Abstract
Staphylococcus aureus (SA) is a leading cause of bacterial infection and antibiotic resistance globally. Therefore, development of an effective vaccine has been a major goal of the SA field for the past decades. With the wealth of understanding of pathogenesis, the failure of all SA vaccine trials has been a surprise. We argue that experimental SA vaccines have not worked because vaccines have been studied in naïve laboratory animals, whereas clinical vaccine efficacy is tested in immune environments reprogrammed by SA. Here, we review the failed SA vaccines that have seemingly defied all principles of vaccinology. We describe major SA evasion strategies and suggest that they reshape the immune environment in a way that makes vaccines prone to failures. We propose that appropriate integration of concepts of host-pathogen interaction into vaccine study designs could lead to insight critical for the development of an effective SA vaccine.
Keywords: Staphylococcus aureus, vaccine, original antigenic sin, evasion mechanisms, T cells, B cells, antibodies, pathogenesis
Graphical Abstract
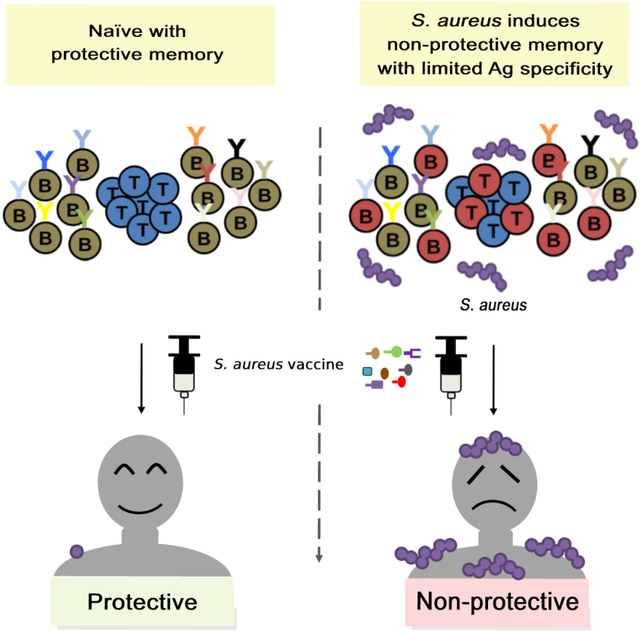
The failure of all S. aureus vaccine trials has been a conundrum. Tsai et al. propose that staphylococcal vaccine failures result from underappreciation of how the pathogen preprograms the host immune system for vaccine failures. Understanding S. aureus modulation of vaccine responses may hold the key to developing successful vaccines.
Introduction
Staphylococcus aureus (SA) is a versatile pathobiont that is exceptionally well-adapted to co-exist with the human host. The Centers for Disease Control estimated that 120,000 cases of SA bloodstream infections and 20,000 associated deaths occurred in the United States in 2017 (Kourtis et al., 2019). Persistence of methicillin-resistant SA (MRSA) infections in hospital and community settings has increased the use of once restricted antibiotics and led to inevitable acceleration and spread of antibiotic resistance (Chambers and Deleo, 2009). The importance of SA to human health as well as its versatility as a pathogen have drawn abundant research interest. As a result, much knowledge has been garnered on how SA interacts with the host in experimental models. In comparison, translational SA research has lagged even though differences in SA interaction with humans and mice have been appreciated for a long time. When called upon to address human relevant issues in the past two decades, the field has repeatedly failed to meet the challenge. For example, mouse studies of Panton-Valentine Leukocidin elaborated by Community-Associated MRSA proved to be unhelpful, and the pathogenic role of the toxin was debated for a decade until it was determined that rodents are not the best tool to study the human-tropic toxin (Spaan et al., 2013). In response to calls for a vaccine to control the expanding MRSA crisis, more than twenty human active and passive vaccine trials have been conducted (Armentrout et al., 2020; Miller et al., 2020). Yet, to date, an effective vaccine remains elusive. Although diverse opinions remain on why the vaccines failed, experts have come closer to an agreement that small tweaks in vaccine designs are unlikely to solve the vaccine conundrum.
It is noteworthy that most SA vaccine studies have been carried out in naïve laboratory animals. The commonly used mouse models show little evidence of prior human SA interaction, apart from the occasional encounter with murine SA that do not produce many of the virulence factors expressed by the human-tropic SA (Holtfreter et al., 2013). In comparison, humans are exposed to SA during the first months of life and show evidence of continued exposure with increasing anti-SA titers into adulthood (Lebon et al., 2008; Li et al., 2021). SA uses diverse strategies to evade both innate and adaptive immune responses and maintain coexistence with the human host. Many experts have pointed to these mechanisms as likely reasons for the vaccine failures, but direct evidence is lacking. However, there is growing momentum to seek out more fundamental reasons for the failure of the vaccines (Teymournejad and Montgomery, 2021). In this review, we argue that a translational approach that fully integrates the prior SA experience of the host with testing of vaccine efficacy is an important step towards building that translational bridge. We provide an overview of the unsuccessful SA vaccine trials and discuss the inability of traditional vaccine approaches to solve the SA vaccine conundrum. We review the primary SA T and B cell evasion mechanisms that conceivably could alter vaccine efficacy by altering the existing immune environment. We then reimagine what vaccine response would look like in that environment, drawing on the concept of original antigenic sin that has been applied to viral pathogens to explain vaccine failures, but underlining the fine differences that exist with immune imprinting as it applies to SA and viral pathogens. Due to space limitation we apologize to many contributors in the field whose important work are not adequately presented.
S. aureus vaccine failures
SA has assumed notoriety as the leading cause of bacterial infections with the emergence of MRSA, first in intensive care units within healthcare settings in the 1980s, and subsequently in communities (Chambers and Deleo, 2009). SA infections range from the common soft tissues infections to invasive diseases that carry significant mortality even with appropriate antibiotic treatment. Increased MRSA burden over the past decades have also broadened the use of second and last line antibiotics, thus driving antibiotics such as vancomycin to the edge of obsolescence (Chambers and Deleo, 2009). Combined, these events have led to the consensus on the urgency of developing a SA vaccine in the Threats Report developed by the CDC (2019).
Following the successes of capsular polysaccharide vaccines against H. influenzae and S. pneumoniae and prompted by promising pre-clinical vaccine studies against SA capsular polysaccharides (Yoshida et al., 1987; Lee et al., 1988; Fattom et al., 1996), early phase II and III SA vaccine trials targeted the Type 5 and Type 8 staphylococcal capsular polysaccharides (Fattom et al., 2015; Rupp et al., 2007; Robbins et al., 2004). Despite demonstrating a trend towards protection at the 40-week timepoint, neither the one-dose regimen in the first trial, nor two-dose approach in the second trial provided protective efficacy in end-stage renal disease hemodialysis patients (Fattom et al., 2015). Among suggested reasons are low capsule expression during human infections and interference with capsular binding by another antibody to surface polysaccharide PNAG through idiotypic binding (Skurnik et al., 2010). Additionally, the target patient population could have decreased complement and phagocyte functions and might not have been an optimal population to study for an initial vaccine trial (Fattom et al., 2015).
With the unsuccessful anti-capsular vaccine approach, several of the next trials targeted cell-wall anchored proteins, in part, motivated by the success of a large number of experimental vaccines against surface antigens (Stranger-Jones et al., 2006). Unexpectedly, none of the trials were successful. The most glaring of the failures was the Merck V710 double-blinded randomized vaccine trial for the prevention of SA infection after cardiothoracic surgery (Fowler et al., 2013). The vaccine that targeted the iron-regulated surface determinant protein B (IsdB) showed significant promise in several murine models of SA infection (Brown et al., 2009; Kuklin et al., 2006; Stranger-Jones et al., 2006). Notably, beyond failing to reduce the incidence of SA in vaccinated subjects, those who were infected with SA after receiving the vaccine had a five-fold increase in mortality (Fowler et al., 2013). Almost all vaccine recipients who died of SA infection had undetectable levels of serum IL2 and IL17A prior to vaccination, suggesting the potential contribution of specific host factors to infection severity (McNeely et al, 2014). A subsequent phase IIB vaccine trial targeted 4 SA surface antigens (MntC, ClfA, CPS5 and 8), based on the concept that immunizing against multiple antigens would be more efficient, but the trial was stopped despite inducing persistent and robust titers against all the antigens over 36 months because of low probability that efficacy objectives, protection against infection over 180 days from time of surgery, would be achieved (Gurtman et al., 2019).
With the failure of vaccines targeting cell surface antigens, efforts turned to neutralization of toxins that are direct source of immunopathology. It was argued that while this approach would not directly eliminate SA, anti-toxin vaccine strategy has been successful against pathogens such as diphtheria, tetanus, and pertussis. One ongoing trial targets seven toxins, including α-toxin, PVL, SEA, SEB, and TSST-1 (Aman, 2018), with several other trials concurrently targeting α-toxin and other cell surface proteins (Miller et al., 2020).
Since active vaccination is a multi-step process involving collaboration between many immune cell types, human and mouse differences at any step have the potential to lead to differences in vaccine efficacy. Hence, passive application of monoclonal antibodies that have been preselected for affinity and antigen neutralization could bypass these potential pitfalls. To date, SA vaccine trials have tested efficacy of monoclonal antibodies against a variety of SA antigens, including toxins (α-toxin), PAMPs (LTA) and surface antigens (ClfA and CP5/CP8), in prophylactic or treatment settings (Francois et al., 2021; Rupp et al., 2007; Weems et al., 2006; Weisman et al., 2011; Yu et al., 2017). Disappointingly, once again, none of the trials achieved their efficiency targets. In the case of the anti-α-toxin monoclonal antibody Suvratoxumab, application of a. 5-gram dose that achieved neutralizing activity of 156 IU/ml on day 2 and 33 IU/ml at 90 days failed to significantly reduce the incidence of pneumonia in ICU ventilated patients at 30 days (Francois et al., 2021; Wu et al., 2018), although promising sub-analysis of patients less than 65 years of age has led to a planned phase III trial.
A different vaccine approach aimed at promoting T effector functions to control SA diseases. Th1 and Th17 immunity appears to be important for protection against a variety of SA infections (Brown et al., 2015; Lee et al., 2020; Paterson et al., 2020). One promising experimental vaccine promoting Th1/Th17 immune response was a vaccine that targeted candida antigen Als3p-N that bears structural similarity to SA ClfA (Lin et al., 2009). The vaccine advanced to a phase II trial that aimed to prevent SA nasal colonization among military recruits (Schmidt et al., 2012). Results of the trial are currently pending.
Overall, repeated failures of the human trials have eroded confidence of the field in traditional vaccine approaches and have fostered a growing consensus that more fundamental understanding of SA vaccination in humans and animals is needed to overcome the current impasse in vaccine development. SA’s versatility as a pathogen and its commensal relationship with the human host likely need to be accounted for in rethinking SA vaccine approaches.
Clinical evidence of protective humoral and cell mediated immunity
Human SA colonization occurs from early childhood (Lebon et al., 2008). Between colonization and occasional infections, children develop levels of SA-specific antibodies and T cells that increase with the age (Li et al., 2021). Despite that, only moderate level of immunity develops. Studies have shown that individuals chronically colonized with SA are more frequently infected, but the severity of invasive SA disease if that occurs is reduced compared to non-colonizers, suggesting some level of acquired protection (Wertheim et al., 2004). Experiments in mice with skin or bloodstream reinfection, to a large extent, reflect the human observation, although Major Histocompatibility Complex (MHC)-restriction of the mouse strain appears to influence protection to reinfection (Si et al., 2020).
When further dissected, the role of B cells in protection is thought to be limited, since individuals with B cells deficiency are not more susceptible to SA infections compared to normal individuals (Fowler and Proctor, 2014). However, there is support for the protective role of some toxin-specific antibodies against select SA diseases. For example, individuals with staphylococcal toxic shock syndrome (TSS) have significantly lower titers of antibody to TSS Toxin (TSST) (Bonventre et al., 1984), which is the rationale behind the use of IVIG, pooled immunoglobulins obtained from at least one thousand donors, in the treatment of patients with TSS (Darenberg et al., 2004). Likewise, convalescent antibodies to exotoxin α-toxin in patients with SA infection correlate with protection against subsequent SA infections (Fritz et al., 2013), while higher antibody titers against several secreted toxins collectively correlate with protection against sepsis syndrome in patients with SA bacteremia (Adhikari et al., 2012). In a model of SA-mediated septic arthritis, B-cell-deficient agammaglobulinemic mice did not develop more severe disease or increased bacterial burden compared to wild-type mice (Gjertsson et al., 2000). Similarly, experiments using Rag2-deficient mice demonstrated that, while adaptive immune cells are activated during SA infection and are required for prolific inflammatory cytokine secretion, the absence of mature B- and T-cells only had a local and temporal effect on bacterial clearance in the liver during early sepsis and had no significant effect altogether in late sepsis.
In contrast to B cells, T cells appear to have a more substantial role in containing SA infections in humans and experimental mouse models (Cho et al., 2010; Levy et al., 2016; Miller and Cho, 2011). Robust evidence supports the central role of CD4+T cells, particularly Th1 and Th17 cells, in mediating protection against SA infections (Brown et al., 2015; Lee et al., 2020; Paterson et al., 2020). Notably, the importance of Th17 cells can be appreciated in individuals with STAT3 loss-of-function mutations or hyper-IgE syndrome that interferes with the differentiation of Th17 cells (Ma et al., 2008; Milner et al., 2008), which thus leads to recurrent and severe mucocutaneous SA infections. Likewise, HIV-positive patients who experience significant depletion of Th17 cells early in the course of their HIV infection are poor controllers of SA skin and soft tissue infections (Brenchley et al., 2008; El Hed et al., 2010; Hidron et al., 2010). Lastly, patients with atopic dermatitis have increased skin colonization and superinfection with SA, with decreased IL-17 pathway cytokines and increased Th2 cytokines, including IL-4 and IL-13, in lesional skin (Guttman-Yassky et al., 2008). Along with Th17, IFN-γ produced by Th1 cells has also been implicated in protection against SA skin and bloodstream infections (Beekhuizen and van de Gevel, 2007; Brown et al., 2015). Another CD3+T cell, which displays neither CD4 nor CD8 coreceptor on its surface, is the gamma-delta T cell that has been reported to mediate protection against skin and soft-tissue SA infections in an IL-17 dependent manner (Dillen et al., 2018; Leyva-Castillo et al., 2021; Marchitto et al., 2019). The specific roles of each subset of CD4+T cell during SA infections is reviewed elsewhere (Armentrout et al., 2020) and is beyond the scope of this review.
S. aureus manipulation of humoral responses
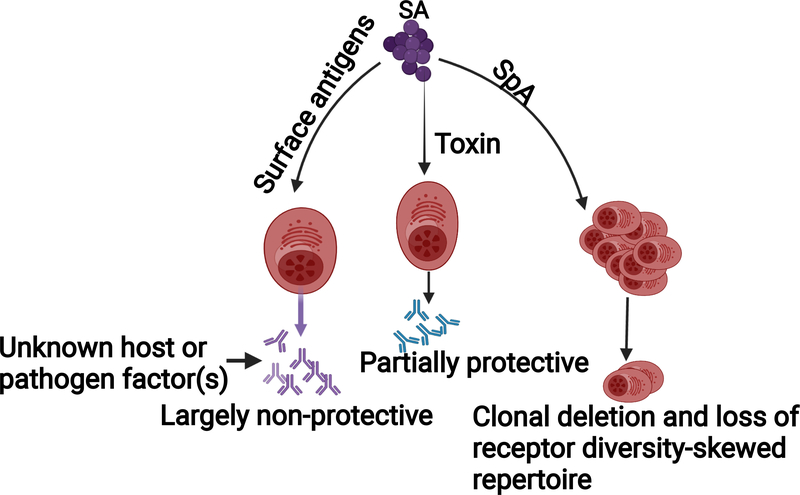
SA evades host humoral defenses through factors that subvert antibody functions and appropriate B-cell development (Figure 1). Principally, the well-characterized SA protein A (SpA) antagonizes humoral immunity through interactions with both the antibody constant fragment (Fc), and the B-cell receptor (BCR) variable domain. Early reports demonstrated that SpA non-specifically binds human gamma-globulins at the Fc receptor to inhibit SA phagocytosis and bacterial killing (Forsgren and Quie, 1974; Forsgren and Sjoquist, 1966). It was later elaborated that SpA binding of antibody Fc fragments can also result in the formation of multi-molecular complexes that mediate intracellular SA survival and lead to systemic dissemination of surgical site infection (Nishitani et al., 2020). This IgG-binding function of SpA is mirrored by a similar virulence factor, Staphylococcal immunoglobulin-binding protein, Sbi, to further expand SA’s capacity to evade antibody-mediated clearance (Zhang et al., 1999; Zhang et al., 1998).
Figure 1. B cell evasion mechanisms.
Antibody responses against immunodominant SA surface antigens are largely non-protective, whereas responses against toxins are partially protective. SpA induces B cell deletion and suppression through its super-antigenic activity, and thus creates “holes” in the B cell repertoire and primes for a skewed specific antibody response. The reason why antibodies against surface antigens are not protective is unclear.
In addition to SpA’s impact on antibody-dependent opsonophagocytosis, Goodyear and Silverman provided evidence of SpA’s superantigenic function by tracking the fate of SpA binding peripheral B-cells and demonstrated that this association promotes supraclonal deletion by apoptosis. By crosslinking with a conserved VH region of the BCR variable domain, SpA induces a B-cell activation state that is driven towards programmed cell death (Goodyear and Silverman, 2003). Consistent with the detrimental effects of SpA, mice and guinea pigs infected with a functionally SpA-deficient mutant show improved phagocytosis of SA and mount a protective B-cell response against lethal SA challenge (Falugi et al., 2013; Fattom et al., 2015).
Amidst its deleterious effects, SpA can also trigger vigorous B-cell proliferation. By cross-linking BCRs, SpA sensitizes B-cells for the recognition of TLR2 ligands to promote expansion of intracellular IgM-expressing B-cells. Further investigation, however, revealed that these B-cells fail to induce significant secretion of the SA-targeting immunoglobulins (Bekeredjian-Ding et al., 2007). SpA-mediated cell activation selectively triggers the expansion of IL-10-secreting regulatory B-cell subsets. In cooperation with plasmacytoid dendritic cells, SpA interaction with B-cells strengthens the characteristic immunosuppressive IL-10 response associated with SA infections (Parcina et al., 2013). Thus, whether by clonal deletion or non-productive cellular expansion, the impact of SpA on B-cells allows SA to efficiently evade humoral immunity. As further evidence for these mechanisms, Schneewind and Missiakas developed a nontoxigenic SpA vaccine that overcomes SpA’s dual functions by antagonizing the effects of SpA on both antibodies and B-cells. Vaccination with SpAKKAA, a variant that does not bind Fcγ or Fab VH3, promoted opsonophagocytic clearance, as well as a more robust antibody response against many SA antigens in mice (Kim et al., 2010).
Many studies have examined the implication of these findings in humans by assessing the presence and efficacy of anti-SA antibodies naturally circulating in human serum and induced after SA infection. A study of plasmablasts from SA-infected subjects showed a focused immunodominant response to SpA and a more limited response to other SA virulence factors, consistent with the proposed superantigenic mechanisms of SpA (Pauli et al., 2014). However, antibody profiling studies from other groups have shown the near ubiquity and high abundance of anti-SA antibodies in the healthy population, which further increase during SA infections (Dryla et al., 2005; Radke et al., 2018; Romero Pastrana et al., 2018). Addressing the functionality of these antibodies, we showed that adoptive transfer of sera from healthy children are largely non-protective in a murine SA challenge model. In comparison, forty percent of convalescent serum samples from children with invasive SA disease are able to reduce SA burden at 4–6 weeks post-infection, but not at 6 months (Tsai et al., 2021). These data are consistent with the relatively non-protective role of humoral immunity in humans, particularly in children, despite abundant antibody production. It remains unclear how SA manages to make most SA-specific antibodies non-protective.
In addition to SpA, a recent study showed that SA leukocidins also play a role in modulating host humoral responses (Tam et al., 2020). The authors showed that infection of mice with a ΔlukED ΔhlgACB double mutant SA resulted in increased anti-SA antibody levels compared to infection with the isogenic WT SA. Hence targeting of SA leukocidins could improve efficacy of vaccines.
S. aureus manipulation of T cell responses
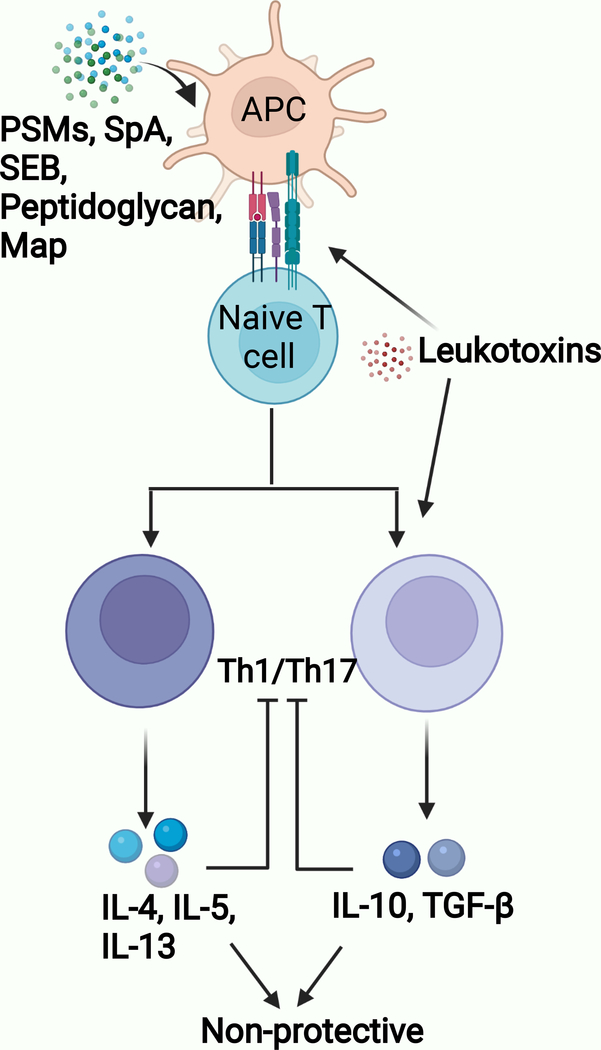
SA targets effector functions of CD4+T cells mainly with two classes of virulence factors: superantigens and toxins (Goldmann and Medina, 2018; Thammavongsa et al., 2015; Xu and McCormick, 2012) (Figure 2). Superantigens like TSST-1 and staphylococcal enterotoxin B bind class II MHC molecules and to the variable region of a specific Vβ chain of the TCR to induce potent activation of about twenty percent of all peripheral T cells (Xu and McCormick, 2012). This non-specific superantigen-mediated T cell activation prevents the development of a focused and coordinated immune response and leads to the loss of overall receptor diversity and lack of antigen-specific protective T cell responses.
Figure 2. T cell evasion mechanisms.
Th1 / Th17 lymphocytes mediate anti-SA immunity. Staphylococcal toxins (α-toxin and LukED) induce cytolysis of mature and memory T cells. Various SA virulence determinants affect T cell priming by modulating antigen-presenting cell – naïve T cell interaction: PSM (induction of tolerogenic DC); SEB (Vβ-specific T cell activation), Peptidoglycan modification (suppression of Th17-related cytokines), Map (induction of Th2 cells), SpA (induction of Treg cells), LukED and LukAB (killing of DC). APC: Antigen Presenting Cells.
SA additionally produces an array of functionally diverse toxins, including leukocidins, hemolysins, and phenol soluble modulins (PSMs), which launch a fierce attack on the host immune system to subvert protective T cell responses (Berends et al., 2019; Richardson et al., 2018; Spaan et al., 2017). Leukocidins are secreted factors that specifically target human and mouse lymphocytes. For example, LukED targets and kills CCR5-positive expressing Th1 and Th17 cells (Alonzo et al., 2013). Similarly, α-toxin induces programmed cell death of human and mouse IFN-γ expressing T cells during MRSA infection (Bonifacius et al., 2020; Nygaard et al., 2012). The detrimental effect of α-toxin on memory T cells is evidenced by the development of enhanced specific memory T cell response in mice born to HlaH35L-immunized dams and subsequently challenged with SA (Lee et al., 2020).
In addition to toxin-mediated killing of T cells, SA leukotoxins LukAB and LukED, induce direct killing of human dendritic cells (DCs) (Alonzo et al., 2013; Berends et al., 2019), which are a central player in priming of adaptive immune responses. PSMs also disturb the adaptive immune response via the induction of tolerogenic dendritic cells (DCs) (Richardson et al., 2018; Schreiner et al., 2013). PSM-treated DCs produce high level of IL-10 and increase the frequency of FOXP3+ regulatory T cells, which have been shown to suppress both Th1 and Th17 cellular immune responses (Mondal et al., 2012; Schreiner et al., 2013). Furthermore, SA infection expands the myeloid-derived suppressor cell (MDSC) population, which are well-known to suppress effector T cell functions through the secretion of IL-10 (Heim et al., 2015; Peng et al., 2017). Consistently, a report showed that SA peptidoglycan-induced IL-10 prevents Th1 and Th17 cellular immune responses (Frodermann et al., 2011). This finding was recently corroborated by a study from our group demonstrating more specifically that O-acetylation of peptidoglycan suppresses Th17 cell responses in an IL-10 dependent manner, and that compared to wild-type mice, IL-10 deficient mice immunized with live SA vaccine had improved Th17 response, and thus, bacterial clearance upon challenge (Sanchez et al., 2017). Adoptive transfer of CD4+T cells from the immunized IL-10 deficient mice into naïve recipient mice conferred significant protection against SA infection. These results indicate that IL-10 produced during primary SA infection is likely one of the unique and important immune evasion strategies employed by SA that contributes to the lack of protective memory CD4+ T cell responses. Further elucidating the mechanism by which IL-10 subverts CD4+T cell immunity, a study in a Mycobacterium tuberculosis (Mtb) model showed that CD4+ T cells primed in an IL-10-enriched environment are functionally incompetent and unable to control the infection. This non-protective phenotype was stable and maintained even after the IL-10-modulated T-cells were transferred into IL-10-low recipients (Ferreira et al., 2021). Whether a similar effect occurs with SA-induced IL-10 warrants thorough investigation. In line with the demonstrated effect with Mtb, SA-induced IL-10 could be postulated as a survival strategy exploited by the pathogen, whereby it may help the pathogen persist and thrive within the host while serving to limit inflammatory damage to host tissues and organs.
Yet another mechanism employed by SA to interfere with the development of effective T cell response is the molecular mimicry of host immune components. SA secretes a Class II MHC analog protein, MAP, which has been shown to impede T cell proliferative response and induce Th2 cell differentiation. Pertinently, Th2-associated IL-4 cytokine suppresses IL-17 response (Leyva-Castillo et al., 2021).
SA T cell evasion mechanisms are more difficult to study clinically than antibody responses. Overall, staphylococcal superantigen effect on the induction of T cell anergy has been reported in atopic dermatitis and psoriasis (Yarwood et al., 2000), but it unclear if it has a more than a transient effect on the human T cell repertoire to affect SA vaccination. SA modulation of host cytokine responses (i.e. elevation of IL-4 and IL-10) has also been demonstrated in atopic dermatitis, as well as after systemic infections (Rose et al., 2012). Particularly, the association of mortality with IL10 in SA bacteremia is well documented (Leyva-Castillo et al., 2021; Rose et al., 2012). In a SA study of children, infections, irrespective of invasive or non-invasive nature, correlated with global impairment of anti-SA Th17 responses compared to healthy (colonized) children, suggesting a mechanism whereby infection to SA drives non-protective outcome (Li et al., 2021).
Vaccination in a S. aureus reprogrammed host environment
Considering an immune system shaped by the above SA mechanisms, we ask how SA vaccines would perform in such an environment instead of the naïve laboratory mouse environment. Although SA frequently colonizes the human host, it is unlikely that the colonizing SA would interact in a significant way to affect vaccine response directly. Hence, we will not consider direct interaction of the vaccine with SA virulence factors in this discussion.
SA vaccine could be expected do one of two things: It could prime for a new cellular or humoral response, or it could recall an anti-staphylococcal memory response. If the memory response is protective, vaccine would be anticipated to amplify the protective memory response, as is shown when patients who recovered from SARS-CoV-2 infection are vaccinated against SARS-CoV-2 (Stamatatos et al., 2021). Also consistent is the success of vaccine against LukAB, a presumed protective antigen, in SA-colonized minipigs (Fernandez et al., 2021). Conversely, most host antibody responses to SA are presumed to be non-protective (Miller et al., 2020), and vaccine recall of these responses would be expected to be non-protective. Hence, a SA capsular polysaccharide vaccine that was effective in naïve mice was shown to be not protective in SA-colonized minipigs, consistent with human trial findings (Fernandez et al., 2021; Fattom et al., 2015).
This hypothesized vaccine response draws from the concept of Original Antigenic Sin (OAS) proposed by Thomas Francis, Jr in 1960 (Francis, 1960). It describes the recall of a memory response to a primary influenza infection by secondary viral exposures or vaccinations. Because viral antigenic shift in influenza occurs seasonally, the antigen seen by the immune system subtly differs between exposures. The efficacy of the recalled memory response is determined to a large extent by the antigenic distance between the initial and subsequent encounters. Additionally, the recalled response could reduce de novo priming of an effective response against the new antigen. As such, OAS could be protective, non-protective, or even suppressive.
In the case of SA, the antigens seen by the immune system with primary infection and subsequent vaccination could be the same because of the routine practice of selecting conserved targets for immunizations. Because most of the humoral responses are presumed to be non-protective, vaccine reliance on recall of the initial response would suggest that many vaccines to SA would be non-protective. However, various factors could modify this response to alter overall SA vaccine efficacy.
Generation of a protective de novo response
Can OAS be overcome? If the recalled memory response is modest, it is conceivable that the SA vaccine could induce a de novo response from the naïve pool of T and B cells. It would be intuitive to think that this de novo response would be non-protective if the host response to the same antigen is non-protective in the context of infection. Surprisingly available data suggest that effective vaccines are readily made to all types of SA antigens, even those cell-surface antigens that are presumed to be “non-protective” (Stranger-Jones et al., 2006). This observation would thus suggest that the context in which the antigen is presented, either infection or adjuvant, could lead to opposite protective outcomes. To rationalize this outcome, it has been well established that both Fab and Fc domains contribute to protective function of antibodies through their interaction with SA antigens and host immunocytes (Bennett et al., 2019; Chen et al., 2020). Hence, changes, for example, in Fc glycosylation as a result of priming with adjuvant or through infection could conceivably lead to differences in protection. It is less obvious how a protective and a non-protective Fab response could be mounted to the identical antigen. However, there are examples of how pathogens have directed immune responses to immunodominant but non-protective subdomains on the microbial cell-surface, with unmasking of protective epitopes only by selective subdomain vaccines (Novotny and Bakaletz, 2003; Wrightsman et al., 1994). Irrespective, protective antibodies to the “non-protective” antigen could conceivably be made through vaccination, although there are additional factors within the SA reshaped environment that could limit their efficacy as discussed below.
Interference with priming
Hypothetically, recalled memory cells could limit de novo T or B cell priming through direct competition for space and cytokines. The extent of interference would depend on the magnitude of the recalled response, the vaccine antigen concentration, and the antigen presenting cell numbers. Prior colonization or infection with SA could also modify the T and B cell repertoire through depletion or other suppressive mechanisms associated with SA T and B cell superantigens. Silverman described the generation of holes in the B cell repertoire as a result of interaction with SpA antigen. When SpA treatment is stopped, conventional B-2 repertoire normalized, but the “hole” in B-1 repertoire persisted (Silverman et al., 2000). It is less clear to what extent SA superantigens affect T and B cell repertoires in humans.
Pre-existence of non-protective specific antibodies can also interfere with de novo T or B cell priming by two distinct mechanisms (Bergstrom et al., 2017; Getahun and Heyman, 2009). The antibodies could bind the vaccine antigen and facilitate its clearance. Alternatively, antibody binding to the vaccine antigen could mask and thereby dampen de novo priming of naïve T or B cells. Modeling both mechanisms using specific IgG to sheep red blood cells, Heyman and colleagues showed suppression of naïve and memory B and T cell activation via both mechanisms although epitope masking was predominant.
Cytokine modulation of T and B cell development
Assuming that the vaccine antigens successfully initiate priming of potentially protective naïve T or B cells, exposure to appropriate cytokines is still required for the development of effective anti-SA immunity. Elevated IL-4 and IL-10, in association with several types of SA infections, have the potential to undermine development of protective T and B cells (Leyva-Castillo et al., 2021; Rose et al., 2012). If the recalled SA-specific T or B cells turn out to be the primary producer of IL10 or IL4 (Sanchez et al., 2017), then the recall response has the potential to further reduce vaccine efficacy through the cytokines’ suppressive properties. By the same argument, it might be possible to drive T or B cells towards a protective phenotype using Th1/Th17 adjuvants. Plasticity might even allow for the conversion of non-protective memory cells as shown in a study where a protective vaccine Th1/Th17 memory response to SA TSST-1 is shown to be lost because of memory cell conversion to a IL10 regulatory phenotype (Narita et al., 2019).
Direct antibody competition
Assuming that vaccination is able to induce de novo protective antibodies, efficacy of the vaccine would be determined by the outcome of competition between the protective and recalled non-protective antibody responses. In support of this mechanism interference, a study in a rodent malaria model demonstrated the capacity of a non-neutralizing monoclonal antibody to interfere with a protective antibody in vitro and in vivo even though they bind non-overlapping regions of the same sporozoite antigen. The authors pointed to the data as proof of principle demonstration that pre-existing non-protective antibodies could make malaria vaccines less efficacious for malaria-exposed individuals in endemic areas (Vijayan et al., 2021). Unlike influenza vaccine antigens which recall cross-reactive antibodies that are not expected to bind with similar affinity as the de novo primed antibody response, vaccine- and infection- induced antibodies target the same SA antigen. Hence, the likelihood of interference is greater. This mechanism of vaccine suppression could have significant implications on SA vaccinology because it could explain not only the pervasive failure in active vaccination, but may also explain the disappointing outcomes in passive immunization platforms.
Post-vaccine effect
Even after vaccination, SA has the potential to further modulate anti-SA effector functions. In a murine model of SA re-infection, Keener et al. showed that SpA alters the fate of plasmablasts and plasma cells by enhancing the short-lived extrafollicular response and reducing the pool of long-lived plasma cells (Keener et al., 2017). The effect of this modulation on antibody production is corroborated by evidence of lower specific antibody titers to several SA antigens after infection with SpA-expressing wild-type compared to a functionally SpA-deficient strain (Falugi et al., 2013). SA also secretes the toxin LukED that specifically targets and kills the predominant CCR5-positive effector memory T cell population (Alonzo et al., 2013). In another mouse study, a mutant TSST-1 vaccine induced a protective specific Th17 response one week after vaccination. However, anti-TSST immunity was lost after 12 weeks and was shown to be related to IL10 secretion by the memory cells which suppresses IL17 production (Narita et al., 2019).
Other human-specific considerations
Microbiome in laboratory animals also has the potential to confound vaccine data as highlighted by a recent discovery that laboratory mice with “wild” microbiome have immune characteristics that are more aligned with human immune responses. The particular effect of microbiome on vaccine-induced immunity was shown in laboratory mice cohoused with pet-store mice (Fiege et al., 2021). These mice showed dampened influenza vaccine-induced humoral responses and poor control of influenza infection compared to the laboratory mice. Additionally, heterosubtypic protective T cell responses were compromised in co-housed mice, indicating the influence of microbiome on vaccine-induced immunity. This has helped to further focus attention on the limited translational potential of current mouse models.
Unrelated to the human environment, SA tropism for human-encoded immune factors is another feature that could drive discrepant results in the murine and human hosts (Spaan et al., 2017). As a human pathogen, SA elaborates many virulence factors, particularly toxins, that interact poorly with the murine host (Spaan et al., 2017). It is unclear how the absence of the full functional complement of SA factors would affect SA vaccines in mice.
Conclusions and outlook
In contemplating the remarkable efficacy of the SARS-CoV-2 vaccines, one might almost be forgiven to think that developing vaccines is easy, and that the tried-and-true vaccinology toolbox still holds the key to successful SA vaccines. Yet vaccines have remained ineffective in repeated trials. Pathogenesis studies have informed us in so many ways that SA is different as a master of immune evasion strategies, and that induction of non-protective immunity is largely the rule. As suggested by leaders of the SA field, a more than incremental approach is now needed to address the fundamental root of vaccine failures. Bridging the translational divide will require simulation of the human host experience in animal models. For example, SA vaccines could be studied in mice that have been infected or colonized previously with SA. Likewise, human anti-SA monoclonal antibodies could be tested in laboratory animals or humanized mice that have been infused with anti-SA antibodies purified from human sera. Our prediction is that, in both cases, vaccine efficacy would be significantly dampened in the modified experimental settings. If our hypothesis is validated, the findings would pave the way for more direct studies of the role of SA virulence determinants in vaccine modulation. Admittedly, no published studies to date have demonstrated a causal relationship between SA prior exposure and vaccine failures. We propose that these studies are urgently needed since understanding of SA reprogramming of the host environment and its effect on vaccination likely holds the clues to development of the next successful SA vaccine.
Acknowledgments
The research of G.Y.L. is supported by grants from the National Institute of Allergy and Infectious Diseases Institute (R01AI144694 and R01AI127406).
Footnotes
Declaration of Interests
The authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 2019, T.r. Centers for Disease Control and Prevention: Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: Centers for Disease Control and Prevention; Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. [Google Scholar]
- Adhikari RP, Ajao AO, Aman MJ, Karauzum H, Sarwar J, Lydecker AD, Johnson JK, Nguyen C, Chen WH, and Roghmann MC (2012). Lower antibody levels to Staphylococcus aureus exotoxins are associated with sepsis in hospitalized adults with invasive SA infections. J Infect Dis 206, 915–923. [DOI] [PubMed] [Google Scholar]
- Alonzo F 3rd, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, Landau NR, Unutmaz D, and Torres VJ (2013). CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman MJ (2018). Integrated BioTherapeutics. Hum Vaccin Immunother 14, 1308–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armentrout EI, Liu GY, and Martins GA (2020). T Cell Immunity and the Quest for Protective Vaccines against Staphylococcus aureus Infection. Microorganisms 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekhuizen H, and van de Gevel JS (2007). Gamma interferon confers resistance to infection with Staphylococcus aureus in human vascular endothelial cells by cooperative proinflammatory and enhanced intrinsic antibacterial activities. Infect Immun 75, 5615–5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekeredjian-Ding I, Inamura S, Giese T, Moll H, Endres S, Sing A, Zahringer U, and Hartmann G (2007). Staphylococcus aureus protein A triggers T cell-independent B cell proliferation by sensitizing B cells for TLR2 ligands. J Immunol 178, 2803–2812. [DOI] [PubMed] [Google Scholar]
- Bennett MR, Dong J, Bombardi RG, Soto C, Parrington HM, Nargi RS, Schoeder CT, Nagel MB, Schey KL, Meiler J, et al. (2019). Human VH1–69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends ETM, Zheng X, Zwack EE, Menager MM, Cammer M, Shopsin B, and Torres VJ (2019). Staphylococcus aureus Impairs the Function of and Kills Human Dendritic Cells via the LukAB Toxin. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom JJ, Xu H, and Heyman B (2017). Epitope-Specific Suppression of IgG Responses by Passively Administered Specific IgG: Evidence of Epitope Masking. Front Immunol 8, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacius A, Goldmann O, Floess S, Holtfreter S, Robert PA, Nordengrun M, Kruse F, Lochner M, Falk CS, Schmitz I, et al. (2020). Staphylococcus aureus Alpha-Toxin Limits Type 1 While Fostering Type 3 Immune Responses. Front Immunol 11, 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonventre PF, Linnemann C, Weckbach LS, Staneck JL, Buncher CR, Vigdorth E, Ritz H, Archer D, and Smith B (1984). Antibody responses to toxic-shock-syndrome (TSS) toxin by patients with TSS and by healthy staphylococcal carriers. J Infect Dis 150, 662–666. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, et al. (2008). Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112, 2826–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AF, Murphy AG, Lalor SJ, Leech JM, O’Keeffe KM, Mac Aogain M, O’Halloran DP, Lacey KA, Tavakol M, Hearnden CH, et al. (2015). Memory Th1 Cells Are Protective in Invasive Staphylococcus aureus Infection. PLoS Pathog 11, e1005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Kowalski R, Zorman J, Wang XM, Towne V, Zhao Q, Secore S, Finnefrock AC, Ebert T, Pancari G, et al. (2009). Selection and characterization of murine monoclonal antibodies to Staphylococcus aureus iron-regulated surface determinant B with functional activity in vitro and in vivo. Clin Vaccine Immunol 16, 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF, and Deleo FR (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shi M, Tong X, Kim HK, Wang LX, Schneewind O, and Missiakas D (2020). Glycosylation-dependent opsonophagocytic activity of staphylococcal protein A antibodies. Proc Natl Acad Sci U S A 117, 22992–23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, Magorien JE, Blauvelt A, Kolls JK, Cheung AL, et al. (2010). IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest 120, 1762–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darenberg J, Soderquist B, Normark BH, and Norrby-Teglund A (2004). Differences in potency of intravenous polyspecific immunoglobulin G against streptococcal and staphylococcal superantigens: implications for therapy of toxic shock syndrome. Clin Infect Dis 38, 836–842. [DOI] [PubMed] [Google Scholar]
- Dillen CA, Pinsker BL, Marusina AI, Merleev AA, Farber ON, Liu H, Archer NK, Lee DB, Wang Y, Ortines RV, et al. (2018). Clonally expanded gammadelta T cells protect against Staphylococcus aureus skin reinfection. J Clin Invest 128, 1026–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryla A, Prustomersky S, Gelbmann D, Hanner M, Bettinger E, Kocsis B, Kustos T, Henics T, Meinke A, and Nagy E (2005). Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin Diagn Lab Immunol 12, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hed A, Khaitan A, Kozhaya L, Manel N, Daskalakis D, Borkowsky W, Valentine F, Littman DR, and Unutmaz D (2010). Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis 201, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falugi F, Kim HK, Missiakas DM, and Schneewind O (2013). Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 4, e00575–00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattom AI, Sarwar J, Ortiz A, and Naso R (1996). A Staphylococcus aureus capsular polysaccharide (CP) vaccine and CP-specific antibodies protect mice against bacterial challenge. Infect Immun 64, 1659–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattom A, Matalon A, Buerkert J, Taylor K, Damaso S, and Boutriau D (2015). Efficacy profile of a bivalent Staphylococcus aureus glycoconjugated vaccine in adults on hemodialysis: Phase III randomized study. Hum Vaccin Immunother 11, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Sanders H, Henn J, Wilson JM, Malone D, Buoninfante A, Willms M, Chan R, DuMont AL, McLahan C, et al. (2021). Vaccination with Detoxified Leukocidin AB Reduces Bacterial Load in a Staphylococcus aureus Minipig Deep Surgical Wound Infection Model. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira CM, Barbosa AM, Barreira-Silva P, Silvestre R, Cunha C, Carvalho A, Rodrigues F, Correia-Neves M, Castro AG, and Torrado E (2021). Early IL-10 promotes vasculature-associated CD4+ T cells unable to control Mycobacterium tuberculosis infection. JCI Insight 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiege JK, Block KE, Pierson MJ, Nanda H, Shepherd FK, Mickelson CK, Stolley JM, Matchett WE, Wijeyesinghe S, Meyerholz DK, et al. (2021). Mice with diverse microbial exposure histories as a model for preclinical vaccine testing. Cell Host Microbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren A, and Quie PG (1974). Effects of staphylococcal protein A on heat labile opsonins. J Immunol 112, 1177–1180. [PubMed] [Google Scholar]
- Forsgren A, and Sjoquist J (1966). “Protein A” from SA. I. Pseudo-immune reaction with human gamma-globulin. J Immunol 97, 822–827. [PubMed] [Google Scholar]
- Fowler VG, Allen KB, Moreira ED, Moustafa M, Isgro F, Boucher HW, Corey GR, Carmeli Y, Betts R, Hartzel JS, et al. (2013). Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA 309, 1368–1378. [DOI] [PubMed] [Google Scholar]
- Fowler VG Jr., and Proctor RA (2014). Where does a Staphylococcus aureus vaccine stand? Clin Microbiol Infect 20 Suppl 5, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T (1960). On the doctrine of original antigenic sin. Proc Am Philos Soc 572–578. [Google Scholar]
- Francois B, Jafri HS, Chastre J, Sanchez-Garcia M, Eggimann P, Dequin PF, Huberlant V, Vina Soria L, Boulain T, Bretonniere C, et al. (2021). Efficacy and safety of suvratoxumab for prevention of Staphylococcus aureus ventilator-associated pneumonia (SAATELLITE): a multicentre, randomised, double-blind, placebo-controlled, parallel-group, phase 2 pilot trial. Lancet Infect Dis 21, 1313–1323. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Tiemann KM, Hogan PG, Epplin EK, Rodriguez M, Al-Zubeidi DN, Bubeck Wardenburg J, and Hunstad DA (2013). A serologic correlate of protective immunity against community-onset Staphylococcus aureus infection. Clin Infect Dis 56, 1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodermann V, Chau TA, Sayedyahossein S, Toth JM, Heinrichs DE, and Madrenas J (2011). A modulatory interleukin-10 response to staphylococcal peptidoglycan prevents Th1/Th17 adaptive immunity to Staphylococcus aureus. J Infect Dis 204, 253–262. [DOI] [PubMed] [Google Scholar]
- Getahun A, and Heyman B (2009). Studies on the mechanism by which antigen-specific IgG suppresses primary antibody responses: evidence for epitope masking and decreased localization of antigen in the spleen. Scand J Immunol 70, 277–287. [DOI] [PubMed] [Google Scholar]
- Gjertsson I, Hultgren OH, Stenson M, Holmdahl R, and Tarkowski A (2000). Are B lymphocytes of importance in severe Staphylococcus aureus infections? Infect Immun 68, 2431–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann O, and Medina E (2018). Staphylococcus aureus strategies to evade the host acquired immune response. Int J Med Microbiol 308, 625–630. [DOI] [PubMed] [Google Scholar]
- Goodyear CS, and Silverman GJ (2003). Death by a B cell superantigen: In vivo VH-targeted apoptotic supraclonal B cell deletion by a Staphylococcal Toxin. J Exp Med 197, 1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtman A, Begier E, Mohamed N, Baber J, Sabharwal C, Haupt RM, Edwards H, Cooper D, Jansen KU, and Anderson AS (2019). The development of a staphylococcus aureus four antigen vaccine for use prior to elective orthopedic surgery. Hum Vaccin Immunother 15, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Zaba LC, Cardinale I, Nograles KE, Khatcherian A, Novitskaya I, Carucci JA, Bergman R, et al. (2008). Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. J Immunol 181, 7420–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim CE, Vidlak D, and Kielian T (2015). Interleukin-10 production by myeloid-derived suppressor cells contributes to bacterial persistence during Staphylococcus aureus orthopedic biofilm infection. J Leukoc Biol 98, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidron AI, Kempker R, Moanna A, and Rimland D (2010). Methicillin-resistant Staphylococcus aureus in HIV-infected patients. Infect Drug Resist 3, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtfreter S, Radcliff FJ, Grumann D, Read H, Johnson S, Monecke S, Ritchie S, Clow F, Goerke C, Broker BM, et al. (2013). Characterization of a mouse-adapted Staphylococcus aureus strain. PLoS One 8, e71142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener AB, Thurlow LT, Kang S, Spidale NA, Clarke SH, Cunnion KM, Tisch R, Richardson AR, and Vilen BJ (2017). Staphylococcus aureus Protein A Disrupts Immunity Mediated by Long-Lived Plasma Cells. J Immunol 198, 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Cheng AG, Kim HY, Missiakas DM, and Schneewind O (2010). Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J Exp Med 207, 1863–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, Nadle J, Kainer MA, Dumyati G, Petit S, et al. (2019). Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections - United States. MMWR Morb Mortal Wkly Rep 68, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, Noble L, Brown MJ, Zorman JK, Wang XM, et al. (2006). A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun 74, 2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebon A, Labout JA, Verbrugh HA, Jaddoe VW, Hofman A, van Wamel W, Moll HA, and van Belkum A (2008). Dynamics and determinants of Staphylococcus aureus carriage in infancy: the Generation R Study. J Clin Microbiol 46, 3517–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Olaniyi R, Kwiecinski JM, and Wardenburg JB (2020). Staphylococcus aureus toxin suppresses antigen-specific T cell responses. J Clin Invest 130, 1122–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Perez NE, Hopkins CA, and Pier GB (1988). Purified capsular polysaccharide-induced immunity to Staphylococcus aureus infection. J Infect Dis 157, 723–730. [DOI] [PubMed] [Google Scholar]
- Levy R, Okada S, Beziat V, Moriya K, Liu C, Chai LY, Migaud M, Hauck F, Al Ali A, Cyrus C, et al. (2016). Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci U S A 113, E8277–E8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Castillo JM, Das M, Kane J, Strakosha M, Singh S, Wong DSH, Horswill AR, Karasuyama H, Brombacher F, Miller LS, et al. (2021). Basophil-derived IL-4 promotes cutaneous Staphylococcus aureus infection. JCI Insight 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Beesetty P, Gerges G, Kleinhenz M, Moore-Clingenpeel M, Yang C, Ahmed LB, Hensley J, Steele L, Chong AS, et al. (2021). Impaired T lymphocyte responses during childhood Staphylococcus aureus infection. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Ibrahim AS, Xu X, Farber JM, Avanesian V, Baquir B, Fu Y, French SW, Edwards JE Jr., and Spellberg B (2009). Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog 5, e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, Fulcher DA, Tangye SG, and Cook MC (2008). Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med 205, 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitto MC, Dillen CA, Liu H, Miller RJ, Archer NK, Ortines RV, Alphonse MP, Marusina AI, Merleev AA, Wang Y, et al. (2019). Clonal Vgamma6(+)Vdelta4(+) T cells promote IL-17-mediated immunity against Staphylococcus aureus skin infection. Proc Natl Acad Sci U S A 116, 10917–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely TB, Shah NA, Fridman A, Joshi A, Hartzel JS, Keshari RS, Lupu F, and DiNubile MJ (2014). Mortality among recipients of the Merck V710 Staphylococcus aureus vaccine after postoperative S. aureus infections: an analysis of possible contributing host factors. Hum Vaccin Immunother 10, 3513–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LS, and Cho JS (2011). Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol 11, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LS, Fowler VG, Shukla SK, Rose WE, and Proctor RA (2020). Development of a vaccine against Staphylococcus aureus invasive infections: Evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol Rev 44, 123–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, et al. (2008). Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452, 773–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S, Martinson JA, Ghosh S, Watson R, and Pahan K (2012). Protection of Tregs, suppression of Th1 and Th17 cells, and amelioration of experimental allergic encephalomyelitis by a physically-modified saline. PLoS One 7, e51869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, and Bakaletz LO (2003). The fourth surface-exposed region of the outer membrane protein P5-homologous adhesin of nontypable Haemophilus influenzae is an immunodominant but nonprotective decoying epitope. J Immunol 171, 1978–1983. [DOI] [PubMed] [Google Scholar]
- Narita K, Hu DL, Asano K, and Nakane A (2019). Interleukin-10 (IL-10) Produced by Mutant Toxic Shock Syndrome Toxin 1 Vaccine-Induced Memory T Cells Downregulates IL-17 Production and Abrogates the Protective Effect against Staphylococcus aureus Infection. Infect Immun 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Ishikawa M, Morita Y, Yokogawa N, Xie C, de Mesy Bentley KL, Ito H, Kates SL, Daiss JL, and Schwarz EM (2020). IsdB antibody-mediated sepsis following SA surgical site infection. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard TK, Pallister KB, DuMont AL, DeWald M, Watkins RL, Pallister EQ, Malone C, Griffith S, Horswill AR, Torres VJ, et al. (2012). Alpha-toxin induces programmed cell death of human T cells, B cells, and monocytes during USA300 infection. PLoS One 7, e36532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcina M, Miranda-Garcia MA, Durlanik S, Ziegler S, Over B, Georg P, Foermer S, Ammann S, Hilmi D, Weber KJ, et al. (2013). Pathogen-triggered activation of plasmacytoid dendritic cells induces IL-10-producing B cells in response to Staphylococcus aureus. J Immunol 190, 1591–1602. [DOI] [PubMed] [Google Scholar]
- Paterson MJ, Caldera JR, Nguyen C, Sharma P, Castro AM, Kolar SL, Tsai CM, Limon JJ, Becker CA, Martins GA, et al. (2020). Harnessing antifungal immunity in pursuit of a Staphylococcus aureus vaccine strategy. PLoS Pathog 16, e1008733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli NT, Kim HK, Falugi F, Huang M, Dulac J, Henry Dunand C, Zheng NY, Kaur K, Andrews SF, Huang Y, et al. (2014). Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J Exp Med 211, 2331–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng KT, Hsieh CC, Huang TY, Chen PC, Shih HN, Lee MS, and Chang PJ (2017). Staphylococcus aureus biofilm elicits the expansion, activation and polarization of myeloid-derived suppressor cells in vivo and in vitro. PLoS One 12, e0183271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke EE, Brown SM, Pelzek AJ, Fulmer Y, Hernandez DN, Torres VJ, Thomsen IP, Chiang WK, Miller AO, Shopsin B, et al. (2018). Hierarchy of human IgG recognition within the Staphylococcus aureus immunome. Sci Rep 8, 13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JR, Armbruster NS, Gunter M, Henes J, and Autenrieth SE (2018). Staphylococcus aureus PSM Peptides Modulate Human Monocyte-Derived Dendritic Cells to Prime Regulatory T Cells. Front Immunol 9, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JB, Schneerson R, Horwith G, Naso R, and Fattom A (2004). Staphylococcus aureus types 5 and 8 capsular polysaccharide-protein conjugate vaccines. Am Heart J 147, 593–598. [DOI] [PubMed] [Google Scholar]
- Romero Pastrana F, Neef J, Koedijk D, de Graaf D, Duipmans J, Jonkman MF, Engelmann S, van Dijl JM, and Buist G (2018). Human antibody responses against non-covalently cell wall-bound Staphylococcus aureus proteins. Sci Rep 8, 3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose WE, Eickhoff JC, Shukla SK, Pantrangi M, Rooijakkers S, Cosgrove SE, Nizet V, and Sakoulas G (2012). Elevated serum interleukin-10 at time of hospital admission is predictive of mortality in patients with Staphylococcus aureus bacteremia. J Infect Dis 206, 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp ME, Holley HP Jr., Lutz J, Dicpinigaitis PV, Woods CW, Levine DP, Veney N, and Fowler VG Jr. (2007). Phase II, randomized, multicenter, double-blind, placebo-controlled trial of a polyclonal anti-Staphylococcus aureus capsular polysaccharide immune globulin in treatment of Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 51, 4249–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Kolar SL, Muller S, Reyes CN, Wolf AJ, Ogawa C, Singhania R, De Carvalho DD, Arditi M, Underhill DM, et al. (2017). O-Acetylation of Peptidoglycan Limits Helper T Cell Priming and Permits Staphylococcus aureus Reinfection. Cell Host Microbe 22, 543–551 e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner J, Kretschmer D, Klenk J, Otto M, Buhring HJ, Stevanovic S, Wang JM, Beer-Hammer S, Peschel A, and Autenrieth SE (2013). Staphylococcus aureus phenol-soluble modulin peptides modulate dendritic cell functions and increase in vitro priming of regulatory T cells. J Immunol 190, 3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y, Zhao F, Beesetty P, Weiskopf D, Li Z, Tian Q, Alegre ML, Sette A, Chong AS, and Montgomery CP (2020). Inhibition of protective immunity against Staphylococcus aureus infection by MHC-restricted immunodominance is overcome by vaccination. Sci Adv 6, eaaw7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman GJ, Cary SP, Dwyer DC, Luo L, Wagenknecht R, and Curtiss VE (2000). A B cell superantigen-induced persistent “Hole” in the B-1 repertoire. J Exp Med 192, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik D, Merighi M, Grout M, Gadjeva M, Maira-Litran T, Ericsson M, Goldmann DA, Huang SS, Datta R, Lee JC, et al. (2010). Animal and human antibodies to distinct Staphylococcus aureus antigens mutually neutralize opsonic killing and protection in mice. J Clin Invest 120, 3220–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CS, White CJ, Ibrahim AS, Filler SG, Fu Y, Yeaman MR, Edwards JE Jr., and Hennessey JP Jr. (2012). NDV-3, a recombinant alum-adjuvanted vaccine for Candida and Staphylococcus aureus, is safe and immunogenic in healthy adults. Vaccine 30, 7594–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan AN, Henry T, van Rooijen WJM, Perret M, Badiou C, Aerts PC, Kemmink J, de Haas CJC, van Kessel KPM, Vandenesch F, et al. (2013). The staphylococcal toxin Panton-Valentine Leukocidin targets human C5a receptors. Cell Host Microbe 13, 584–594. [DOI] [PubMed] [Google Scholar]
- Spaan AN, van Strijp JAG, and Torres VJ (2017). Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat Rev Microbiol 15, 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Czartoski J, Wan YH, Homad LJ, Rubin V, Glantz H, Neradilek M, Seydoux E, Jennewein MF, MacCamy AJ, et al. (2021). mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger-Jones YK, Bae T, and Schneewind O (2006). Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A 103, 16942–16947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam K, Lacey KA, Devlin JC, Coffre M, Sommerfield A, Chan R, O’Malley A, Koralov SB, Loke P, and Torres VJ (2020). Targeting leukocidin-mediated immune evasion protects mice from Staphylococcus aureus bacteremia. J Exp Med 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teymournejad O, and Montgomery CP (2021). Evasion of Immunological Memory by SA Infection: Implications for Vaccine Design. Front Immunol 12, 633672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammavongsa V, Kim HK, Missiakas D, and Schneewind O (2015). Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 13, 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CM, Soper N, Bennett M, Fallon JK, Michell AR, Alter G, Liu GY, and Thomsen I (2021). Adoptive Transfer of Serum Samples From Children With Invasive Staphylococcal Infection and Protection Against Staphylococcus aureus Sepsis. J Infect Dis 223, 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan K, Visweswaran GRR, Chandrasekaran R, Trakhimets O, Brown SL, Watson A, Zuck M, Dambrauskas N, Raappana A, Carbonetti S, et al. (2021). Antibody interference by a non-neutralizing antibody abrogates humoral protection against Plasmodium yoelii liver stage. Cell Rep 36, 109489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems JJ Jr., Steinberg JP, Filler S, Baddley JW, Corey GR, Sampathkumar P, Winston L, John JF, Kubin CJ, Talwani R, et al. (2006). Phase II, randomized, double-blind, multicenter study comparing the safety and pharmacokinetics of tefibazumab to placebo for treatment of Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 50, 2751–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman LE, Thackray HM, Steinhorn RH, Walsh WF, Lassiter HA, Dhanireddy R, Brozanski BS, Palmer KG, Trautman MS, Escobedo M, et al. (2011). A randomized study of a monoclonal antibody (pagibaximab) to prevent staphylococcal sepsis. Pediatrics 128, 271–279. [DOI] [PubMed] [Google Scholar]
- Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, and Verbrugh HA (2004). Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364, 703–705. [DOI] [PubMed] [Google Scholar]
- Wrightsman RA, Dawson BD, Fouts DL, and Manning JE (1994). Identification of immunodominant epitopes in Trypanosoma cruzi trypomastigote surface antigen-1 protein that mask protective epitopes. J Immunol 153, 3148–3154. [PubMed] [Google Scholar]
- Wu Y, Liu X, Akhgar A, Li JJ, Mok H, Sellman BR, Yu L, Roskos LK, Esser MT, and Ruzin A (2018). Prevalence of IgG and Neutralizing Antibodies against Staphylococcus aureus Alpha-Toxin in Healthy Human Subjects and Diverse Patient Populations. Infect Immun 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SX, and McCormick JK (2012). Staphylococcal superantigens in colonization and disease. Front Cell Infect Microbiol 2, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, Leung DY, and Schlievert PM (2000). Evidence for the involvement of bacterial superantigens in psoriasis, atopic dermatitis, and Kawasaki syndrome. FEMS Microbiol Lett 192, 1–7. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Umeda A, and Ohshima Y (1987). Induction of resistance in mice by the capsular polysaccharide antigens of Staphylococcus aureus. Microbiol Immunol 31, 649–656. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Robbie GJ, Wu Y, Esser MT, Jensen K, Schwartz HI, Bellamy T, Hernandez-Illas M, and Jafri HS (2017). Safety, Tolerability, and Pharmacokinetics of MEDI4893, an Investigational, Extended-Half-Life, Anti-Staphylococcus aureus Alpha-Toxin Human Monoclonal Antibody, in Healthy Adults. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jacobsson K, Strom K, Lindberg M, and Frykberg L (1999). Staphylococcus aureus expresses a cell surface protein that binds both IgG and beta2-glycoprotein I. Microbiology (Reading) 145 ( Pt 1), 177–183. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jacobsson K, Vasi J, Lindberg M, and Frykberg L (1998). A second IgG-binding protein in Staphylococcus aureus. Microbiology (Reading) 144 ( Pt 4), 985–991. [DOI] [PubMed] [Google Scholar]