late 1990s, the pseudodipeptide (2S,3R)-3-amino-2-hydroxy-5-methyl-hexanoyl-proline (AHMH-Pro; Fig. 2b, called CQ04 or 1 here), which contains the 3-amino-2-hydroxy acid pharmacophore of the natural product metalloprotease inhibitor bestatin, was reported to potently inhibit PEPD42, but this compound was not studied in detail. We next synthesized both AHMH-Pro (CQ04) and the corresponding methyl ester (CQ31, 2; Fig. 2b). We found that CQ04 and CQ31 inhibited recombinant human PEPD with IC50 values of 160 and 675 nM, respectively (Fig. 2c), and that both compounds completely blocked prolidase activity (that is, Ala-Pro dipeptide cleavage) in THP-1 cell lysates in a dose-dependent manner. Consistent with the idea that PEPD is the only enzyme that can cleave Xaa-Pro dipeptides, we found that THP-1 PEPD knockout cell lysates, like inhibitor-treated lysates, were unable to cleave the Ala-Pro dipeptide (Fig. 2d). CQ04 and CQ31 both had at least some selectivity for PEPD, as they did not inhibit a number of bestatin-sensitive aminopeptidases nor DPP9 (Supplementary Table 1). In addition, both CQ04 and CQ31 stabilized PEPD, but not DPP9 nor the bestatin-sensitive aminopeptidase NPEPPS, in a cellular thermal shift assay (CETSA) in HEK 293T lysates (Fig. 2e).
CQ31 selectively activates CARD8.
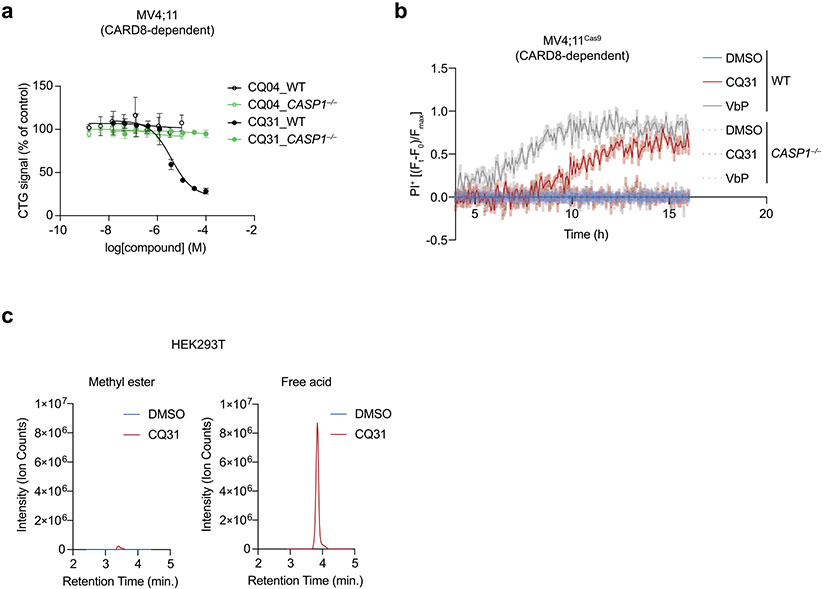
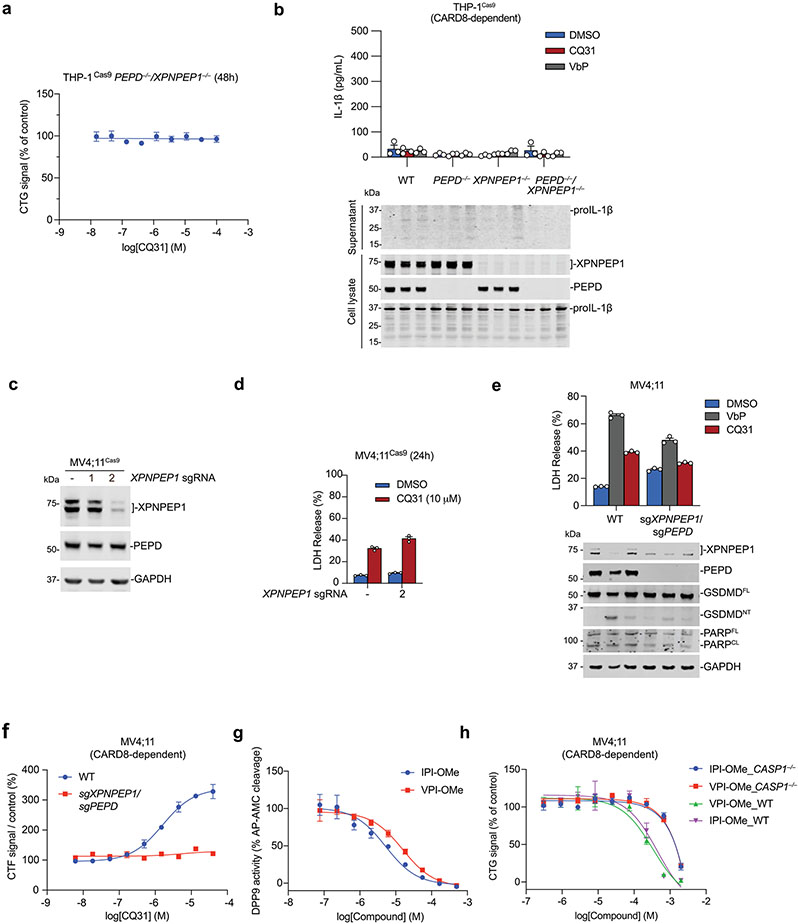
We next treated WT and CASP1−/− MV4;11 cells with either CQ31 or CQ04 to determine whether either compound induced cell death (Extended Data Fig. 3a,b). We found that CQ04 was not cytotoxic to either cell line. However, CQ31 killed wild-type (WT), but not CASP1−/−, cells, consistent with the induction of pyroptosis. These data strongly indicated that the methyl ester was necessary for cell penetrance of this scaffold, similar to the Xaa-Pro dipeptides investigated above. Such methyl esters are often rapidly removed by intracellular esterases to release the free acid39, and we found that CQ31 was completely converted into CQ04 within cells (Extended Data Fig. 3c). Thus, CQ31 is a prodrug that enables the delivery and release of the more potent acid inside cells.
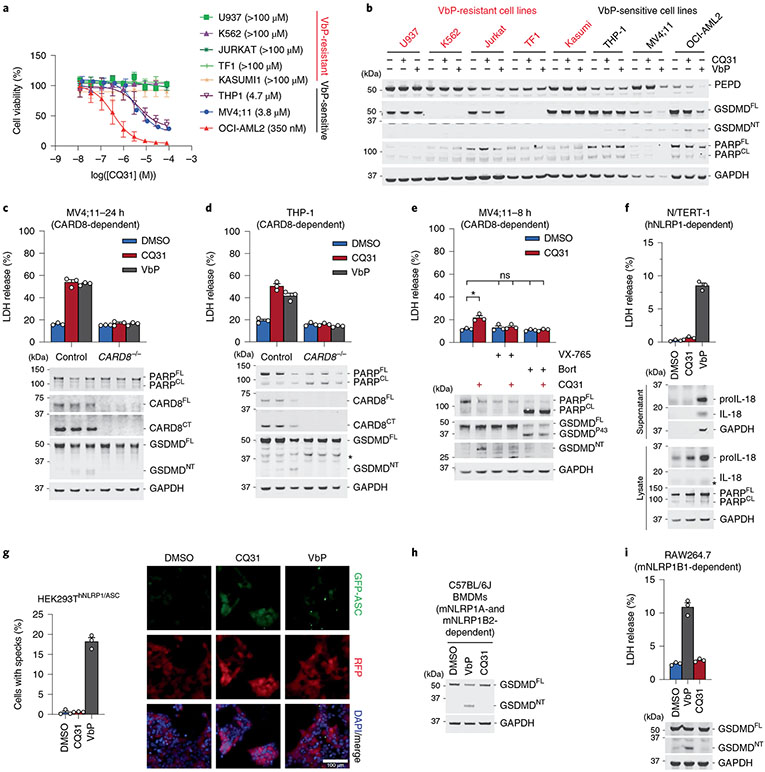
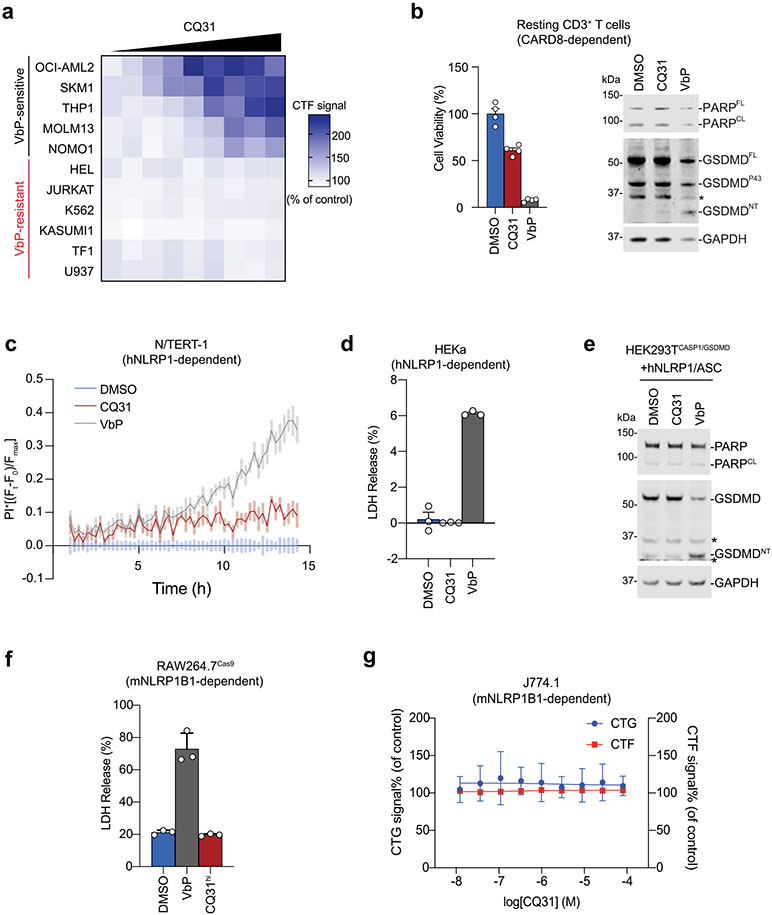
We next wanted to evaluate the impact of CQ31 against additional cell types. Like the XP-OMes, we found that CQ31-induced pyroptosis, as evidenced by GSDMD cleavage, LDH release and the loss of cell viability, in several VbP-sensitive AML cancer cell lines, including in THP-1 and OCI-AML2 cells (Fig. 3a-e and Extended Data Fig. 4a). By contrast, CQ31 did not kill any of the VbP-resistant cancer cell lines (Fig. 3a,b and Extended Data Fig. 4a). Notably, CQ31 killed the sensitive cell lines with IC50 values in the high nanomolar to low micromolar concentration range, and therefore is at least two orders of magnitude more potent than the esterified Xaa-Pro dipeptides (Extended Data Fig. 2f). As expected, CQ31 did not induce cell death in CARD8−/− MV4;11 and THP-1 cells (Fig. 3c,d), and both VX-765 and bortezomib blocked CQ31-induced cell death in WT MV4;11 cells (Fig. 3e). In addition to these cancer cell lines, we found that CQ31, like VbP, also induced pyroptosis in resting primary T cells (Extended Data Fig. 4b). Thus, CQ31 activates the CARD8 inflammasome in a manner consistent with ‘functional degradation’.
Fig. 3 ∣. CQ31 selectively activates the CARD8 inflammasome.
a, Viability of cell lines after treatment with CQ31 (24 h), assessed by CTG. b, Immunoblots of lysates from the indicated cell lines treated with CQ31 (10 μM) or VbP (10 μM) for 24 h. c,d, The indicated MV4;11 (c) and THP-1 (d) cells were treated with CQ31 (10 μM) or VbP (10 μM) for 24 h before LDH release and immunoblot analyses. e, MV4;11 cells were treated with CQ31 (10 μM) for 2 h before treating with bortezomib (1 μM) or VX-765 (50 μM) for 6 h before LDH release and immunoblot analyses. f, N/TERT-1 keratinocytes were treated with CQ31 (10 μM) or VbP (10 μM) for 16 h before LDH release and immunoblot analyses. *P < 0.05 by two-sided Students t-test. NS, not significant. The indicated significant P value is as follows: CQ31 versus DMSO = 0.0223. g, HEK 293T cells were transfected with plasmids expressing FLAG-tagged NLRP1 and GFP-tagged ASC and treated with CQ31 (10 μM) or VbP (10 μM) for 24 h. ASC speck formation was assessed by fluorescence microscopy. Representative images and average cells with specks (%) ± s.e.m. are shown. Scale bar, 100 μm. h,i, BMDMs from C57BL/6J mice (h) or RAW264.7 cells (i) were treated with CQ31 (10 μM) or VbP (10 μM) for 24 h or 6 h, respectively, before LDH release and/or immunoblot analyses. Data in a (n = 4), c–g (n = 3) and i (n = 3) are presented as means ± s.e.m. of replicates. All data, including immunoblots, are representative of three or more independent experiments.
By contrast, CQ31, unlike VbP, did not activate the human NLRP1 inflammasome in immortalized N/TERT-1 keratinocytes (Fig. 3f and Extended Data Fig. 4c) or in primary epidermal keratinocytes (HEKa; Extended Data Fig. 4d). Similarly, we found that CQ31 did not trigger ASC speck formation (Fig. 3g) or GSDMD cleavage (Extended Data Fig. 4e) in HEK 293T cells ectopically expressing human NLRP1 inflammasome components. To test whether CQ31 could activate rodent NLRP1 inflammasomes, we applied CQ31 to primary bone marrow macrophages (BMDMs) from C57BL/6J mice, which express both NLRP1 alleles A and B2, as well as on RAW264.7 and J774.1 macrophages, both of which express NLRP1B1. CQ31 did not appear to induce any pyroptosis in any of these rodent cells, even at high (100 μM) concentrations (Fig. 3h,i and Extended Data Fig. 4f,g). These data show that CQ31, like VP- and IP-OMe, is a selective CARD8 activator, but notably acts at much more pharmacologically relevant concentrations.
CQ31 acts via inhibition of DPP8/9 activity.
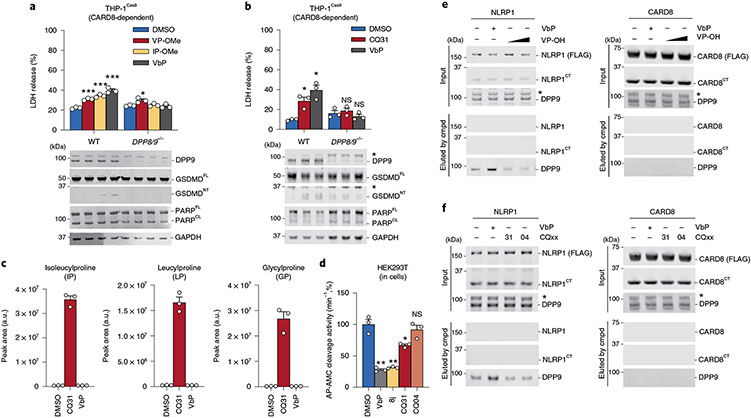
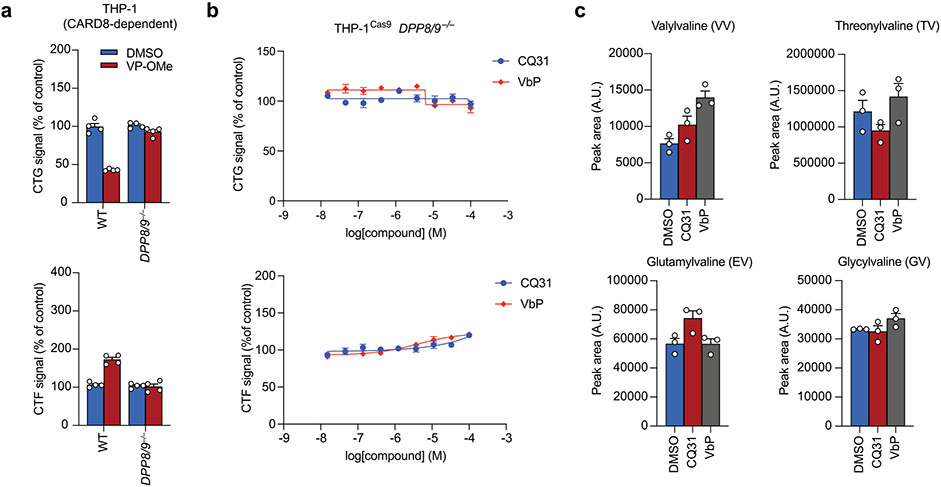
We reasoned that the XP-OMes and CQ31 activated the CARD8 inflammasome by inhibiting DPP8/9 activity in cells. Consistent with this mechanism, we found that VP-OMe, IP-OMe and CQ31, like VbP, did not induce any additional pyroptosis in DPP8/9−/− THP-1 cells11, as determined by LDH release, GSDMD cleavage and CTG and CTF assays (Fig. 4a,b and Extended Data Fig. 5a,b). CQ31, unlike the other compounds, does not directly inhibit DPP8/9 (Supplementary Table 1). As described above, we instead hypothesized that it induced the accumulation of endogenous Xaa-Pro dipeptides in cells and thereby indirectly inhibited DPP8/9. To explore this premise, we next treated HEK 293T cells (which do not express inflammasome components and therefore do not undergo pyroptosis) with CQ31 or VbP before extracting and analyzing metabolites by liquid chromatography-mass spectrometry (LC-MS). Indeed, we found that CQ31, but not VbP, induced the accumulation of several Xaa-Pro dipeptides (Fig. 4c), including Ile-Pro, in these cells. As expected, CQ31 did not induce the accumulation of dipeptides, including Val-Val and Gly-Val, that lack CT proline residues (Extended Data Fig. 5c). To directly confirm that CQ31 treatment causes DPP8/9 inhibition in cells, we treated living cells with VbP, 8j, CQ31 or CQ04 (as a negative control) for 6 h before adding the cell-permeable, fluorogenic DPP8/9 substrate AP-AMC. We discovered that CQ31, but not CQ04, significantly reduced the rate of AP-AMC cleavage (Fig. 4d). Taken together, these data indicate that CQ31 inhibits PEPD activity in cells, resulting in the build-up of Xaa-Pro dipeptides, the inhibition of DPP8/9’s enzymatic activity and activation of the CARD8 inflammasome.
Fig. 4 ∣. CQ31 indirectly inhibits DPP8/9 activity.
a,b, The indicated THP-1 cells were treated with VP-OMe (1 mM), IP-OMe (1 mM) (a) or CQ31 (10 μM) (b) and VbP (2 μM) for 24 h before LDH release and immunoblot analyses. c, HEK 293T cells were treated with vehicle control (DMSO), CQ31 (10 μM) or VbP (10 μM) for 24 h. Intracellular metabolites were extracted and dipeptide concentrations were measured by LC-MS. d, HEK 293T cells were treated with the indicated compounds (10 μM, 6 h) before treatment with the DPP4 inhibitor sitagliptin (1 μM, to block any DPP4 activity in the media) for 1 h and assaying for AP-AMC (2.5 μM) cleavage. Data in a–d are presented means ± s.e.m. of three replicates. ***P < 0.001, **P < 0.01, *P < 0.05 by two-sided Students t-test. NS, not significant. Indicated significant P values versus DMSO are as follows: VbP = 0.0087, 8j = 0.0121, CQ31 = 0.0449. e,f, FLAG-tagged NLRP1 and CARD8 from HEK 293T cells (input) were immobilized on anti-FLAG beads and treated with VbP (10 μM) or VP dipeptide (0.5, 5 mM) (e) or CQ31 or CQ04 (10 μM) (f). DPP9 displaced from the complex was evaluated by immunoblotting. Cmpd, compound. All LDH, enzymatic activity, immunoprecipitation and immunoblot data are representative of three or more independent experiments.
We next wanted to determine the mechanistic basis for the selective activation of the CARD8 inflammasome. As described above, VbP not only inhibits DPP8/9’s enzymatic activity, but also directly disrupts the NLRP1FL–NLRP1CT–DPP9 ternary complex. We hypothesized that weak-binding Xaa-Pro dipeptides might be unable to compete with the NLRP1CT for binding to the DPP9 active site and disrupt the ternary complex. To explore this premise, we expressed FLAG-tagged NLRP1 or CARD8 in HEK 293T cells, immobilized the FLAG-tagged protein on anti-FLAG beads, incubated the beads with the various compounds, and assessed DPP9 elution by immunoblotting. As expected, VbP disrupted the NLRP1–DPP9 interaction and caused DPP9 to be eluted from the solid support (Fig. 4e,f). By contrast, the Val-Pro dipeptide (as well as CQ04 and CQ31, which do not directly interact with DPP9) had no impact on the NLRP1 ternary complex (Fig. 4e,f). No compounds disrupted the CARD8–DPP9 interaction, consistent with the lack of interaction between the DPP9 active site and CARD819. Overall, these data indicate that XP-OMes, and consequently CQ31, probably cannot activate NLRP1 because of an inability to disrupt the repressive DPP9–NLRP1 ternary complex.
CQ31 engages a target in addition to PEPD.
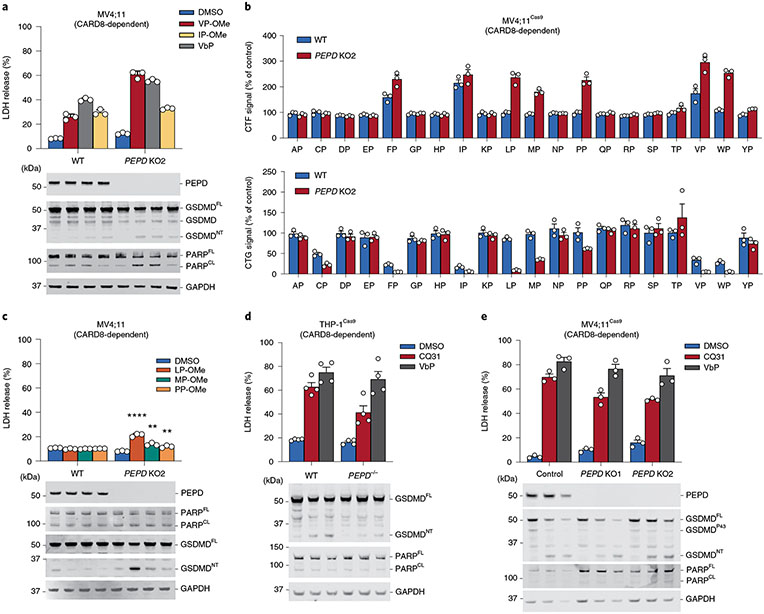
Our results strongly indicate that PEPD plays a critical role in catabolizing XP dipeptides and thereby preventing the activation of the CARD8 inflammasome. Based on this reasoning, we predicted that PEPD−/− cells would be more sensitive to esterified XP dipeptides, as they would not be catabolized into free amino acids intracellularly. Indeed, we found that VP-OMe and IP-OMe induced more LDH release and GSDMD cleavage in PEPD−/− MV4;11 cells than in control cells (Fig. 5a). In addition, we found that several esterified dipeptides (including LP-, PP- and MP-OMe) that were inactive in control cells induced pyroptosis in PEPD−/− knockout cells (Fig. 5b,c). Thus, PEPD cleaves and deactivates XP dipeptides in cells.
Fig. 5 ∣. CQ31 engages a target in addition to PEPD.
a, The indicated MV4;11 cells were treated with VbP (10 μM) or the indicated XP-OMe (1 mM) for 6 h before LDH release and immunoblot analyses. b, WT and PEPD knockout MV4;11 cells were treated with XP-OMes (1 mM, 14 h) before CTG and CTF analyses. c–e, The indicated THP-1 and MV4;11 cells were treated with CQ31 (10 μM), VbP (10 μM) or XP-OMes (1 mM) for 14 h (c) or 24 h (d,e) before LDH release and immunoblot analyses. Data are presented as means ± s.e.m. of three replicates. All data, including immunoblots, are representative of three or more independent experiments.
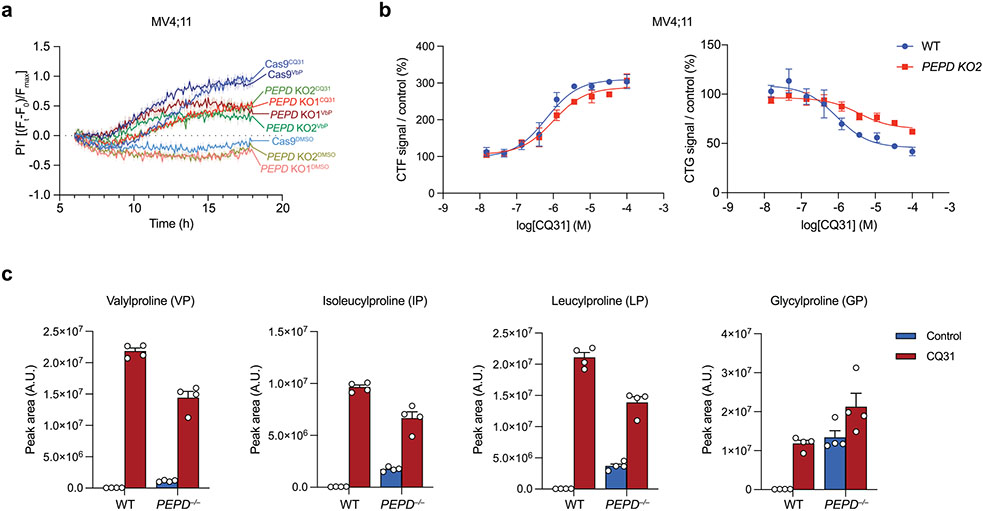
We next wanted to definitively establish that PEPD was the only key target of CQ31. In particular, we hypothesized that PEPD−/− cells would be completely resistant to CQ31, as CQ31 should have no effect when the key target was absent. Surprisingly, however, we found that CQ31 still induced LDH release, PI uptake and GSDMD cleavage in these knockout cells (Fig. 5d,e and Extended Data Fig. 6a,b). Moreover, PEPD−/− MV4;11 cells had higher basal levels of several XP dipeptides relative to control cells, as expected, but CQ31 nevertheless increased these levels even further (Extended Data Fig. 6c). Together, these data revealed that CQ31 inhibits at least one enzyme in addition to PEPD that contributes to the accumulation of proline-containing peptides, DPP8/9 inhibition and CARD8 activation. Also, these results show that, even if PEPD is the primary enzyme that cleaves XP dipeptides (Fig. 2d), at least one other enzyme can catalyze this reaction in living cells, at least to some extent.
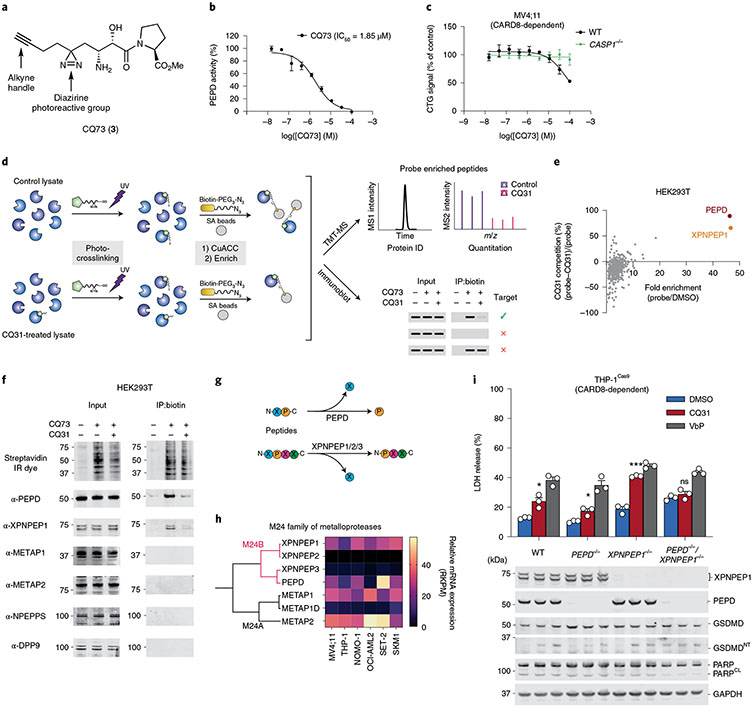
CQ31 inhibits additional M24B aminopeptidases.
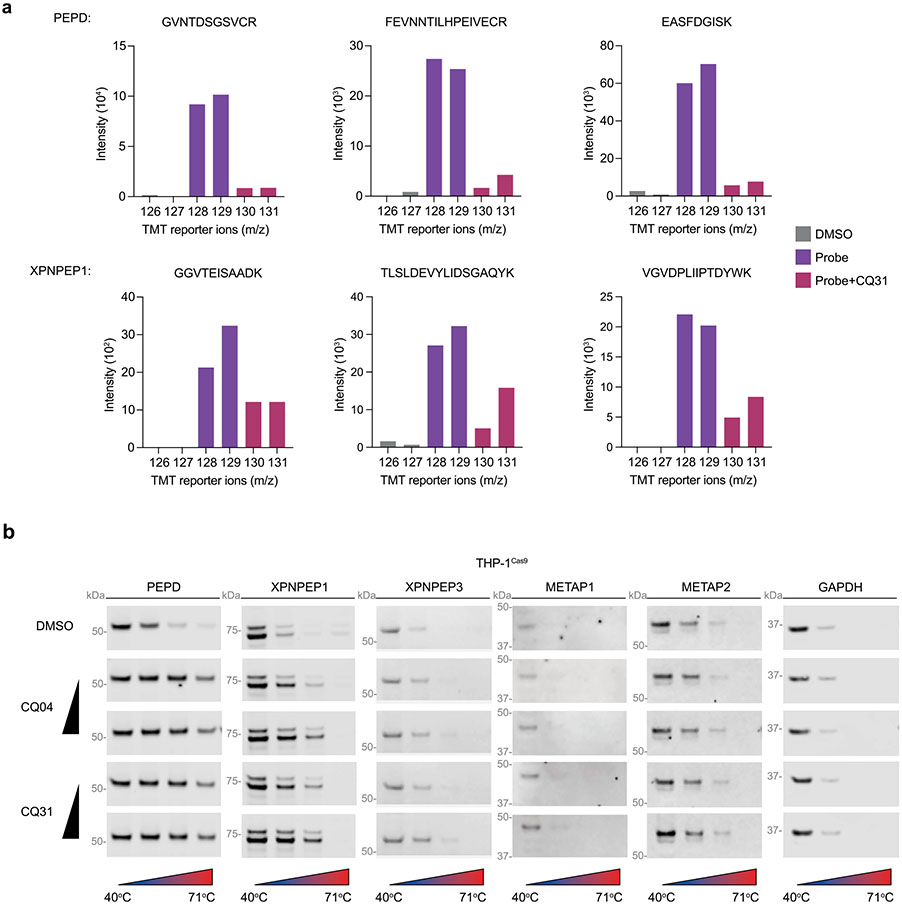
To unbiasedly identify all protein targets of CQ31, we synthesized a CQ31 analog containing a photoactivatable diazirine group and a clickable alkyne handle, which together enable the covalent modification, enrichment and identification of target proteins (CQ73, 3; Fig. 6a). We confirmed that CQ73 retained the ability to inhibit recombinant PEPD (Fig. 6b) and induce caspase-1-dependent pyroptosis in MV4;11 cells (Fig. 6c), demonstrating that this modified structure still engages the key targets in cells. We next assessed the overall interaction profile of CQ73 in cell lysates. Briefly, we incubated HEK 293T lysates with CQ73 in the presence or absence of CQ31 before exposing the samples to UV light, coupling CQ73-modified proteins to an azide-biotin enrichment tag by azide-alkyne cycloaddition chemistry, and enriching labeled proteins on streptavidin-agarose beads. We then performed on-bead trypsinization, tandem mass tag (TMT) labeling, and quantitative mass spectrometry analysis to identify these proteins (Fig. 6d). Strikingly, this assay revealed that CQ73 interacted with just two targets, PEPD and XPNPEP1, and that both interactions were competed with CQ31 (Fig. 6e, Extended Data Fig. 7a and Supplementary Data 1). We confirmed these two interactions by immunoblotting the CQ73-enriched fractions (Fig. 6d,f). Moreover, we found that CQ31 and CQ04 stabilize both PEPD and XPNPEP1 in a CETSA assay in THP-1 cell lysates (Extended Data Fig. 7b), and that CQ04 inhibits XPNPEP1 enzymatic activity (Supplementary Table 2).
Fig. 6 ∣. Dual inhibition of XPNPEP1 and PEPD activates CARD8.
a, Structure of CQ73. b, CQ73 inhibits recombinant PEPD activity. The IC50 value is shown in parentheses. c, The indicated MV4;11 cells were treated with CQ73 for 24 h before CTG analysis. d, Schematic of the target identification protocol using the CQ73 probe. e, Scatter plots depicted the proteins enriched by CQ73 and competed by CQ31 as determined by TMT-MS. f, Immunoblots of lysates from HEK 293T cells before (input) and after streptavidin enrichment (IP:biotin) following the scheme in d. g, Schematic of the known enzymatic function of PEPD and XPNPEPs. h, Cladogram depicting all M24 aminopeptidases and their messenger RNA (mRNA) abundances in the indicated cell lines. i, The indicated THP-1 cells were treated with control (DMSO), CQ31(10 μM) or VbP (10 μM) for 48 h before LDH release and immunoblot analyses. Data in b, c and i are presented as means ± s.e.m. of three replicates. Data in b, c and i, including immunoblots, are representative of three or more independent experiments.
PEPD, XPNPEP1, XPNPEP2 and XPNPEP3 are the four members of the M24B family of Mn2+-dependent metalloenzymes that share the rare ability to cleave the NT amino acid from peptides with proline in the second position (Fig. 6g,h). It is thought that PEPD can only cleave dipeptides41, whereas XPNPEP1, 2 and 3 cleave longer substrates42,43. Notably, XPNPEP2 is not expressed and XPNPEP3 is only weakly expressed in the CQ31-sensitive AML cells (Fig. 6h), perhaps accounting for their absence among the CQ73-enriched proteins. Regardless, CQ04 slightly stabilized XPNPEP3 in a CETSA analysis (Extended Data Fig. 7b) and inhibited XPNPEP3 in an enzymatic assay (Supplementary Table 2), indicating that this probe probably targets the entire M24B family. The M24A aminopeptidases, which include METAP1 and METAP2 (Fig. 6h), are the next most similar enzymes to the M24B aminopeptidases. We confirmed that the M24A enzymes were not targets of CQ31 as determined by competition assays with CQ73 (Fig. 6f), CETSA analyses (Extended Data Fig. 7b) and enzymatic activity assays (Supplementary Table 2). Overall, these data not only reveal XPNPEP1 as the likely other key target of CQ31, but also importantly demonstrate proteome-wide selectivity of CQ31 for just the M24B aminopeptidases.
To show that XPNPEP1 was indeed the only other important target of CQ31, we generated XPNPEP1 single knockouts and XPNPEP1/PEPD dual knockouts in THP-1 cells. We found that both PEPD and XPNPEP1 single knockouts were sensitive to CQ31 (Fig. 6i), but that the XPNPEP1/PEPD double knockout was completely resistant (Fig. 6i and Extended Data Fig. 8a,b). We next used single-guide RNAs (sgRNAs) to target XPNPEP1 in MV4;11 cells (Extended Data Fig. 8c-f). Although we were unable to isolate complete XPNPEP1/PEPD double knockouts in MV4;11 cells (we could only achieve a knockdown of XPNPEP1), probably due to the propensity of these cells to spontaneously undergo CARD8-dependent pyroptosis13, we found that PEPD knockout/XPNPEP1-deficient cells, unlike either knockout alone, were highly resistant to CQ31 (Extended Data Fig. 8c-f). Together, these data show that dual inhibition of PEPD and XPNPEP1 is responsible for CARD8 inflammasome activation in these cells. It should be noted that, based on the known substrate specificity of XPNPEP1, the dual inhibition of these enzymes probably increases XP dipeptides as well as longer proline-containing peptides, which similarly inhibit DPP9 and induce caspase-1-dependent pyroptosis (Extended Data Fig. 8g,h). We speculate that the accumulation of these longer peptides in XPNPEP1−/− cells renders them even more reliant on PEPD and thereby more sensitive to CQ31 (Fig. 6i and Extended Data Fig. 8d), but this requires further study.
Discussion
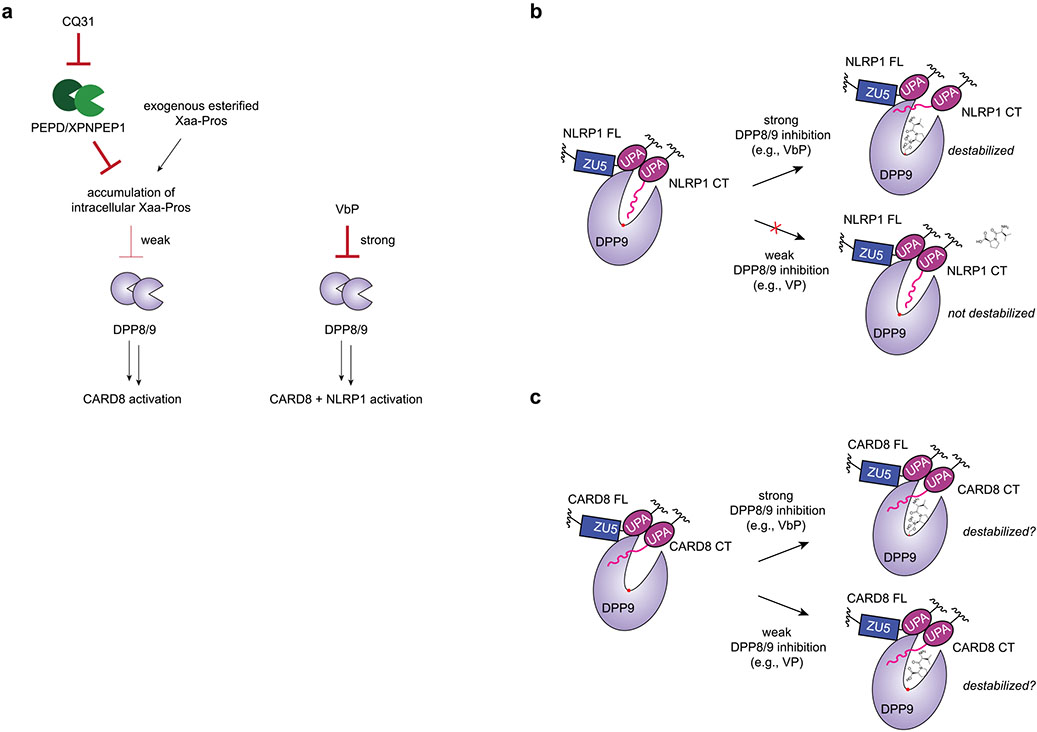
Potent small-molecule inhibitors of DPP8/9, including VbP, were recently discovered to activate the related NLRP1 and CARD8 inflammasomes11-13, and have propelled a number of recent studies of these important innate immune signaling platforms14,20,22,23,25. However, no chemical probes have yet been discovered that selectively stimulate just one of these inflammasomes without the other. Here we have identified and characterized a small molecule called CQ31 that only activates the CARD8 inflammasome. CQ31 inhibits M24B aminopeptidases PEPD and XPNPEP1, which causes the intracellular accumulation of peptides that contain proline in the second position. It should be noted that while CQ31 is highly selective for PEPD and XPNPEP1 in the cells tested, it will probably also inhibit the other two M24B aminopeptidases (XPNPEP2 and 3) in cell types in which they are expressed. These proline-containing peptides then weakly inhibit DPP8/9 in a way that is sufficient to activate the CARD8, but not the NLRP1, inflammasome. We speculate that NLRP1, which is more inflammatory than CARD8, evolved to directly bind to the DPP8/9 active site to prevent this danger signal (that is, proline-containing peptide accumulation) alone from activating this inflammasome.
Notably, our work here reveals important principles regarding the discovery and development of DPP8/9 inhibitors (Extended Data Fig. 9). Briefly, there are at least two ‘functions’ of the DPP8/9 active site: (1) to bind and cleave some poorly defined polypeptide substrates that regulate a proteasome pathway and (2) to bind to the neo-N terminus of the NLRP1CT fragment as part of the repressive NLRP1CT–NLRP1FL–DPP9 ternary complex. Potent DPP8/9 inhibitors, like VbP, interfere with both functions and activate both NLRP1 and CARD8. Weak inhibitors, like proline-containing peptides, only interfere with DPP8/9’s enzymatic activity (and not NLRP1CT fragment binding) and only activate CARD8. Thus, it appears that DPP8/9 inhibitors must be potent enough to interfere with NLRP1CT fragment binding to activate the NLRP1 inflammasome (Extended Data Fig. 9b). It remains unclear whether DPP8/9 inhibitors must also destabilize the CARD8–DPP9 ternary complex to activate the CARD8 inflammasome, but, if they do, the same level of potency is not required (Extended Data Fig. 9c). We anticipate that future drug discovery efforts will be able to build on these insights to develop direct ‘weak’ DPP8/9 inhibitors that selectively activate CARD8. We speculate that it might also be possible to create DPP8/9-binding small molecules that selectively activate NLRP1, for example, by displacing the NLRP1CT fragment while stabilizing the CARD8CT fragment.
In addition to establishing these concepts regarding DPP8/9 inhibitor design, this Article also shows that the M24B metalloenzymes, and in particular prolidase (PEPD) and XPNPEP1, restrain the innate immune system. On that note, germline mutations in the PEPD gene that reduce prolidase activity cause prolidase deficiency, a disorder that is characterized by splenomegaly, lupus-like symptoms, skin lesions and mental retardation40,44. Patients with prolidase deficiency accumulate Xaa-Pro dipeptides in the blood and urine (imidopeptiduria), as expected, but the mechanistic basis for this autoinflammatory condition, and whether it is linked to these Xaa-Pros, has not yet been established. It is tempting to speculate that Xaa-Pro dipeptide accumulation leads to CARD8 inflammasome activation and, possibly, in this chronic condition, NLRP1 activation. The molecular basis of prolidase deficiency and its connection to the NLRP1 and CARD8 inflammasomes warrants future study.
Projecting forward, we expect that the selective CARD8 activators reported here will not only be useful tools to study CARD8 biology, but also have the potential form the bases of new therapeutics. For example, the activation of the CARD8 inflammasome by VbP directly kills AML cells and slows AML progression in animal models13. We predict that selective CARD8 activators will retain these beneficial anticancer effects, but will not be accompanied by the inflammation associated with concomitant NLRP1 activation. Finally, the CARD8 inflammasome is present in a number of critical immune cell types, including T cells and B cells. It is possible that selective CARD8 activators like CQ31 will alter these populations in tumors or in autoimmune disorders for therapeutic benefit. Overall, this work provides an important new tool to explore the function of the CARD8 inflammasome, and provides the foundation for a new class of inflammasome-activating drugs with potential to treat human disease.
Methods
Antibodies and reagents.
The antibodies (Abs) used include GSDMD rabbit polyclonal Ab (Novus Biologicals, NBP2-33422), CASP1 rabbit polyclonal Ab (Cell Signaling Tech, 2225S), CARD8 CT rabbit polyclonal Ab (Abcam, Ab24186), PARP rabbit polyclonal Ab (Cell Signaling Tech, 9542), GAPDH rabbit monoclonal Ab (Cell Signaling Tech, 14C10), IL-18 goat polyclonal Ab (R&D Systems, AF-2548), mouse GSDMD rabbit monoclonal Ab (EPR19828; Abcam, ab209845), NLRP1 sheep polyclonal Ab (R&D Systems, AF-6788), ASC goat polyclonal Ab (R&D Systems, AF-3805), PEPD rabbit monoclonal Ab (EPR16959; Abcam, ab197890), DPP9 rabbit polyclonal (Abcam, ab42080), NPEPPS rabbit polyclonal (Abcam, ab96066), FLAG M2 monoclonal Ab (Sigma, F3165), XPNPEP1 (Abcam, ab123929), XPNPEP3 (Millipore-Sigma, HPA000527), METAP1 (Novus Biologicals, NBP1-53088), METAP2 (R&D Systems, AF-3795-SP), GSDMD rabbit monoclonal Ab (EPR20829-408; Abcam, ab215203), IL-1β goat polyclonal Ab (R&D systems, AF201NA), IRDye 800CW donkey anti-rabbit (LICOR, 925-32211), IRDye 680RD donkey anti-rabbit (925-68073), IRDye 800CW donkey anti-mouse (925-32212), IRDye 680RD donkey anti-mouse (925-68072) and IRDye 800CW donkey anti-goat (925-32214). The other reagents used include Val-boroPro (VbP; Tocris 3719), compound 8j11, bortezomib (Millipore-Sigma, 504314), bestatin (MP biomedicals, 152844), CHR 2797 (Tocris 3595), bestatin methyl ester (MeBs; Sigma, 200485), batimastat (Tocris, 2691) and FuGENE HD (Promega, E2311).
Cell culture.
HEK 293T, THP-1, RS4;11, JURKAT, U937, RAW264.7, HeLa and HEKa cells were purchased from ATCC. MV4;11, MOLM13, NOMO1, OCI-AML2, SET2, SKM1, KG1, NB4, OCI-AML3, HEL, K562, KASUMI1 and TF1 cells were purchased from DSMZ. N/TERT-1 cells were a gift from the Rheinwald Laboratory45. HEK 293T, J774.1 and RAW264.7 cells were grown in DMEM with l-glutamine and 10% FBS. HEKa cells were cultured in dermal cell basal medium supplemented with the recommended keratinocyte growth kit (ATCC). N/TERT-1 cells were grown in keratinocyte serum-free medium (KSFM) supplemented with 1× penicillin/streptomycin, bovine pituitary extract (25 μg ml−1) and epidermal growth factor (EGF; 0.2 ng ml−1). All other cell lines were grown in RPMI medium 1640 with l-glutamine and 10% FBS. All cells were grown at 37 °C in a 5% CO2 atmosphere incubator. Cell lines were regularly tested for mycoplasma using the MycoAlert mycoplasma detection kit (Lonza).
Mouse and rat BMDM isolation and culture.
Bone marrow was collected from the femurs and tibias of 7–12-week-old mice and rats. Briefly, femurs and tibias were collected from mice and crushed with a mortar and pestle in cold 1× PBS supplemented with 2.5% FBS. The mixture was strained through a 70-μm nylon cell strainer. Red blood cells (RBCs) were lysed for 4–5 min on ice in 1× RBC lysis buffer (Biolegend) and cells were centrifuged at 300g for 5 min at 4 °C. The cell pellet was washed in cold 1× PBS supplemented with 2.5% FBS before being strained in a 70-μm nylon cell strainer and counted. Counted cells were plated on 10-cm non-tissue-culture plates at 5 to 10 × 106 cells per plate in DMEM supplemented with 10% FBS and 15–20% L-cell medium. The cells were incubated at 37 °C for six days before assaying as indicated.
Human primary cell isolation and culture.
Isolated human primary cells were obtained from Astarte Biologics. All cells were >90% purity, as validated by flow cytometry by Astarte Biologics. T cells were thawed in RPMI-1640 medium supplemented with 10% FBS and cultured in RPMI-1640 medium, 10% FBS and 30 U ml−1 IL-2 (Peprotech). T cells were activated for 48 h using human T-activator CD3/CD28 Dynabeads for T-cell expansion and activation (Gibco) according to the manufacturer’s protocol.
Cloning.
Plasmids for NLRP1 and ASC were cloned as described previously13. Complementary DNA (cDNA) encoding human prolidase (PEPD) was purchased from GenScript (OHu24790) and cloned into a pDEST42 vector containing a 6× His tag. cDNA for XPNPEP3 was purchased from GenScript (OHu22210) in a pcDNA3.1+/C-(K)DYK vector. The sgRNAs were designed using the Broad Institute’s web portal46 (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design) and cloned into the lentiGuide-puro vector (Addgene 52963) or pXPR016 (Addgenehttps://www.addgene.org/107143/107143) as described previously47. The following sgRNA sequences were used: sgPEPD_1, 5′- ACTCACCGCCCATGTCGAAC-3′, used in MV4;11 PEPD−/− clone 1; sgPEPD_2, 5′- CATGGCACCCATGACGGCAC-3′, used in MV4;11 PEPD−/− clone 2, THP-1 PEPD−/−, THP-1 PEPD/XPNPEP1−/− and MV4;11 sgPEPD/sgXPNPEP1; sgXPNPEP1, 5′-GATGTAGGCCTGGATCGGTT-3′, used in THP-1 XPNPEP1−/−, THP-1 PEPD/XPNPEP1−/−, MV4;11 XPNPEP1−/− 2, and MV4;11 PEPD/XPNPEP1−/−. DPP8/9−/− THP-116, CARD8−/− THP-1 and MV4;11 cells13, and CASP1−/− THP-1, MV4;11 and RAW246.7 cells12 were generated previously.
CTG cell viability and CTF cell death assays.
Cells were plated (2,000 cells per well) in white, 384-well clear-bottom plates (Corning) using an EL406 microplate washer/dispenser (BioTek) in a 25-μl final volume of medium. To the cell plates were added compounds at different concentrations using a pintool (CyBio) and the plates were allowed to incubate in the incubator. After incubation for the indicated times, CTF reagent (Promega, G9262) was added according to the manufacturer’s protocol. The assay plates were then incubated for another 30 min before fluorescence was recorded using a Cytation 5 cell imaging multi-mode reader (BioTek). CTG reagent (Promega, G7573) was subsequently added to the assay plates following the manufacturer’s protocol. The assay plates were shaken on an orbital shaker for 2 min and incubated at 25 °C for 10 min. Luminescence was then read using a Cytation 5 cell imaging multi-mode reader (BioTek).
LDH cytotoxicity and immunoblotting assays.
HEK 293T cells were transiently transfected and treated with inhibitors as indicated. MV4;11, THP-1, RAW264.7 and J774.1 cells were plated in 12-well culture plates at 5 × 105 cells per well and treated as indicated. N/TERT-1 and HEKa cells were plated in 12-well culture plates at 1 × 105 cells per well until they reached 70% confluence and were treated as indicated. Supernatants were analyzed for LDH activity using the Pierce LDH cytotoxicity assay kit (Life Technologies). LDH activity was quantified relative to a lysis control where cells were lysed in 80 μl of a 9% Triton X-100 solution. For immunoblotting, cells were washed twice in PBS (pH 7.4), resuspended in PBS and lysed by sonication. Protein concentrations were determined and normalized using the DCA protein assay kit (Bio-Rad). The samples were separated by SDS–PAGE, immunoblotted, and visualized using the Odyssey imaging system (Li-Cor).
IL-1β ELISA assays.
Supernatants from the indicated cell lines were collected and pre-diluted fourfold before quantification of cytokines using an ELISA assay targeting IL-1β (R&D systems) following the manufacturer’s recommendations. The samples were analyzed on a Cytation 5 multimodal plate reader (BioTek) and the data exported to spreadsheet software for statistical analysis.
CRISPR/Cas9 gene editing.
To generate knockouts in MV4;11 and THP-1 cells, 1.5 × 106 cells stably expressing Cas913 were infected with lentivirus containing sgRNA plasmids. After 48 h, cells were selected with puromycin (1 μg ml−1) or hygromycin (100 μg ml−1) until control cells were all dead. Single-cell clones were isolated by serial dilution and confirmed by western blotting.
Propidium iodide flux analysis.
MV4;11 or THP-1 cells (2 × 104) were plated in a 384-well, black clear-bottom plate (Corning) in RPMI medium. For N/TERT-1 cells, 2.5 × 104 cells were plated in a 96-well, white clear-bottom plate (Corning) in 150 μl of KSFM medium for 24 h. The cells were then treated as indicated and PI was added at a final concentration of 10 μM. PI fluorescence was measured at an excitation/emission of 535/617 nm recorded at 37 °C every 5 min using a Cytation 5 cell imaging multi-mode reader (BioTek). The obtained measurements were baseline-corrected to vehicle/DMSO and normalized to maximum values (MV4;11 THP-1) or to measurements from cells lysed with 9% Triton X (N/TERT-1).
Transient transfections.
HEK 293T cells were plated in six-well culture plates at 5.0 × 105 cells per well in DMEM. The next day, the indicated plasmids were mixed with an empty vector to a total of 2.0 μg of DNA in 125 μl of Opti-MEM and transfected using FuGENE HD (Promega) according to the manufacturer’s protocol. Unless indicated otherwise, 0.02 μg NLRP1 and 0.01 μg ASC were used. After 16–20 h, the cells were treated as described. For microscopy experiments, HEK 293T cells were seeded into 12-well tissue culture (TC)-treated plates at 2.5 × 105 cells per well. After 24 h, the cells were transfected with 10 ng of plasmids encoding C-terminally FLAG-tagged NLRP1, 5 ng of a plasmid encoding N-terminally V5-GFP-tagged ASC, and 985 ng of a plasmid encoding RFP using FuGene as the transfection agent, and given 24 h to express protein and then treated with the indicated agent for 24 h, Hoechst stain (1 μg ml−1) was added.
Fluorescence microscopy and analysis.
Cells transfected as described above were imaged on a Zeiss Axio Observer.Z1 inverted wide-field microscope using a ×10/0.95NA air objective. For each well, ten positions were imaged in the bright-field, Hoescht (4′,6-diamidino-2-phenylindole (DAPI)), red fluorescent protein (RFP) and GFP channels. Data were analyzed using a custom macro written in ImageJ/Fiji. The number of cells containing GFP-ASC specks was quantified by setting threshold values on the GFP channel and performing the ‘Analyze Particles’ algorithm (size = 0–∞ and circularity = 0.50–1.00). The data were then exported to spreadsheet software and analyzed to compute the ratio of specks.
Protein purification.
Human prolidase (PEPD) in pDEST42 containing a 6× His tag was expressed in Escherichia coli Rosetta DE3 cells. Cells were induced with IPTG for 4 h at 37 °C, pelleted and lysed in 20 mM Tris-HCl pH 8.0, 300 mM NaCl, 5% glycerol, 0.5 mM β-mercaptoethanol by sonication. Affinity purification was performed using TALON resin and according to previously published protocols48. Human XPNPEP3 in pcDNA3.1+/C-(K)DYK vector was transfected into HEK 293T cells and overexpressed for 48 h, before performing FLAG-immunoprecipitation and using in substrate assays.
Substrate assays.
For the PEPD assay, a solution of substrate (Ala-Pro) was prepared in DMSO. A 24-μl volume of lysates (normalized to 5 mg ml−1 by the DC assay kit (Bio-Rad)) or 50 nM recombinant human PEPD were added to a 384-well, black clear-bottom plate (Corning) with 1 μl of dipeptide Ala-Pro (final concentration of 40 μM). The alanine liberated was measured as an increasing fluorescence signal (resorufin, excitation/emission: 535/587 nm) recorded at 25 °C using an l-alanine assay kit (Abcam, ab83394) at 25 °C according to the manufacturer’s instructions. For the AMC reporter assays, experiments were performed with recombinant enzyme, in lysates or in cells. For recombinant enzyme assays, 25 μl of enzyme solution (RNPEP 1.4 nM, DPP3 0.6 nM, LTA4H 114.3 nM, NPEPPS 1 nM, ANPEP 1.2 nM, DPP9 1 nM, MeTAP2 20 nM) was added to a 384-well, black clear-bottom plate (Corning) according to the manufacturer’s protocol, and compounds at different concentrations were added using a pintool (CyBio), followed by AMC substrate to initiate the reaction (RNPEP, 200 μM R-AMC; DPP3, 100 μM RR-AMC; LTA4H, 400 μM R-AMC; NPEPPS, 10 μM l-AMC; ANPEP, 100 μM A-AMC; DPP9, 25 μM AP-AMC; MeTAP2, 25 μM M-AMC). For lysate assays, a solution of substrate (1 mM peptide-AMC as indicated) was prepared in DMSO. A 19-μl volume of PBS or the indicated cell lysate was added to a 384-well, black clear-bottom plate (Corning), followed by 1 μl of 1 mM substrate to initiate the reaction. Substrate cleavage was measured as an increasing fluorescence signal (excitation/emission: 380/460 nm) recorded at 25 °C for 30 min. For in-cell assays, 8.0 × 104 cells were seeded per well in a 96-well, black clear-bottom plate (Corning) overnight in Opti-MEM and treated with compounds for 5 h, and then with sitagltiptin (1 μM) to block DPP4 activity for 1 h before substrate (Ala-Pro-AMC, final concentration of 5 μM) was added to the medium to initiate the reaction. AMC fluorescence was recorded at 25 °C for 40 min. For XPNPEP1 and XPNPEP3, enzymes (XPNPEP1 3.5 nM, XPNPEP3 obtained by FLAG-immunoprecipitation) were plated on a black 384-well clear-bottom plate and treated with the indicated doses of compounds. H-Lys(abz)-Pro-Pro-pNA substrate was added to a final concentration of 100 μM, and fluorescence was monitored (excitation/emission: 320/410 nm) for 30 min.
CETSA analysis.
HEK 293T or THP-1 cells were homogenized by sonication and cleared of debris by centrifugation at 10,000g for 10 min. Clarified lysates were then incubated with the indicated inhibitors for 30 min, before heating at the indicated temperatures (Fig. 2e: 38.7 °C, 45.5 °C, 53.2 °C, 61.4 °C and 70.9 °C; Extended Data Fig. 7b: 40.6 °C, 50.8 °C, 60.1 °C and 70.9 °C) for 30 min. Aggregated proteins were then precipitated by centrifugation at 18,000g for 20 min. The supernatant was then collected and then subjected to immunoblotting.
Metabolite analysis using LC-MS.
HEK 293T (0.25 × 106) or the indicated MV4;11 (0.75 × 106) cells were seeded on six-well tissue culture dishes in 2 ml of DMEM or RPMI, respectively, supplemented with 10% FBS per well. The next day, cells were treated with the indicated compounds. Metabolism was quenched and metabolites were extracted by aspirating the medium and adding 1 ml of 80:20 methanol:water chilled to −80 °C. After overnight incubation at −80 °C, cells were collected and centrifuged at 20,000g for 20 min at 4 °C. The supernatants were dried in a vacuum evaporator (Genevac EZ-2 Elite) for 3 h. Dried extracts were resuspended in 40 μl of 60% acetonitrile in water. Samples were vortexed, incubated on ice for 20 min, followed by addition of 40 μl of methanol. After briefly vortexing, the dried extracts were clarified by centrifugation at 20,000g for 20 min at 4 °C.
Dipeptide analysis was achieved using an Agilent 6545 Q-TOF mass spectrometer with a Dual Jet Stream source in positive ionization, coupled to an Acquity UPLC BEH amide column (150 mm × 2.1 mm, 1.7-μm particle size, Waters) kept at 40 °C. The composition of mobile phase A consisted of 10 mM ammonium acetate in 10:90 acetonitrile:water with 0.2% acetic acid at pH 4. Mobile phase B consisted of 10 mM ammonium acetate in 90:10 acetonitrile:water with 0.2% acetic acid at pH 4. The gradient was as follows at an initial flow rate of 0.4 ml min−1 at 95% B; 9 min, 70% B; 13 min, 30% B; 14 min, 30% B; 14.5 min, 95% B. The flow rate was increased to 0.6 ml min−1 from 15 min, 95% B to 20 min, 95% B. The injection volume was 5 μl for each sample. MS parameters included the following: gas temperature, 300 °C; gas flow, 10 l min−1; nebulizer pressure, 35 psi; sheath gas temp, 350 °C; sheath gas flow, 12 l min−1; VCap, 4,000 V; fragmentor, 125 V. Data were acquired from 50 to 1,700 m/z with reference mass correction (m/z: 121.05087 and 922.00980). Identification of dipeptides was achieved by running synthesized dipeptide standards to confirm retention time matching as well as performing tandem mass spectrometry (MS/MS) for spectral matching to SIRIUS49.
Diazirine crosslinking and click chemistry.
HEK 293T cells were collected and pelleted at 400g, washed three times in cold PBS, resuspended in 1 ml of PBS and lysed by sonication. Lysates were then clarified at 20,000g for 15 min. The soluble fraction was retained, and protein concentrations were determined using the DC protein assay kit (Bio-Rad) and adjusted to 1 mg ml−1. Aliquots of 1.0 ml were treated with probe (10 μM) for 30 min. For competition experiments, the lysates were pretreated with 100 μM inhibitor CQ31 (30 min) and then probe (30 min). Samples were then irradiated for 30 min in the photochemical reactor equipped with 350-nm lamps at 4 °C. A 10-μl volume of 4% SDS was added to each sample and heated at 60 °C for 30 min. A mix of click reagents (1 mM CuSO4, 100 μM BTTAA ligand and 100 μM biotin-PEG3 azide, 1 mM TCEP) was added and incubated for 60 min at room temperature. The proteins were then precipitated with acetone, washed three times and then spun at 3,500g for 5 min at 4 °C. The pellets were air-dried and re-solubilized in 100 μl 4% SDS PBS by sonication and gentle heat, PBS (2.0 ml) was added, then dilution to 0.2% SDS was performed. They were then enriched with streptavidin beads (100 μl per sample), incubated at room temperature for 1 h. Beads were then pelleted by centrifugation (1,400g, 2 min), washed three times with 1% SDS, three times with 2 M urea PBS and three times with PBS. These samples were then split for immunoblot and proteomic analysis.
Tandem mass tag labeling for mass spectrometry.
To a solution of the probe-bound streptavidin beads (from above), ammonium bicarbonate (ABC; 25 mM) and 10 mM DTT were added, then the solution was placed in a 42 °C heat block for 30 min. Iodoacetamide (20 mM) was added and allowed to react at 37 °C for 30 min (protected from light). It was then spun down and washed with 10 mM DTT in 200 μl ABC, followed by washing three times with 25 mM ABC. The beads were pelleted by centrifugation (1,300g, 2 min) and resuspended in 25 mM ABC, 1 mM CaCl2 and trypsin 2 μg. The digestion was allowed to proceed overnight at 37 °C with shaking. Digested peptides were collected and dried using the Genevac EZ-2 evaporator. TMTsixplex isobaric label reagents (Thermo Fisher Scientific), 0.8 mg per label, were equilibrated to room temperature, dissolved in dry acetonitrile and mixed by vortexing briefly before use. Each TMT label reagent was carefully added to each sample (126 and 127 for the blank control, 128 and 129 for the probe, 130 and 131 for the competition) and incubated at room temperature for 1 h. 5% hydroxylamine was then added to each sample and incubated for 15 min to quench the labeling reaction. Samples were then combined in equal quantities (~100 μg), purified using a high-pH reversed-phase peptide fractionation kit (Pierce) and divided into two fractions (CQ73TMT1 and CQ73TMT2), and dried with a Genevac EZ-2 evaporator.
Tandem LC-MS and MS/MS.
Mass spectrometry data were collected on an Orbitrap Fusion Lumos mass spectrometer coupled to an Easy-nLC 1200 instrument (Thermo Fisher Scientific). Peptides were separated over a 180-min gradient of 0–50% acetonitrile in water with 0.1% formic acid at a flow rate of 300 nl min−1 on a 50-cm-long PepMap RSLC C18 column (2 mm, 100 Å, 75 mm × 50 cm). The full MS spectra were acquired in the Orbitrap at a resolution of 120,000. The ten most intense MS1 ions were selected for MS2 analysis. The isolation width was set at 0.7 m/z and isolated precursors were fragmented by collision-induced dissociated (CID) (35% collision energy). Following acquisition of each MS2 spectrum, a synchronous precursor selection (SPS) MS3 scan was collected on the top ten most intense ions in the MS2 spectrum. The isolation width was set at 1.2 m/z and isolated precursors were fragmented using higher-energy C-trap dissociation (HCD). The mass spectrometry proteomics data will be made available at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository50.
Proteomic analysis.
MS raw files were analyzed using MaxQuant v1.6.17.0 by searching against the UniProt human database supplemented with common contaminant protein sequences and quantifying according to SPS MS3 reporter ions. MaxQuant was run using the following parameters: reporter ion MS3—6plex TMT; variable modifications—methionine oxidation (+15.995 Da), N-terminal protein acetylation (+42.011 Da), asparagine or glutamine deamidation (+0.984 Da); fixed modification—carbamidomethylation (+57.021 Da) of cysteine; digestion—trypsin/P; and a 1% false discovery rate. For each identified protein, mean reporter ion intensity was calculated for DMSO, probe and competition. Proteins were excluded from further analysis if they were identified as reverse or contaminants and/or had zero reporter ion intensity. Fold enrichment was calculated by dividing the probe average reporter ion intensity by the DMSO average reporter ion intensity. The CQ31 competition percentage was calculated by subtracting the competition average reporter ion intensity from the probe average reporter ion intensity and dividing by the probe average reporter ion intensity. A summary of the identified proteins and their quantitation is provided in Supplementary Data 1.
Statistical analysis.
Two-sided Student’s t-tests were used for significance testing unless stated otherwise. P values of less than 0.05 were considered to be significant. Graphs and error bars represent means ± s.e.m. of a single experiment representative of three or more independent experiments, unless stated otherwise. The investigators were not blinded in all experiments. All statistical analysis was performed using GraphPad Prism 9.
Data availability
The proteomics dataset is available in the PRIDE database: accession code, PXD027707; project name, ‘Target identification of CQ31, a selective activator of the CARD8 inflammasome’ (https://www.ebi.ac.uk/pride/archive/projects/PXD027707). All other data in this study are available within the paper, the Supplementary Information and the Extended Data, and/or from the corresponding author upon reasonable request. Source data are provided with this paper.
Extended Data
Extended Data Fig. 1 ∣. Activation of the NLRP1 and CARD8 inflammasomes.
The proteasome-mediated degradation of the NT fragments of NLRP1 and CARD8 release the CT fragments from autoinhibition. DPP8/9 inhibitors and several other danger signals (for example, pathogen proteases) accelerate the rate of NT degradation. If the rate of degradation is slow (top), CT fragments are restrained in ternary complexes consisting of the CT fragment, a full-length (FL) PRR, and DPP9. Disruption of the ternary complex (for example, by VbP) can release the CT fragments to form an inflammasome. If the rate of degradation is fast (bottom), sufficient CT fragments are released to overwhelm the DPP9 ternary complex checkpoint and to form an inflammasome. NLRP1 is activated by a similar overall mechanism, but only CARD8 is shown for clarity. Key differences between NLRP1 and CARD8 are discussed in the text.
Extended Data Fig. 2 ∣. Certain Xaa-Pro dipeptides inhibit DPP9 and activate CARD8.
(a) Inhibition of recombinant DPP9 by the twenty XP dipeptides in an AP-AMC cleavage assay. (b) Inhibition of AP-AMC cleavage activity in HEK 293 T cell lysates (pretreated with 10 μM sitagliptin to inhibit any DPP4 activity) by indicated compounds. (c) The indicated cell types were immunoblotted for proteins involved in VbP-induced pyroptosis. MV4;11 and THP-1 cells express CARD8, whereas N/TERT-1 and HEKa cells express NLRP1. Asterisks indicate background bands. (d,e) The indicated MV4;11 cells were treated with compounds (1 mM) and monitored for PI uptake over 12 h (d) or analyzed by CTG and CTF after 24 h (e). (f) MV4;11 cells were treated with XP-OMes (1 mM) for 14 h before CTG and CTF analyses. (g) The indicated AML cell lines were treated with VP-OMe (dose range = 5mM-19.5 μM, 2-fold dilution) for 24 h before CTG analysis. (h) Primary resting CD3+ T-cells were treated with VP-OMe (1 mM), IP-OMe (1 mM) or VbP (10 μM) for 18 h before immunoblot analysis. Data is representative of two independent experiments. (i) J774.1 macrophages were treated with VP-OMe (1 mM) or VbP (2 μM) for 24 h before assaying for LDH release. (j) The indicated RAW264.7 cells were treated with VP-OMe or IP-Ome (5 mM, 24 h) before CTF and CTG analyses. Data in d, e, and g (n = 4) and a,b, f, i, and j (n = 3) are means ± SEM of replicates. All data except where indicated, including immunoblots, are representative of three or more independent experiments.
Extended Data Fig. 3 ∣. CQ31 releases CQ04 in cells and causes pyroptosis.
(a) The indicated MV4;11 cells were treated with CQ04 or CQ31 for 24 h before assessing cell viability by CTG. Data are means ± SEM of 3 biological replicates. (b) The indicated MV4;11 cells were treated with CQ31 (16 μM) or VbP (16 μM), incubated for 4 h, and stained with PI. PI uptake was recorded for 12 h. Data are means ± SEM of 10 replicates. a and b are representative of three or more independent experiments. (c) HEK 293T cells were treated with vehicle control (DMSO) or CQ31(10 μM) for 24 h before intracellular metabolites were extracted. Methyl ester and free acid of CQ31 were measured by LC-MS and confirmed against pure standards.
Extended Data Fig. 4 ∣. CQ31 selectively activates the CARD8 inflammasome.
(a) Viability of cell lines after treatment with CQ31 for 24 h as assessed by CTF. (b) Human resting T-cells were treated with CQ31 (10 μM) or VbP (10 μM) for 24 h before assessing cell viability by CTG and pyroptotic and apoptotic markers by immunoblotting. Data is representative of two independent experiments. (c) N/TERT-1 immortalized keratinocyte cells were treated with CQ31 (10 μM) or VbP (10 μM), incubated for 1 h, and stained with PI. PI uptake was assessed over 14 h. (d) HEKa immortalized keratinocyte cells were treated with CQ31 (10 μM) or VbP (10 μM), before assaying for LDH release. (e) HEK 293 T cells stably expressing CASP1 and GSDMD were transfected with plasmids expressing NLRP1 and ASC and treated with CQ31 (10 μM) or VbP (10 μM) for 24 h. GSDMD cleavage was assessed by immunoblotting. (f) RAW264.7 cells were treated with CQ31 (100 μM) or VbP (2 μM) for 24 h, before assaying for LDH release. (n = 3) (g) J774.1 cells were treated with various doses of CQ31 before cell viability was measured by CTG and CTF. Data in a (n = 4), b,d,f (n = 3), and c,g (n = 6) are means ± SEM of the indicated replicates. All data except where indicated, including immunoblots, are representative of three or more independent experiments.
Extended Data Fig. 5 ∣. CQ31-induced pyroptosis is DPP8/9 dependent.
(a,b) Indicated THP-1 cells were treated with VP-OMe (1 mM, 14 h) (a) or varying doses of CQ31 or VbP (b) before assaying cell viability by CTG and CTF assays. (c) HEK 293T cells were treated with vehicle control (DMSO), CQ31 (10 μM), or VbP (10 μM) for 6 h before intracellular metabolites were extracted, and dipeptide concentrations were measured by LC-MS. Data in a (n = 4) and b,c (n = 3) are means ± SEM of the indicated replicates. Data in a and b are representative of three or more independent experiments.
Extended Data Fig. 6 ∣. PEPD knockout cells are sensitive to CQ31.
(a) The indicated MV4;11 cells were treated with compounds (16 μM) for 4 h before monitoring for PI uptake. Data is mean ± SEM of 10 biological replicates. (b) The indicated MV4;11 cells were treated with varying doses of CQ31 before assessing cell viability by CTG and CTF assays. (c) The indicated MV4;11 cells were all treated with VX-765 (50 μM) to prevent pyroptosis. Cells were then co-treated with DMSO (control) or CQ31 (10 μM). Intracellular metabolites were extracted and dipeptide concentrations were measured by LC-MS. Data are means ± SEM of the 3 biological replicates, unless indicated otherwise. Data in a and b are representative of three or more independent experiments.
Extended Data Fig. 7 ∣. CQ31 also targets XPNPEP1.
(a) Representative TMT-labeled peptides used to quantify the enrichment of the indicated proteins by CQ73. (b) CETSA analysis of CQ04 (0.5, 5 μM) and CQ31 (0.5, 5 μM) in THP-1Cas9 lysates is representative of three or more independent experiments.
Extended Data Fig. 8 ∣. Dual PEPD and XPNPEP1 inhibition induces pyroptosis.
(a) XPNPEP1/PEPD knockout THP-1 cells were treated with the indicated concentrations of CQ31 for 48 h prior to CTG analysis. (b) The indicated THP-1 cells were treated with VbP (10 μM) or CQ31 (20 μM) for 48 h prior to evaluating supernatants for IL-1β levels by ELISA and evaluating lysates and supernatants for IL-1β cleavage by immunoblotting. (c) Evaluation of XPNPEP1 and PEPD levels in MV4;11 cells stably expressing Cas9 and treated with sgRNAs targeting XPNPEP1. (d) The indicated cells were treated with CQ31 for 24 h before LDH release was evaluated. (e) The indicated MV4;11 cells were treated with VbP (10 μM) or CQ31 (20 μM) for 48 h before LDH release and immunoblot analyses. PEPD/XPNPEP1 knockout MV4;11 cells were generated with sgPEPD_2 and sgXPNPEP1. Data are means ± SEM of biological replicates. *** p < 0.001, ** p < 0.01 by two-sided Students t-test. (f) The indicated MV4;11 cells were treated with CQ31 for 48 h before assessing cell viability by CTF. (g) Inhibition of AP-AMC cleavage in HEK 293 T lysates by methyl esters of IPI or VPI tripeptides. (h) The indicated MV4;11 cells were treated with methyl esters of IPI or VPI for 24 h before CTG analysis. Data in b-h are means ± SEM of 3 replicates. All data, including immunoblots, are representative of three or more independent experiments.
Extended Data Fig. 9 ∣. Schematic comparing CQ31-induced and VbP-induced activation inflammasome activation.
(a) Weak DPP8/9 inhibition selectively activates CARD8, whereas strong DPP8/9 inhibition activates both NLRP1 and CARD8. (b) Strong DPP8/9 inhibitors (for example, VbP) are required to compete with the neo-N-terminus of the NLRP1CT fragment for the DPP8/9 active site and destabilize the ternary complex. (c) CARD8 does not directly interact with the DPP8/9 active site, and thus strong inhibitors are not needed to disrupt that interaction. It has not yet been established precisely how DPP8/9 inhibitors impact the CARD8-DPP9 ternary complex in cells. Red dots in b and c represent the DPP9 active site serine.
Supplementary Material
Acknowledgements
This work was supported by the Pew Charitable Trusts (D.A.B. is a Pew-Stewart Scholar in Cancer Research), the NIH (R01 AI137168 and R01 AI163170 to D.A.B.; T32 GM007739-Andersen to A.R.G.; NIH T32 GM115327-Tan to E.L.O.-H.; F30 CA008748 to A.R.G.; the MSKCC Core Grant P30 CA008748; R25AI140472 to J.R.C.), Gabrielle’s Angel Foundation (D.A.B.), Mr William H. and Mrs Alice Goodwin, the Commonwealth Foundation for Cancer Research, and The Center for Experimental Therapeutics of Memorial Sloan Kettering Cancer Center (D.A.B.), the Emerson Collective (D.A.B.) and a Marie-Josée Kravis Women in Science Endeavor (WISE) fellowship (S.D.R.).
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this Article.
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41589-021-00964-7.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41589-021-00964-7.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41589-021-00964-7.
References
- 1.Broz P & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Lamkanfi M & Dixit VM Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Rathinam VA & Fitzgerald KA Inflammasome complexes: emerging mechanisms and effector functions. Cell 165, 792–800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Osualdo A et al. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS ONE 6, e27396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finger JN et al. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem 287, 25030–25037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frew BC, Joag VR & Mogridge J Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 8, e1002659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chui AJ et al. N-terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandstrom A et al. Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taabazuing CY, Griswold AR & Bachovchin DA The NLRP1 and CARD8 inflammasomes. Immunol. Rev 297, 13–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachovchin DA NLRP1: a jack of all trades, or a master of one? Mol. Cell 81, 423–425 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Okondo MC et al. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat. Chem. Biol 13, 46–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okondo MC et al. Inhibition of Dpp8/9 activates the Nlrp1b inflammasome. Cell Chem. Biol 25, 262–267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson DC et al. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat. Med 24, 1151–1156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong FL et al. Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J. Biol. Chem 293, 18864–18878 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gai K et al. DPP8/9 inhibitors are universal activators of functional NLRP1 alleles. Cell Death Dis. 10, 587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griswold AR et al. DPP9’s enzymatic activity and not its binding to CARD8 inhibits inflammasome activation. ACS Chem. Biol 14, 2424–2429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chui AJ et al. Activation of the CARD8 inflammasome requires a disordered region. Cell Rep. 33, 108264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollingsworth LR et al. DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature 592, 778–783 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharif H et al. Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. Immunity 54, 1392–1404 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang M et al. Structural and biochemical mechanisms of NLRP1 inhibition by DPP9. Nature 592, 773–777 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Goethem S et al. Inhibitors of dipeptidyl peptidase 8 and dipeptidyl peptidase 9. Part 2: isoindoline containing inhibitors. Bioorg. Med. Chem. Lett 18, 4159–4162 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Linder A et al. CARD8 inflammasome activation triggers pyroptosis in human T cells. EMBO J. 39, e105071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson DC et al. DPP8/9 inhibitors activate the CARD8 inflammasome in resting lymphocytes. Cell Death Dis. 11, 628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong FL et al. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Robinson KS et al. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370, eaay2002 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Drutman SB et al. Homozygous NLRP1 gain-of-function mutation in siblings with a syndromic form of recurrent respiratory papillomatosis. Proc. Natl Acad. Sci. USA 116, 19055–19063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandemange S et al. A new autoinflammatory and autoimmune syndrome associated with NLRP1 mutations: NAIAD (NLRP1-associated autoinflammation with arthritis and dyskeratosis). Ann. Rheum. Dis 76, 1191–1198 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Ball DP et al. Caspase-1 interdomain linker cleavage is required for pyroptosis. Life Sci. Alliance 3, e202000664 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollingsworth LR et al. Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes. Nat. Commun 12, 189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Q et al. Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8. Nat. Commun 12, 188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang HK et al. Biochemical properties and expression profile of human prolyl dipeptidase DPP9. Arch. Biochem. Biophys 485, 120–127 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Lee HJ et al. Investigation of the dimer interface and substrate specificity of prolyl dipeptidase DPP8. J. Biol. Chem 281, 38653–38662 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Geiss-Friedlander R et al. The cytoplasmic peptidase DPP9 is rate-limiting for degradation of proline-containing peptides. J. Biol. Chem 284, 27211–27219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griswold AR et al. A chemical strategy for protease substrate profiling. Cell Chem. Biol 26, 901–907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandt W et al. A model of the active site of dipeptidyl peptidase IV predicted by comparative molecular field analysis and molecular modelling simulations. Int. J. Pept. Protein Res 46, 494–507 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Hikida A, Ito K, Motoyama T, Kato R & Kawarasaki Y Systematic analysis of a dipeptide library for inhibitor development using human dipeptidyl peptidase IV produced by a Saccharomyces cerevisiae expression system. Biochem. Biophys. Res. Commun 430, 1217–1222 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Lan VT et al. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 175, 66–73 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Sekine K, Fujii H, Abe F & Nishikawa K Augmentation of death ligand-induced apoptosis by aminopeptidase inhibitors in human solid tumor cell lines. Int. J. Cancer 94, 485–491 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Krige D et al. CHR-2797: an antiproliferative aminopeptidase inhibitor that leads to amino acid deprivation in human leukemic cells. Cancer Res. 68, 6669–6679 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Lupi A, Tenni R, Rossi A, Cetta G & Forlino A Human prolidase and prolidase deficiency: an overview on the characterization of the enzyme involved in proline recycling and on the effects of its mutations. Amino Acids 35, 739–752 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Wilk P et al. Substrate specificity and reaction mechanism of human prolidase. FEBS J. 284, 2870–2885 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Maggiora LL, Orawski AT & Simmons WH Apstatin analogue inhibitors of aminopeptidase P, a bradykinin-degrading enzyme. J. Med. Chem 42, 2394–2402 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Singh R et al. Structure of the human aminopeptidase XPNPEP3 and comparison of its in vitro activity with Icp55 orthologs: insights into diverse cellular processes. J. Biol. Chem 292, 10035–10047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bissonnette R et al. Prolidase deficiency: a multisystemic hereditary disorder. J. Am. Acad. Dermatol 29, 818–821 (1993). [DOI] [PubMed] [Google Scholar]
- 45.Dickson MA et al. Human keratinocytes that express hTERT and also bypass a p16INK4a-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell Biol 20, 1436–1447 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupi A et al. Characterization of a new PEPD allele causing prolidase deficiency in two unrelated patients: natural-occurrent mutations as a tool to investigate structure–function relationship. J. Hum. Genet 49, 500–506 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Duhrkop K et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics dataset is available in the PRIDE database: accession code, PXD027707; project name, ‘Target identification of CQ31, a selective activator of the CARD8 inflammasome’ (https://www.ebi.ac.uk/pride/archive/projects/PXD027707). All other data in this study are available within the paper, the Supplementary Information and the Extended Data, and/or from the corresponding author upon reasonable request. Source data are provided with this paper.