Abstract
The specific rates of growth, substrate utilization, and ethanol production as well as yields of biomass and ethanol production on xylose for the recombinant Zymomonas mobilis ZM4(pZB5) were shown to be much less than those on glucose or glucose-xylose mixtures. Typical fermentations with ZM4(pZB5) growing on glucose-xylose mixtures followed two-phase growth kinetics with the initial uptakes of glucose and xylose being followed by slower growth and metabolic uncoupling on xylose after glucose depletion. The reductions in rates and yields from xylose metabolism were considered in the present investigation and may be due to a number of factors, including the following: (i) the increased metabolic burden from maintenance of plasmid-related functions, (ii) the production of by-products identified as xylitol, acetate, lactate, acetoin, and dihydroxyacetone by 13C-nuclear magnetic resonance (NMR) spectroscopy and high-performance liquid chromatography, (iii) growth inhibition due to xylitol by the putative inhibitory compound xylitol phosphate, and (iv) the less energized state of ZM4(pZB5). In vivo 31P-NMR studies have established that the levels of NTP and UDP sugars on xylose were less than those on glucose, and this energy limitation is likely to restrict the growth of the recombinant strain on xylose media.
Zymomonas mobilis has attracted widespread interest for fuel ethanol production because of its higher specific rates of sugar uptake and ethanol production, higher ethanol tolerance, and higher ethanol conversion efficiencies when compared to the traditionally used yeasts (10, 14, 21, 22, 26). However, wild-type strains of Z. mobilis can only utilize glucose, fructose, and sucrose, and they lack the pentose metabolism pathway necessary to ferment such sugars as xylose or arabinose. The cloning of enzymes for xylose assimilation and metabolism in Z. mobilis has now been reported (30), and a subsequent study has resulted in the successful integration of the requisite genes into the Zymomonas genome (M. Zhang, Y. C. Chou, X. K. Lai, S. Milstrey, N. Danielson, K. Evans, A. Mohagheghi, and M. Finkelstein, Abstr. 21st Symp. Biotechnol. Fuels Chem., abstr. 2–16, 1999). These genetically engineered strains can now convert xylose to ethanol by the combined use of the Entner-Doudoroff and pentose pathways facilitated by the cloned enzymes xylose isomerase and xylulokinase for xylose assimilation and by transketolase and transaldolase for pentose metabolism. In a further study, the cloning of three additional enzymes for arabinose utilization has been reported (4). However, when xylose is the sole carbon source, lower biomass yields and slower growth rates as well as lower ethanol yields for recombinant strains have been reported (9, 11, 12, 23; H. G. Lawford and J. D. Rousseau, Abstr. 21st Symp. Biotechnol. Fuels Chem., abstr. 2–24, 1999).
In this study, the fermentation characteristics of the recombinant Z. mobilis ZM4(pZB5) on xylose or glucose alone, or on xylose-glucose mixtures, have been investigated to determine possible reasons for decreased cell and ethanol yields. By-product formation has been evaluated by 13C nuclear magnetic resonance (NMR) spectroscopy as well as the energy status of the recombinant strain by in vivo 31P-NMR spectroscopy. The latter noninvasive technique provides information on the energy status of the cells by virtue of its ability to determine the various intracellular nucleotide phosphates and other energy-rich compounds, as well as on changes in intracellular pH, from the chemical shifts of internal phosphate and other phosphorylated intermediates (16, 17). Studies on wild-type strains of Z. mobilis with similar NMR spectroscopy techniques have been reported previously (2, 25), with more recent work on a recombinant strain growing in xylose-fed continuous culture now reported. Interestingly, the results of the latter analysis with 13C-NMR spectroscopy have identified a metabolic bottleneck in the recombinant xylose-fermenting Z. mobilis strain at the level of heterologous xylulokinase (5).
MATERIALS AND METHODS
Organism and culture maintenance.
The xylose-fermenting recombinant Z. mobilis ZM4(pZB5) and host strain Z. mobilis ZM4 (ATCC 31821) were used in this work, with the recombinant strain being kindly provided by Min Zhang, National Renewable Energy Laboratory, Golden, Colo., under a Material Transfer Agreement (30). For long-term storage, these strains were kept at −70°C in 150 g of glycerol per liter. For use in experiments, the strains were maintained on a rich agar medium containing (per liter) 20 g of xylose for ZM4(pZB5) (20 g of glucose for ZM4), 10 g of yeast extract (Oxoid), and 20 g of agar (agar no. 1; Oxoid) at pH 5.4. Ten milligrams of tetracycline per liter was added to the media as a selective pressure for the recombinant strain. Colonies were grown on this medium for 3 days at 30°C and then stored at 4°C for no longer than 2 weeks before use as inocula in liquid media.
Media composition and preparation.
First seed medium contained (per liter) 25 g of xylose for ZM4(pZB5) (25 g of glucose for ZM4), 10 g of yeast extract, 1 g of MgSO4 · 7H2O, 1 g of (NH4)2SO4, and 2 g of KH2PO4. Second seed culture medium was identical in composition to the main culture medium. Main culture medium was identical to first seed medium except for the reduced yeast extract concentration (5 g/liter) and the range of sugar concentrations indicated. The sugars and phosphate were autoclaved separately from the other media components.
Preparation of inocula.
All inocula were prepared at 30°C. A single colony of ZM4(pZB5) or ZM4 was transferred from the stock culture plate to 10 ml of first seed culture medium in a 15-ml cap tube and incubated statically for 24 h. The culture was transferred to 140 ml of second seed medium in a 250-ml flask. After 15 h of static incubation, the culture was inoculated into the main culture medium to yield an optical density (660 nm, 1-cm light path) of approximately 0.05, which corresponded to approximately 15 mg of dry cell weight per liter.
Batch fermentations.
For kinetic analysis of ZM4(pZB5), controlled batch experiments were conducted in a 2-liter fermentor (LH Engineering, Maidenhead, Berkshire, United Kingdom) with a working volume of 1.5 liters. The main culture was carried out under nonaerated conditions, with mild agitation of 200 rpm provided to maintain a homogeneous culture. All the cultures were maintained at 30°C and pH 5.0 (by addition of 2 M NaOH). For evaluation of the effect of xylitol on the growth of Zymomonas strains, flask fermentation experiments were conducted in 0.5-liter screw-cap Erlenmeyer flasks with working volumes of 0.2 liters. These flask cultures were incubated in a shaking incubator with mild shaking at 100 rpm. Samples for sugar, ethanol, and by-product determinations were collected at various times and stored at −20°C prior to analysis.
Identification and quantification of by-products.
By-products of the xylose fermentations were analyzed by 13C-NMR spectroscopy and high-performance liquid chromatography (HPLC). 13C-NMR measurements were performed on a Bruker DMX-600 spectrometer, operating at 175 MHz for the 13C nucleus. Spinning sample tubes with a 5-mm outside diameter were used at a temperature of 30°C. Spectra were obtained with a sweep width corresponding to 240 ppm, using a 90° pulse with a 2-s repetition rate. The carbon nuclei were proton decoupled by broad band irradiation with a noise-modulated band width equivalent to 10 ppm for the 1H nucleus. Chemical shifts were expressed as parts per million downfield from tetramethylsilane (TMS), with the primary reference being dimethyl sulfoxide (DMSO), which has a resonance at 39.5 ppm relative to that of TMS. DMSO also served as an internal standard for quantification purposes. Quantification was carried out by measuring the height of the by-product peaks normalized to the height of DMSO resonance, relative to a standard curve obtained under identical spectrometer conditions by measurements of the heights of the peaks after the addition of known amounts of pure standards. The by-products identified by 13C-NMR spectroscopy were verified and quantified by HPLC analysis. An Aminex HPX-87H column (Bio-Rad) was used for HPLC to identify and quantify by-products for the same operating conditions as for sugar and ethanol analysis, as described in the following analytical methods.
Evaluation of xylitol as a possible substrate.
Strains were grown to mid-logarithmic phase and washed twice with saline phosphate buffer. The washed cells were used then to inoculate media containing xylitol (20 or 50 g/liter) as a possible carbon source. Any growth or xylitol uptake was monitored by changes in optical densities at 660 nm and ethanol concentrations.
Evaluation of xylitol as a possible growth inhibitor.
Various concentrations of xylitol were added to the main culture media, as indicated in Results. The growth of cells, sugar utilization, and ethanol production was measured under these conditions.
Analytical methods.
Cell growth was determined by optical density measurements (at 660 nm). Biomass concentration was measured by dry cell weight after centrifuging the cells (1700 × g, 10 min) from a known volume of culture sample, washing twice with distilled water, and then drying to constant weight at 105°C.
The concentrations of sugars, ethanol, and by-products were determined from sample supernatants with a Waters high-performance liquid chromatograph and an Aminex HPX-87H column (Bio-Rad) with 5 mM H2SO4 (65°C, 0.6 ml/min) as the mobile phase. For quantification of sugars, ethanol, xylitol, glycerol, acetic acid, and acetoin, a refractive index detector (Waters) was used, and for quantification of dihydroxyacetone, a UV detector (Waters) at 220 nm was used. Standards containing known concentrations of mixed components were run periodically to verify calibration accuracy. Lactic acid concentrations were analyzed with a YSI 2300 STAT Plus analyzer (Yellow Springs Instrument Co., Yellow Springs, Ohio).
In vivo 31P-NMR.
Cells were grown in batch culture to the late-exponential growth phase, as determined by optical density and sugar concentrations. They were harvested by centrifugation at 3,000 × g for 15 min at 4°C and then washed in 100 mM MES (morpholineethanesulfonic acid) buffer (pH 5.5) containing 0.1% KH2PO4 and 0.1% MgCl2 · 6H2O. In vivo 31P-NMR spectroscopy studies are generally limited by the relatively low sensitivity of this technique (17). To overcome this limitation, the cells were concentrated to approximately 1.7 × 1011 cells/ml. The resulting cell suspension was kept on ice until used for NMR spectroscopy experiments. Samples for in vivo 31P-NMR measurements contained the following (final volume, 4.0 ml): 2.92 ml of cell suspension, 0.31 ml of D2O, 0.05 ml of 0.24 M triethylphosphate (TEP), and 0.72 ml of 1.54 M glucose or 1.54 M xylose (equivalent to 277 mM concentrations of each sugar). All 31P-NMR measurements were performed at 30°C. Spectra were obtained with a Bruker DMX-500 spectrometer, operating in the Fourier transform mode and using a 10-mm broad band multinuclear probe. 31P-NMR spectra were recorded at 202.46 MHz with a recycle time of 1.0 s and a flip angle of 60°. NMR spectra were acquired in 5-min blocks of 300 scans using composite pulse 1H decoupling in a bilevel scheme with 2-W decoupler power during acquisition.
Calculation of kinetic parameters.
The maximum specific growth rates were calculated at the exponential phase of growth on xylose, glucose, or glucose-xylose mixtures or at the phase of growth when glucose was fully utilized and only xylose remained. The maximum specific sugar uptake rates (qmax,s) and maximum specific ethanol production rates (qmax,p) were calculated over the exponential phase of growth and based on the following formulae: qmax,s = (1/x) (ds/dt) and qmax,p = (1/x) (dp/dt), where x, s, and p are the concentrations of biomass, sugars, and ethanol, respectively.
For the xylose utilization phase after the complete depletion of glucose during glucose and xylose cofermentation, during which growth was very limited, the formulae were modified to the following: qs = (1/xav) (Δs/Δt) and qp = (1/xav) (Δp/Δt), where Δs and Δp are the changes in the xylose and ethanol concentrations, respectively, over the time period Δt (usually the first 4 to 6 h), and xav is the average biomass concentration over Δt. The overall yields for biomass (Yx/s) and ethanol (Yp/s) production on sugar mixture media were based on the initial and final concentrations of biomass, sugars (combined sugars for cofermentation), and ethanol.
RESULTS
Comparative fermentation performances of Z. mobilis ZM4(pZB5) on xylose, glucose, and mixed glucose-xylose media.
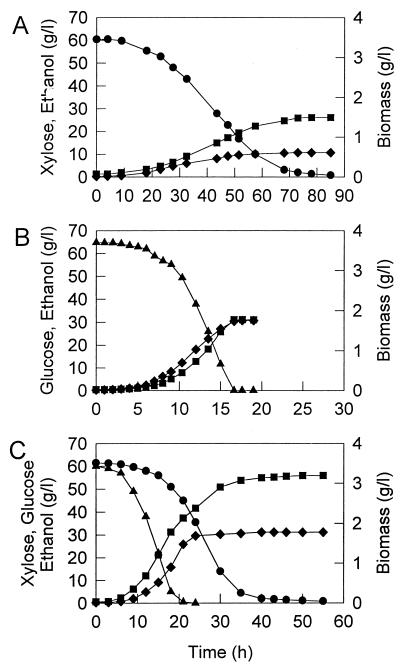
Figure 1 shows time courses of batch fermentation of ZM4(pZB5) with xylose, glucose, and a glucose-xylose mixture under controlled environmental conditions. On the xylose medium, cell growth occurred at a maximum specific rate of 0.11 h−1 and ethanol was produced at a yield of 0.42 g/g of xylose consumed, corresponding to an 81% theoretical yield. By comparison, on the glucose medium, the maximum specific growth rate was 0.42 h−1 and the ethanol yield was 0.47 g/g of glucose consumed, corresponding to a 93% theoretical yield. The fermentation of ZM4(pZB5) growing on the glucose-xylose mixture followed two-phase growth kinetics with the initial uptake of glucose and xylose being followed by slower growth on xylose after glucose depletion. Following glucose utilization, the specific growth rate decreased gradually to zero, while the slower uptake of remaining xylose and ethanol production continued with increasingly uncoupled metabolism. A similar kinetic pattern has been reported by Joachimsthal et al. (9).
FIG. 1.
Fermentation characteristics of recombinant strain ZM4(pZB5) using xylose (60.5 g/liter) (A), glucose (64.8 g/liter) (B), and a mixture of glucose (60.0 g/liter) and xylose (61.5 g/liter) (C). ●, Xylose; ▴, glucose; ■, ethanol; ⧫, biomass.
The kinetic parameters of ZM4(pZB5) for different sugar concentrations are given in Table 1. The specific rates of growth, substrate utilization, and ethanol production as well as yields of biomass and ethanol production on xylose were much lower than those on glucose or for glucose-xylose mixtures. The overall yields of biomass were significantly reduced with the increasing sugar levels due to the increased metabolic uncoupling which occurred at the higher ethanol concentrations, with the values obtained with xylose being much lower than those for glucose medium.
TABLE 1.
Kinetic parameters of Z. mobilis ZM4(pZB5) in xylose, glucose, or mixed glucose-xylose media
| Kinetic parameter | Xylose concn (g/liter)
|
Glucose concn (g/liter)
|
Glucose concn-xylose concn (g/liter)
|
||||
|---|---|---|---|---|---|---|---|
| 27.8 | 40.3 | 60.5 | 27.5 | 64.8 | 26.8–32.0 | 60.0–61.5 | |
| Glucose and/or xylose metabolism | |||||||
| μmax (h−1)a | 0.13 | 0.12 | 0.11 | 0.43 | 0.42 | 0.42 | 0.22 |
| qmax,glucose (g/g/h) | 9.34 | 10.63 | 9.10 | 8.60 | |||
| qmax,xylose (g/g/h) | 3.06 | 3.24 | 3.38 | 2.49 | 1.41 | ||
| qmax,p (g/g/h) | 1.39 | 1.37 | 1.40 | 4.48 | 5.11 | 4.73 | 4.62 |
| Xylose metabolismb | |||||||
| qs (g/g/h) | 3.01 | 2.13 | |||||
| qp (g/g/h) | 1.36 | 0.91 | |||||
| Overall yield | |||||||
| Yx/s (g/g) | 0.019 | 0.014 | 0.010 | 0.036 | 0.027 | 0.031 | 0.015 |
| Yp/s (g/g) | 0.45 | 0.42 | 0.41 | 0.48 | 0.47 | 0.46 | 0.46 |
μmax, the maximum specific growth rate.
Kinetic parameters for xylose utilization phase after the complete depletion of glucose during glucose and xylose cofermentation.
Identification of by-products formed during batch fermentation of Z. mobilis ZM4(pZB5) on xylose.
The 13C-NMR spectrum of culture supernatant from a fermentation of ZM4(pZB5) on xylose (50 g/liter) was determined, and the presence of xylose, ethanol, xylitol, glycerol, acetate, lactate, acetoin, and dihydroxyacetone was identified. Table 2 gives the assignments of the resonances associated with each of these components. Additionally, the presence of each of these compounds was confirmed by HPLC. The peaks from the HPLC analysis were assigned by comparing retention times with authentic compounds. From the 13C-NMR and HPLC analyses, xylitol was found to be the major by-product.
TABLE 2.
Chemical shift and assignment of resonances for 13C-NMR spectrum of recombinant strain ZM4(pZB5) culture supernatant from xylose (50 g/liter) fermentationa
| Xylose or product | Resonance at carbon:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Xylose α | 93.1 | 72.5 | 73.9 | 70.4 | 61.9 |
| Xylose β | 97.5 | 75.1 | 76.8 | 70.2 | 66.1 |
| Xylitol | 63.9 | 73.2 | 72.0 | 73.2 | 63.9 |
| Ethanol | 58.3 | 17.8 | |||
| Glycerol | 64.0 | 73.5 | 64.0 | ||
| Acetate | 182.1 | 24.1 | |||
| Lactate | 183.2 | 69.3 | 20.9 | ||
| Acetoin | 216.2 | 73.9 | 25.8 | 19.1 | |
| Dihydroxyacetone | 66.0 | 213.1 | 66.0 | ||
| Dihydroxyacetone hydrate | 64.9 | 96.5 | 64.9 | ||
Chemical shifts expressed as parts per million downfield from TMS. DMSO was used as the internal standard at 39.5 ppm. NMR operation: 90° pulse, 2-s pulse repetition, 16,000 scans accumulated.
Kinetics of by-products formation by Z. mobilis ZM4(pZB5).
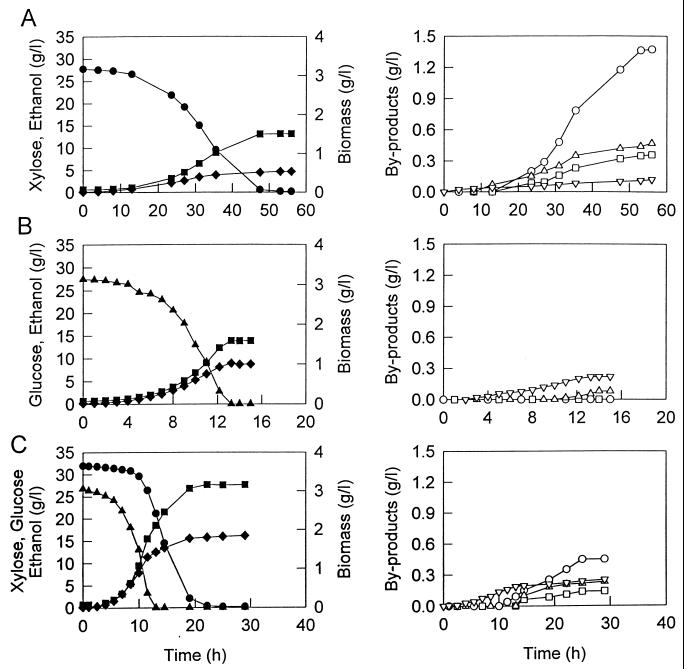
Figure 2 shows time courses of growth, substrate utilization, and products formation for ZM4(pZB5) growing on xylose, glucose, and a glucose-xylose mixture under controlled environmental conditions. For the growth of ZM4(pZB5) with xylose as a sole carbon source, xylitol was produced as the main by-product, with its production closely related to cell growth. Dihydroxyacetone, acetate, and glycerol were also produced at significant levels. Acetoin and lactic acid were minor by-products. With glucose as the sole carbon source, acetoin and acetate were produced as the main by-products; however, xylitol was not produced.
FIG. 2.
Fermentation characteristics of recombinant strain ZM4(pZB5) using xylose (27.8 g/liter) (A), glucose (27.5 g/liter) (B), and a mixture of glucose (26.8 g/liter) and xylose (32.0 g/liter) (C). ●, Xylose; ▴, glucose; ■, ethanol; ⧫, biomass; ○, xylitol; ▵, acetic acid; □, glycerol; ▿, acetoin.
The effects of increasing levels of xylose on by-product formation are summarized in Table 3. Production of xylitol occurred during cell growth in parallel with ethanol production and was approximately proportional to the initial xylose concentrations. The final concentrations of other major by-products, dihydroxyacetone, glycerol, and acetic acid, were also directly related to the initial xylose concentration.
TABLE 3.
The production of by-products and percentage of ATP loss due to by-product production by Z. mobilis ZM4(pZB5) in xylose, glucose, or mixed glucose-xylose media
| Fermentation product | Xylose concn (g/liter)
|
Glucose concn (g/liter)
|
Glucose concn-xylose concn (g/liter)
|
||||
|---|---|---|---|---|---|---|---|
| 27.8 | 40.3 | 60.5 | 27.5 | 64.8 | 26.8/32.0 | 60.0/61.5 | |
| Ethanol | 12.6 | 17.0 | 25.1 | 13.3 | 30.5 | 27.0 | 55.7 |
| Xylitol | 1.37 | 2.68 | 4.86 | 0 | 0 | 0.45 | 0.91 |
| Dihydroxyacetone | 0.68 | 1.34 | 2.10 | 0 | 0.14 | 0.25 | 0.88 |
| Glycerol | 0.36 | 0.80 | 1.26 | 0 | 0.10 | 0.14 | 0.45 |
| Acetic acid | 0.47 | 0.67 | 1.21 | 0.08 | 0.17 | 0.23 | 0.68 |
| Acetoin | 0.12 | 0.15 | 0.18 | 0.22 | 0.32 | 0.25 | 0.52 |
| Lactic acid | 0.017 | 0.016 | 0.022 | 0.008 | 0.014 | 0.022 | 0.035 |
| Theoretical maximum ATP produced from sugar(s) utilized (mmol)a | 185.2 | 268.5 | 403.1 | 152.6 | 359.6 | 361.9 | 742.7 |
| Loss of ATP for by-product production (mmol)b | 32.0 | 64.8 | 106.4 | 0 | 5.3 | 11.6 | 35.3 |
| % ATP loss | 17.3 | 24.1 | 26.4 | 0 | 1.5 | 3.2 | 4.8 |
The sugar concentrations were converted to micromoles per liter by dividing xylose concentration (gram/liter) by 0.150 and glucose concentration (gram/liter) by 0.180.
The amount of ATP lost due to by-product production was calculated from the concentrations of dihydroxyacetone, glycerol, and xylitol produced during fermentation.
Effect of xylitol on growth.
ZM4(pZB5) and its parental strain, ZM4, were tested in a medium containing xylitol as the sole carbon source to see whether growth and/or fermentation on xylitol was possible. Growth was monitored by changes in optical density at 660 nm. However, no growth or increase in ethanol concentration was observed for either strain after incubation for 2 weeks. It was concluded therefore that neither ZM4(pZB5) nor ZM4 could utilize xylitol as a sole carbon source.
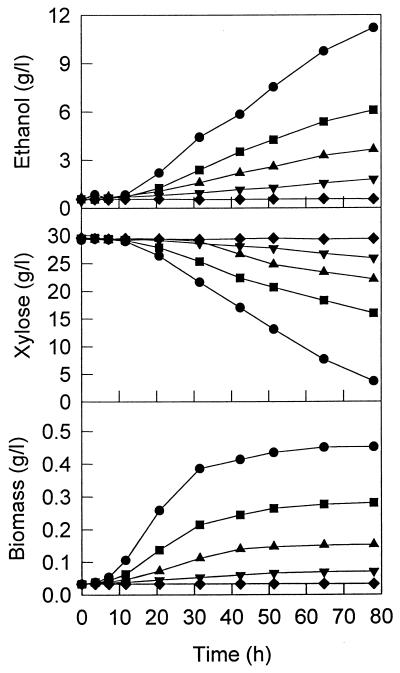
The effect of xylitol on the growth and fermentation of ZM4(pZB5) and ZM4 was tested although it was recognized that intracellular xylitol levels produced by xylose metabolism would be different from added extracellular levels. Addition of xylitol to the 30-g/liter xylose medium in shake flask experiments (at 30°C and an initial pH of 5.7) resulted in growth inhibition of ZM4(pZB5), with the degree of inhibition being dependent on the concentration of added xylitol (Fig. 3). Above concentrations of 5 g of xylitol per liter, ZM4(pZB5) could not grow at all. However, the growth of ZM4 in 30-g/liter glucose medium was not affected by xylitol addition even at concentrations of up to 10 g/liter (data not shown).
FIG. 3.
Effect of xylitol addition on the growth of recombinant ZM4(pZB5) using xylose (30 g/liter) in flask cultures at 30°C. ●, control without xylitol; ■, 0.5 g of xylitol/liter; ▴, 1 g of xylitol/liter; ▾, 2 g of xylitol/liter; ⧫, 5 g of xylitol/liter.
In vivo 31P-NMR analysis of glucose and xylose metabolism.
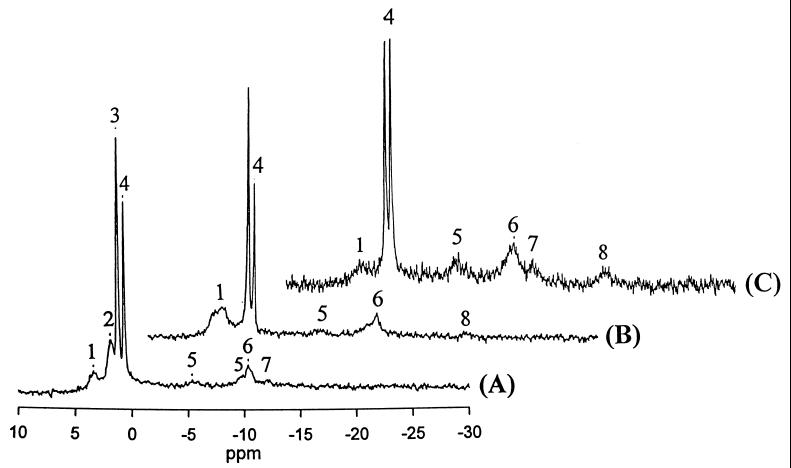
In order to compare the energy status of ZM4(pZB5) growing on xylose and glucose media, in vivo 31P-NMR spectroscopy experiments were conducted. Spectra obtained with suspensions of ZM4(pZB5) cells actively metabolizing xylose or glucose are shown in Fig. 4. Relatively broad intracellular resonances of sugar phosphates, inorganic phosphate, nucleoside diphosphates, NADH, and uridine diphosphosugars were identified before sugar addition (Fig. 4A). The resonances with the smaller line widths were from extracellular inorganic phosphate and TEP standard (0.44 ppm). After the cells had begun to metabolize xylose, there was a rapid buildup of intracellular sugar phosphates (about fourfold), with a concomitant decrease in the intracellular inorganic phosphate level (Fig. 4B). The intracellular resonances of nucleoside triphosphates (NTP) appeared at −18.4 ppm (NTP-β), −10.0 ppm (NTP-α), and −5.0 ppm (NTP-γ), although no NTP resonances were observed before the addition of xylose. However, NTP signals were small. Because in vivo resonances of NTP were broad, it was difficult to differentiate between ATP and the other NTP in the spectra. In most cells, however, it has been reported that more than 90% of NTP consist of ATP (18). In contrast, following addition of a similar concentration of glucose, strong resonances of NTP appeared, with a concomitant increase in resonances of sugar phosphates and UDP sugars (Fig. 4C).
FIG. 4.
31P-NMR spectra of Z. mobilis ZM4(pZB5) cells before the addition of sugars as the control (A) and cells actively metabolizing xylose (B) and glucose (C) at 30°C and pH 5.5. Spectra of actively metabolizing cells were obtained at 2.5 min following the addition of 277 mM xylose or 277 mM glucose. 1, sugar phosphates; 2, intracellular phosphate; 3, extracellular phosphate; 4, TEP as the internal standard; 5, NDP; 6, NAD and NADP; 7, UDP sugars; 8, β-NTP. The resonances of the α- and γ-NTP phosphate groups overlapped the NDP signals.
A chemical shift of intracellular inorganic phosphate [Pi(int)] resonance is an indication of a pH-sensitive resonance and gives information on the cytoplasmic pH. As soon as the cells began to utilize either sugar, the resonance of Pi(int) was shifted upfield and overlapped with the extracellular inorganic phosphate [Pi(ext)] resonance. The disappearance of the Pi(int) resonance during sugar metabolism was probably due to the rapid uptake of inorganic phosphate with the net symport of protons and the synthesis of sugar phosphates and NTP. It was difficult under these conditions to identify any pH changes because of the low intensity of the Pi(int) resonance and the overlapping of the Pi(int) and Pi(ext) resonances.
The chemical shifts of sugar phosphate resonances of glucose-utilizing cells were more upfield in the present investigation than those of xylose-utilizing cells, reflecting the accumulation of different sugar phosphates for the metabolism of the two sugars.
Table 4 provides an estimate of the relative concentrations of the phosphorylated metabolites from ZM4(pZB5) cell suspensions metabolizing either glucose or xylose. From the data, it is evident that the level of NTP in glucose-metabolizing cells was approximately 10-fold higher than that in xylose-metabolizing cells. The levels of UDP sugars, which are precursors for the synthesis of cell wall materials and storage carbohydrates, were also 10-fold higher in glucose-utilizing cells than in xylose-metabolizing cells. Interestingly, the intracellular sugar phosphates for the metabolism of glucose were approximately half those of xylose-metabolizing cells. By dividing the sugar phosphate concentrations by the corresponding specific rates of xylose or glucose utilization obtained at these 31P-NMR experimental conditions (viz., 1.78 and 2.99 g/g/h, respectively), normalized sugar phosphate values were obtained (15). Glucose-utilizing cells, which have higher rates of sugar metabolism, had significantly lower normalized levels of sugar phosphates than xylose-utilizing cells.
TABLE 4.
Relative concentrations of intracellular phosphorylated metabolites and total inorganic phosphates in Z. mobilis ZM4(pZB5) cell suspensionsa
| Carbon source | Relative concentration (arbitrary units) of:
|
|||
|---|---|---|---|---|
| Sugar phosphatesb | UDP sugars | NTP | Inorganic phosphate | |
| Control | 1.00 | NDc | ND | 7.99 |
| Xylose | 4.36 (2.45) | 0.07 | 0.18 | 4.28 |
| Glucose | 2.14 (0.72) | 0.82 | 1.88 | 3.45 |
31P-NMR spectra of Z. mobilis ZM4(pZB5) cells before the addition of sugars were used as controls. Concentrations were estimated by integration to determine the area under the peak of interest.
Values in parentheses are normalized values as described in the text.
ND, not determined (below the detection range).
31P-NMR spectra for glucose fermentation by the parental strain, ZM4, were similar to those of ZM4(pZB5) using glucose (data not shown). When comparing the spectra of recombinant ZM4(pZB5) using xylose with those using glucose, it was clear that xylose-fermenting ZM4(pZB5) cells were significantly less energized than those fermenting glucose.
DISCUSSION
When genetically engineering a microorganism to utilize a wider range of substrates for growth and fermentation, it is essential to understand the major factors which regulate carbon and energy metabolism. Although there have been reports about the fermentation kinetics of recombinant Zymomonas strains using xylose, glucose, and mixed glucose-xylose media (9), less has been discovered about the reason(s) why the specific rates of growth, substrate utilization, and ethanol production as well as yields of biomass and ethanol on xylose are significantly lower than those on glucose or glucose-xylose mixtures (5, 11, 12, 23; Lawford and Rousseau, Abstr. 21st Symp. Biotechnol. Fuels Chem.).
It has been established previously that the growth of Zymomonas on glucose can be significantly uncoupled, which leads to the high conversion efficiency of substrates to ethanol at high sugar concentrations due to the relatively low levels of biomass produced (13). The low biomass yield is partly due to its characteristic catabolism: glucose is converted to ethanol by the Entner-Doudoroff pathway, which yields only 1 mol of ATP/mol of glucose, and partly due to substrate consumption for energy spilling and maintenance reactions (26).
The reduction in biomass yield on xylose can be considered initially on the basis of the assumption for coupled growth and metabolism, which is 10.5 g of biomass/mol of ATP, giving theoretical yields of biomass on glucose and xylose of 0.058 and 0.073 g/g, respectively (Lawford and Rousseau, Abstr. 21st Symp. Biotechnol. Fuels Chem.). However, this theoretical value was never approached on the xylose media, and the biomass yields were much lower than those on glucose. The lower cell yields on xylose compared to those on glucose may be explained by a number of factors, as follows.
(i) Increased metabolic burden resulting from plasmid maintenance.
There have been a number of studies of a “plasmid burden” effect on the metabolism of recombinant bacteria, viz., the effect of an extrachromosomal plasmid(s) in reducing the growth rate, cell yield, and cell viability of these recombinant strains (3, 6, 24). In a recent report, an average biomass yield on xylose of 0.034 g/g of xylose for recombinant Z. mobilis CP4(pZB5) was compared with yields on glucose for the recombinant and wild-type cultures, viz., 0.055 and 0.058 g/g of glucose, respectively (Lawford and Rousseau, Abstr. 21st Symp. Biotechnol. Fuels Chem.). The difference of Yx/glucose between the wild type and the recombinant was explained by the additional plasmid burden effect on the recombinant, whereby energy that would otherwise have been available for growth was diverted for plasmid-related functions. However, this effect is unlikely to provide a complete explanation for the significant difference between Yx/glucose and Yx/xylose for ZM4(pZB5).
(ii) Loss of carbon and energy due to formation of products specific to xylose metabolism.
The metabolism of xylose by the recombinant strains might have an important influence on growth rate and biomass yield resulting from loss of carbon and possible ATP limitation due to formation of by-products. ATP losses (in percentage) due to by-products were calculated (Table 3) based on the following energetics: glucose + ADP + Pi → 2 ethanol + 2CO2 + ATP and 3 xylose + 3ADP + 3Pi →5 ethanol + 5CO2 + 3ATP. From these relationships, the molar yield of ATP from both glucose and xylose is 1.0. Dihydroxyacetone and glycerol are formed from dihydroxyacetone-3-phosphate and glycerol-3-phosphate by the action of phosphatase(s) (27), and as a consequence of these reactions no ATP would be produced, leading to a net loss of 2 mol of ATP per mol of dihydroxyacetone or glycerol produced. Theoretically, in the fermentation of 27.8 g (185.2 mmol) of xylose per liter to ethanol, 185.2 mmol of ATP would be formed. However, due to the formation of dihydroxyacetone and glycerol, net amounts of 15.1 and 7.8 mmol of ATP, respectively, would be wasted. Thus, the total generation of ATP would be only 162.3 mmol, or a 12.4% reduction from the theoretical yield. The production of xylitol does not involve the consumption or production of ATP, but the consequent redirection of xylose accounted for a further loss of 9.1 mmol of ATP, leading to an additional 4.9% reduction from the theoretical ATP yield. Acetoin, acetate, and lactate, by-products originating from metabolic steps after pyruvate formation, do not alter the ATP yield. Thus, a total of 17.3% of the theoretical ATP which could have been produced from the 27.8-g/l xylose was lost due to by-product formation. From the experimental and calculated results, it is evident that the percent ATP loss was significant only on the xylose medium and was increased with increasing concentrations of xylose. These observations have been supported by the 31P-NMR spectroscopy determination of the lower energy status of ZM4(pZB5) on xylose medium. Moreover, as xylitol is converted to xylitol phosphate with concomitant utilization of ATP (8), the loss of ATP on xylose would be further increased.
(iii) Growth inhibition due to metabolite(s) formed from xylose.
Xylitol has been shown to be a major by-product of xylose metabolism, causing significant growth inhibition of ZM4(pZB5). Furthermore, ZM4(pZB5) could not utilize xylitol as a carbon source. However, the growth of the host strain, ZM4, was not inhibited by xylitol, suggesting that xylitol could be converted to an inhibitory compound only in the recombinant strain. Other authors have also reported the conversion of xylose to xylitol by an NADPH-dependant aldose reductase in Z. mobilis CP4 and in a recombinant strain expressing xylose isomerase and xylulokinase from Klebsiella pneumoniae (8). It was found that only the recombinant strain was growth inhibited with addition of xylose, and this resulted from the accumulation of xylitol phosphate due to a side reaction effected by a cloned xylulokinase. A similar phenomenon is likely to have occurred with ZM4(pZB5).
Dihydroxyacetone and acetic acid are also growth-inhibitory compounds (19, 27), although the inhibitory mechanism of the former compound is unknown. These by-products may also inhibit the growth of recombinant strain synergistically in the presence of ethanol, although the effects are likely to be smaller than those of xylitol at the concentrations determined in the present studies.
(iv) The slower rates of xylose assimilation and metabolism and thus less energized state of ZM4(pZB5) cells during xylose fermentation.
The differences between the growth rates and biomass yields on xylose and glucose can be examined on the basis of the fermentation kinetic data on mixtures of these sugars; viz., two-phase growth kinetics, with the initial higher growth rate phase with the uptake of both sugars being followed by slower growth and metabolic uncoupling on xylose after glucose depletion, as shown in Fig. 1. From analysis of the kinetic data given in Table 1, it was evident that qmax,glucose was considerably greater than qmax,xylose in the first phase. In Zymomonas, glucose is transported by a low-affinity, high-velocity, energy-independent, glucose-facilitated diffusion (Glf) transport system (7) that is well suited to the high-sugar plant saps that Z. mobilis inhabits (26). The presumptive glf gene has been isolated and sequenced (1), and the transport kinetics of its gene product have been characterized for recombinant Escherichia coli (20, 28). Kinetic studies on sugar uptake in an heterologous E. coli host have shown that the Glf transporter can take up xylose very rapidly, with a maximum rate of metabolism at 5°C which is twice that for glucose or fructose (29). These results suggest that the Glf transporter may not be limiting for xylose uptake, although evidence exists in the present data (Table 1) that xylose uptake is reduced in the presence of glucose, possibly due to competitive effects between the two substrates. A further possibility for the lower specific xylose uptake rates compared to those for glucose is that one of the cloned heterologous enzymes is rate limiting (viz., xylulokinase), as has been reported in a very recent study on a xylose-fermenting recombinant strain of Z. mobilis by De Graaf et al. (5).
Our in vivo 31P-NMR data have shown that xylose-metabolizing cells are less energized than glucose-metabolizing cells, with cells consuming glucose having higher levels of NTP and UDP sugars than cells consuming xylose (Table 4). NTP (mostly ATP) are cellular energy reserve materials and UDP sugar levels are indicative of cell growth potential; thus, cells metabolizing xylose would have less energy for growth than cells metabolizing glucose. Further support for this is provided by the observation that the total inorganic phosphate resonances of xylose-metabolizing cells were significantly higher than those of glucose-metabolizing cells, indicating that the rate of uptake of inorganic phosphate for production of phosphorylated compounds, NTP, sugar phosphates, and UDP sugars was lower in xylose-metabolizing cells.
From the present kinetic and NMR results for the recombinant Z. mobilis ZM4(pZB5), various factors have been identified as contributing to the slower growth and lower yields of cells and ethanol on xylose compared to those on glucose. Analysis would suggest that xylose uptake rate limitations (possibly due to one of the cloned heterologous enzymes), reduced ATP availability, and formation of additional by-products (particularly xylitol) are likely to be major contributing factors. However, these effects were less in evidence on mixed glucose-xylose substrates, particularly at higher sugar concentrations, at which most of the xylose uptake occurred in a slow or nongrowth uncoupled phase following glucose depletion. Additional quantitative analysis is necessary to apportion the relative contributions of each of these effects on xylose metabolism.
ACKNOWLEDGMENT
This work was partially supported by a grant from the U.S. Department of Energy, National Renewable Energy Laboratory, Golden, Colo. (subcontract no. ACG-8-18029-01).
REFERENCES
- 1.Barnell W O, Yi K C, Conway T. Sequence and genetic organization of a Zymomonas mobilis gene cluster that encodes several enzymes of glucose metabolism. J Bacteriol. 1990;172:7227–7240. doi: 10.1128/jb.172.12.7227-7240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrow K D, Collins J G, Norton R S, Rogers P L, Smith G M. 31P nuclear magnetic resonance studies of the fermentation of glucose to ethanol by Zymomonas mobilis. J Biol Chem. 1984;259:5711–5716. [PubMed] [Google Scholar]
- 3.Cheah U E, Weigand W A, Stark B C. Effect of recombinant plasmid size on cellular processes in Escherichia coli. Plasmid. 1987;18:127–134. doi: 10.1016/0147-619x(87)90040-0. [DOI] [PubMed] [Google Scholar]
- 4.Deanda K, Zhang M, Eddy C, Picataggio S. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl Environ Microbiol. 1996;62:4465–4470. doi: 10.1128/aem.62.12.4465-4470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Graaf A A, Striegel K, Wittig R M, Laufer B, Schmitz G, Wiechert W, Sprenger G A, Sahm H. Metabolic state of Zymomonas mobilis in glucose-, fructose-, and xylose-fed continuous cultures as analysed by 13C- and 31P-NMR spectroscopy. Arch Microbiol. 1999;171:371–385. doi: 10.1007/s002030050724. [DOI] [PubMed] [Google Scholar]
- 6.Diaz-Ricci J C, Tsu M, Bailey J E. Influence of expression of the pet operon on intracellular metabolic fluxes of Escherichia coli. Biotechnol Bioeng. 1992;39:59–65. doi: 10.1002/bit.260390110. [DOI] [PubMed] [Google Scholar]
- 7.DiMarco A A, Romano A H. d-Glucose transport system of Zymomonas mobilis. Appl Environ Microbiol. 1985;49:151–157. doi: 10.1128/aem.49.1.151-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann S D, Sahm H, Sprenger G A. Pentose metabolism in Zymomonas mobilis wild-type and recombinant strains. Appl Microbiol Biotechnol. 1992;38:354–361. [Google Scholar]
- 9.Joachimsthal, E., K. D. Haggett, and P. L. Rogers. 1999. Evaluation of recombinant strains of Zymomonas mobilis for ethanol production from glucose/xylose media. Appl. Biochem. Biotechnol. 77–79:147–157.
- 10.Lawford H G. A new approach to improving the performance of Zymomonas in continuous ethanol fermentation. Appl Biochem Biotechnol. 1988;17:203–219. [Google Scholar]
- 11.Lawford, H. G., J. D. Rousseau, A. Mohagheghi, and J. D. McMillan. 1998. Continuous culture studies of xylose-fermenting Zymomonas mobilis. Appl. Biochem. Biotechnol. 70–72:353–367. [DOI] [PubMed]
- 12.Lawford, H. G., and J. D. Rousseau. 1999. The effect of glucose on high-level xylose fermentations by recombinant Zymomonas in batch and fed-batch fermentation. Appl. Biochem. Biotechnol. 77–79:235–249.
- 13.Lazdunski A, Belaich J P. Uncoupling in bacterial growth: ATP pool variation in Zymomonas mobilis cells in relation to different uncoupling conditions of growth. J Gen Microbiol. 1972;70:187–197. [Google Scholar]
- 14.Lee K J, Skotnicki M L, Tribe D E, Rogers P L. Kinetic studies on a highly productive strain of Zymomonas mobilis. Biotechnol Lett. 1980;2:339–344. [Google Scholar]
- 15.Lohmeier-Vogel E M, Hahn-Hägerdal B, Vogel H J. Phosphorus-31 and carbon-13 nuclear magnetic resonance studies of glucose and xylose metabolism in Candida tropicalis cell suspensions. Appl Environ Microbiol. 1995;61:1414–1419. doi: 10.1128/aem.61.4.1414-1419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loureiro-Dias M, Santos H. Effect of ethanol on Saccharomyces cerevisiae as monitored by in vivo 31P and 31C nuclear magnetic resonance. Arch Microbiol. 1990;153:384–391. doi: 10.1007/BF00249010. [DOI] [PubMed] [Google Scholar]
- 17.Lundberg P, Harmsen E, Ho C, Vogel H J. Nuclear magnetic resonance studies of cellular metabolism. Anal Biochem. 1990;191:193–222. doi: 10.1016/0003-2697(90)90210-z. [DOI] [PubMed] [Google Scholar]
- 18.Moyer J D, Henderson J F. Compartmentation of intracellular nucleotides in mammalian cells. Crit Rev Biochem. 1985;19:45–61. doi: 10.3109/10409238509086787. [DOI] [PubMed] [Google Scholar]
- 19.Pampulha M E, Loureiro V. Interaction of the effects of acetic acid and ethanol on inhibition of fermentation in Saccharomyces cerevisiae. Biotechnol Lett. 1989;11:269–274. [Google Scholar]
- 20.Parker C, Barnell W O, Snoep J L, Ingram L O, Conway T. Characterization of the Zymomonas mobilis glucose facilitator gene product (glf) in recombinant Escherichia coli: examination of the transport mechanism, kinetics and role of glucokinase in glucose transport. Mol Microbiol. 1995;15:795–802. doi: 10.1111/j.1365-2958.1995.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 21.Rogers P L, Lee K J, Tribe D E. Kinetics of alcohol production by Zymomonas mobilis at high sugar concentrations. Biotechnol Lett. 1979;1:165–170. [Google Scholar]
- 22.Rogers P L, Lee K J, Skotnicki M L, Tribe D E. Ethanol production by Zymomonas mobilis. Adv Biochem Eng. 1982;23:37–84. doi: 10.1007/978-1-4684-4142-0_22. [DOI] [PubMed] [Google Scholar]
- 23.Rogers P L, Joachimsthal E L, Haggett K D. Ethanol from lignocellulosics: potential for a Zymomonas-based process. Australian Biotechnol. 1997;7:304–309. [Google Scholar]
- 24.Satyagal V N, Agrawal P. Cellular plasmid content and cloned-gene expression: some useful equations. Biotechnol Bioeng. 1990;35:23–30. doi: 10.1002/bit.260350105. [DOI] [PubMed] [Google Scholar]
- 25.Strohhäcker J, De Graaf A A, Schoberth S M, Wittig R M, Sahm H. 31P nuclear magnetic resonance studies of ethanol inhibition in Zymomonas mobilis. Arch Microbiol. 1993;159:484–490. [Google Scholar]
- 26.Swings J, De Ley J. The biology of Zymomonas. Bacteriol Rev. 1977;41:1–46. doi: 10.1128/br.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viikari L, Korhola M. Fructose metabolism in Zymomonas mobilis. Appl Microbiol Biotechnol. 1986;24:471–476. [Google Scholar]
- 28.Weisser P, Kramer R, Sham H, Sprenger G. Functional expression of the glucose transporter of Zymomonas mobilis leads to restoration of glucose and fructose uptake in Escherichia coli mutants and provides evidence for its facilitator action. J Bacteriol. 1995;177:3351–3354. doi: 10.1128/jb.177.11.3351-3354.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisser P, Kramer R, Sprenger G A. Expression of the Escherichia coli pmi gene, encoding phosphomannose-isomerase in Zymomonas mobilis, leads to utilization of mannose as a novel growth substrate, which can be used as a selective marker. Appl Environ Microbiol. 1996;62:4155–4161. doi: 10.1128/aem.62.11.4155-4161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science. 1995;267:240–243. doi: 10.1126/science.267.5195.240. [DOI] [PubMed] [Google Scholar]