Abstract
We identified conditions under which Buffalo green monkey cells grew on the surfaces of cellulose nitrate membrane filters in such a way that they covered the entire surface of each filter and penetrated through the pores. When such conditions were used, poliovirus that had previously been adsorbed on the membranes infected the cells and replicated. A plaque assay method and a quantal method (most probable number of cytopathic units) were used to detect and count the viruses adsorbed on the membrane filters. Polioviruses in aqueous suspensions were then concentrated by adsorption to cellulose membrane filters and were subsequently counted without elution, a step which is necessary when the commonly used methods are employed. The pore size of the membrane filter, the sample contents, and the sample volume were optimized for tap water, seawater, and a 0.25 M glycine buffer solution. The numbers of viruses recovered under the optimized conditions were more than 50% greater than the numbers counted by the standard plaque assay. When ceftazidime was added to the assay medium in addition to the antibiotics which are typically used, the method could be used to study natural samples with low and intermediate levels of microbial pollution without decontamination of the samples. This methodological approach also allowed plaque hybridization either directly on cellulose nitrate membranes or on Hybond N+ membranes after the preparations were transferred.
Animal cells attach to membranes with rough surfaces and pores, such as those of cellulose nitrate, and multiply until they cover the whole surface of the membrane (17). The present study was initiated on the assumption that cells would not only cover the rough surface of the cellulose nitrate membranes but also penetrate through the pores.
On the other hand, viruses adsorb to electronegative membrane filters, such as cellulose nitrate membrane filters, and this property has been used to concentrate viruses from water (7, 8, 12, 20). Certain methods for counting viruses in low-titer viral suspensions are based on adsorption of the viruses on membranes, followed by elution and counting. Moreover, workers have previously described conditions that favor adsorption of viruses on membranes, including low pH and the presence of di- and trivalent salts (8, 19, 20).
Taking into account the information described above, we envisaged a method for counting viruses in low-titer viral suspensions in which cells expanding into the pores reach the viruses adsorbed on a cellulose nitrate membrane. Then each of the adsorbed viruses infects one cell, and virus replication occurs. The viruses released by the infected cells infect adjacent cells, similar to what occurs in techniques based on infecting cells with free viruses. Such a method may have multiple applications, such as counting viruses in natural water samples, testing disinfectants and antiseptics, etc.
Here we describe the conditions under which the new method was used to concentrate and count type 1 polioviruses in Buffalo green monkey (BGM) cells. We also show that (i) this method can be used with various quantification methods, such as a method for determining the number of PFU on a cell monolayer, or with quantal assays, such as an assay to determine the most probable number of cytopathogenic units (MPNCU); (ii) this method efficiently recovers suspended viruses from water; (iii) this method can be used for several volumes of a sample; (iv) this method can be used for viruses suspended in various types of aqueous solutions; and (v) when this method is used to count the PFU on a monolayer, the viruses and viral nucleic acids remaining in the plaques formed on the membrane can be identified by nucleic acid hybridization.
MATERIALS AND METHODS
Viruses, cells, and media.
All experiments were performed with attenuated poliovirus type 1 strain Lsc-2ab. The viruses were propagated by using the BGM kidney continuous cell line. The cells were grown in Eagle's minimum essential medium (MEM) (Gibco, Paisley, United Kingdom) containing 5% fetal bovine serum, 2 mM l-glutamine, 26.8 mM NaHCO3, 100 U of penicillin per ml, and 100 μg of streptomycin per ml.
The overlay medium used for the standard plaque assay was medium 199 (Gibco) supplemented with 2% fetal bovine serum, 26.8 mM NaHCO3, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The overlay medium was prepared as a 2×-concentrated preparation and was mixed with an equal volume of 2% purified agar (Oxoid, Hampshire, United Kingdom).
Extra antibiotics were added to the growth and overlay media, since the method described here is intended to be used for environmental samples that have not been decontaminated and thus may contain bacteria and fungi that would destroy the cell cultures. In addition to the antibiotics typically used (100 U of penicillin ml−1, 100 μg of streptomycin ml−1, 50 μg of gentamicin ml−1, and 50 μg of nystatin ml−1), 20 μg of ceftazidime ml−1 was added. The concentration of ceftazidime used was determined after we tested a significant number of bacterial strains (usually gram-negative oxidative rods) that were isolated from cell cultures that were inoculated with water samples which had not been decontaminated and contained the other antibiotics. All of the antibiotics except ceftazidime were obtained from Gibco. Ceftazidime (Ceftazidime Fortum 1g) was obtained from GlaxoWellcome (Greenford, United Kingdom). Ceftazidime was diluted in sterile distilled water to a concentration of 10,000 μg ml−1 and was stored at −20°C. Defrosted ceftazidime was used within 7 days.
Standard plaque assay.
Virus suspensions were titrated by the plaque assay method on confluent monolayers of BGM cells as described elsewhere (3).
Membrane filters.
Cellulose nitrate filters (pore sizes, 3 and 5 μm; diameter, 47 mm; Millipore, Bedford, Mass.) were used in this study; 3-μm-pore-size membrane filters were used unless indicated otherwise (Table 1).
TABLE 1.
Recovery of viruses added to different aqueous solutions by adsorption to cellulose nitrate membrane filters and a plaque assay performed with a monolayer
| Type of sample | Pore size of the membrane filter (μm) | Treatment of the sample | Vol filtered (ml) | No. of tests | % Recoverya |
|---|---|---|---|---|---|
| Tap water | 3 | pH 3.5 | 500 | 10 | 157.0 (34.5) |
| Tap water | 3 | 0.05 M MgCl2 | 500 | 10 | 151.0 (24.1) |
| Seawater | 3 | pH 3.5 | 500 | 16 | 153.0 (96.5) |
| Seawater | 3 | pH 3.5 | 100 | 6 | 179.0 (94.0) |
| Seawater | 5 | pH 3.5 | 500 | 6 | 154.0 (81.0) |
| Seawater | 5 | pH 3.5 | 100 | 6 | 176.0 (86.0) |
| 0.25 M glycine | 3 | pH 3.5 | 500 | 10 | 51.0 (17.8) |
| 0.25 M glycine | 3 | 0.05 M MgCl2 | 500 | 16 | 73.3 (15.3) |
| 0.25 M glycine | 3 | 0.05 M MgCl2 | 100 | 10 | 166.0 (69.0) |
Percentage of viruses recovered after filtration compared to the number of viruses determined by the standard plaque assay method. The values are means (standard deviations).
Samples tested.
Seawater with low turbidity (less than 2 nephelometric turbidity units) from Cyprus and Barcelona, dechlorinated tap water, and a 0.25 M glycine solution were used as aqueous solutions that contained viruses which could be adsorbed. Seawater was prefiltered through Whatman no. 1 filter paper (Whatman, Kent, United Kingdom). None of the samples was decontaminated. All tests were performed with samples that were spiked with 1-ml portions of viral suspensions containing approximately 20 PFU, as determined by the standard plaque assay method. One aliquot of the suspension used in each experiment was titrated by the standard plaque assay method each time that a concentration experiment was performed. The percentages of viruses recovered were calculated from the values obtained by the two methods in each experiment; 100% recovery was defined as the number of spiked viruses counted by the standard plaque assay method.
Amendment of the sample to favor virus adsorption.
Samples were either acidified to pH 3.5 by adding 1.0 N HCl or amended by adding 4.14 M MgCl2 · 6H2O so that the final concentration of MgCl2 was 0.05 M (19, 20). The effect of the addition on the recovery of viruses was examined by using tap water and a 0.25 M glycine solution.
Adsorption of viruses on the membranes.
The aqueous samples were filtered through the membranes by using filtration systems typically used for bacterial enumeration; both a Fisher Scientific model FDC-300-N system (Fisher Scientific, Weston upon Mare, United Kingdom) and a Millipore model XX2504735 system were used. The filter holder had to be perfectly flat for the method described here to be successful; stainless steel grating filter holders were used. Depending on how the sample was amended, the membrane filter was soaked in sterile pH 3.5 water or sterile 0.05 M MgCl2. The filter was then laid on the filter holder. We made sure that the membrane filter was not deformed when the funnel was secured on top of the filter holder and during filtration. The amended sample was then poured into the funnel, and the filtration apparatus was connected to a water vacuum pump. A vacuum was applied, and the flow rate never exceeded 1 liter · 5 min−1. When all of a sample had been filtered, the membrane filter was washed by passing either 100 ml of sterile pH 3.5 water or 100 ml of sterile 0.05 M MgCl2 through it. The volume filtered was either 100 or 500 ml.
Detection of viruses adsorbed on membrane filters by a plaque assay performed with a monolayer.
After we tested several conditions to ensure that cells completely covered the surfaces of the membrane filter, the following method was developed. The growth medium in a 60-mm-diameter petri dish with a confluent monolayer of BGM cells was discarded. Then 100 μl of a suspension of BGM cells in MEM supplemented with antibiotics containing 1.5 × 107 to 2.0 × 107 cells · ml−1 was placed in the center of the petri dish. A membrane filter was placed upside down on top of the suspension and the cell monolayer. Before the membrane filter was completely laid on the monolayer and while the one side was touching the periphery of the petri dish, the other side was lifted once or twice with flat-end forceps to ensure that the entire surface of the filter was covered with the cell suspension and that no air bubbles were trapped under the filter. Five milliliters of overlay medium was poured slowly onto the center of the membrane filter and spread all over the plate. The whole procedure was performed on a laboratory bench near a Bunsen burner flame, and the plates were protected from light before they were placed in an incubator. The agar was allowed to set, and the petri dishes were incubated at 37°C in the presence of 5% CO2 at a relative humidity of more than 80% for 48 to 72 h.
A wide spatula was used to remove the agar and the membrane filter simultaneously from the monolayer. Subsequently, the monolayer was stained with 0.1% crystal violet.
Detection of viruses adsorbed on membrane filters by a presence-absence test.
A sample was filtered as described above. Then, the 47-mm-diameter membrane filter was immersed right side up in a 60-mm-diameter petri dish containing 5 ml of MEM supplemented with antibiotics. One milliliter of a BGM cell suspension containing approximately 2.0 × 106 cells · ml−1 was then added on top of the membrane filter. The cell suspension was added drop by drop until the whole membrane was covered. The petri dish was incubated at 37°C in a 5% CO2 atmosphere and examined on days 3, 4, and 6. The ring between the filter (diameter, 47 mm) and the edge of the plate (diameter, 57 mm) was then examined with an inverted microscope to determine the presence of healthy growing cells or extensive cytopathic effects on the cell monolayer. To confirm that viruses were present when there were extensive cytopathic effects, 100 μl of medium was removed from each petri dish and added to growing BGM cell monolayers. The appearance of cytopathic effects on the monolayers confirmed that one or more viruses were present in the first plate.
The MPNCU method was used for this presence-absence test. A concentration series consisting of one 50-ml preparation, five 10-ml preparations, and five 1-ml preparations was used.
Scanning electron microscope observation of cells.
Once cells had grown over a cellulose nitrate membrane filter in liquid or semisolid medium, the filter was cut into 1-cm2 fragments, which were fixed with osmium tetroxide (5) and then freeze-dried (6). A sputter coating was then applied (5) to the fragments of the filter before it was observed with a scanning electron microscope (model δ 2300; Hitachi).
Probe preparation.
Oligonucleotide 5′-ATTGTCACCATAAGCAGCCA-3′ specific for poliovirus 1 (14) and oligonucleotide 5′-CGAACCATCAATGCTGCCAA-3′ corresponding to a sequence of the genome of group II RNA coliphage GA (9) were used as the specific probe and the negative control, respectively. These oligonucleotides, which were synthesized by Boehringer (Mannheim, Germany), were labeled to high specific activities by phosphorylation with bacteriophage T4 polynucleotide kinase, as described by Sambrook et al. (16), with minor modifications.
Plaque hybridization.
After the cellulose nitrate membrane filters were incubated for 72 h, the overlays were removed, and the filters were placed, with the side in contact with the cell monolayer up, onto Whatman 3MM filter paper soaked either in a denaturing solution for direct hybridization or in phosphate-buffered saline for transfer to a Hybond N+ membrane.
The viruses and the viral RNA were transferred from the cellulose nitrate membrane filter to a Hybond N+ membrane as follows. The Hybond N+ membrane was gently placed on the surface of the cellulose nitrate membrane filter so that no air bubbles were trapped. Five minutes later, the Hybond N+ membrane was peeled off by using blunt-end forceps (Millipore) and placed on Whatman 3MM filter paper soaked in a denaturing solution, with the side in contact with the cellulose nitrate membrane filter facing up.
For nucleic acid release and denaturation, the membranes were left, as described above, on Whatman 3MM filter paper soaked in 0.1 N NaOH. After 1 min of incubation at room temperature, they were quickly transferred to new Whatman 3MM filter paper soaked in neutralizing solution (0.1 M sodium acetate) and left at room temperature for 5 min. After the membranes were air dried, the RNA was then fixed to the cellulose nitrate filters by baking the filters at 80°C for approximately 2 h (16) and to the Hybond N+ membranes by exposing the membranes to UV light as described by Sambrook et al. (16).
The membranes were then hybridized with the probes described above by using the procedure described by Sambrook et al. (16), with minor modifications. Finally, Medical/Curix RP2 X-ray film (Agfa-Gevaert, Mortsel, Belgium) was exposed to the hybridized membranes at −80°C. The film was then developed and matched with the corresponding stained monolayers.
Statistical analysis.
Statistical computations and tests were performed by using the SAS Statistic Program (2). Differences were considered significant when P was < 0.05 as determined the appropriate comparative test, either analysis of variance (ANOVA) or Student's t test. We calculated the MPNCU by using the formula of De Man (4).
RESULTS
Cell growth on membrane filters.
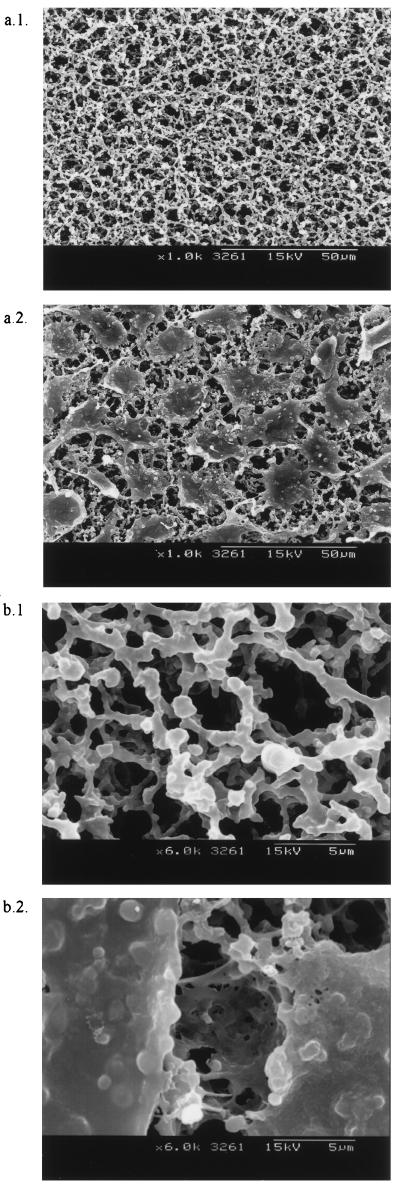
After we tried different approaches to determine the exact procedure and number of cells necessary to cover membrane filters with cells, we chose the conditions described above to provide cellulose nitrate membrane filters that were completely covered with growing BGM cells (Fig. 1). Low-magnification photographs showed that cells completely covered the membranes, forming monolayers on the surfaces, and higher-magnification photographs showed that the cells penetrated into the membrane pores.
FIG. 1.
Scanning electron micrographs showing 3-μm-pore-size cellulose nitrate membranes at a low magnification (a.1) and a high magnification (b.1) and 3-μm-pore-size cellulose nitrate membranes covered by BGM cells at a low magnification (a.2) and a high magnification (b.2).
Detection of viruses adsorbed on membrane filters by a plaque assay performed with monolayers.
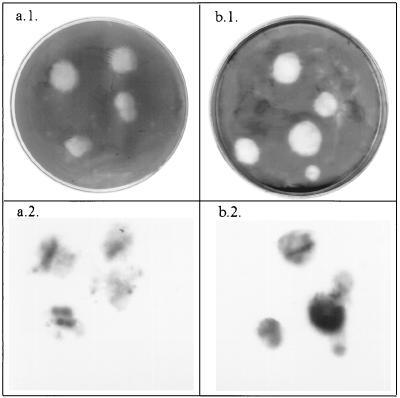
Detection of viruses adsorbed on membrane filters by the plaque assay method revealed that plaques were present both in monolayers on petri dishes and in the cell layers on the surfaces of filters. Plaques in the cell layers on filters were revealed by staining with 0.1% neutral red. The plaques in the monolayers on petri dishes were revealed by staining with 0.1% crystal violet (Fig. 2) and were similar to the plaques obtained by the standard plaque assay method on confluent monolayers of BGM cells. The poliovirus 1 plaques were 3 to 4 mm in diameter after 72 h, like the plaques obtained when the standard plaque assay was used. The plaques on the plate and on the filter matched completely.
FIG. 2.
Plaques obtained on monolayers by the method described here after staining with 0.1% crystal violet (a.1 and b.1) and the corresponding hybridized plaques (plaques hybridized on a cellulose nitrate membrane filter [a.2] and plaques hybridized on a Hybond N+ membrane after transfer [b.2]). The images were scanned with an Epson model GT 9500 scanner, treated by using Adobe Photoshop 4.0, and printed with a Sony model UP-D8800 printer.
Effect of ceftazidime on virus counts.
Adding extra antibiotics, including ceftazidime, to the overlay medium did not have a significant effect on the poliovirus 1 counts.
Efficiency of detection of virus adsorbed to membrane filters by the plaque assay performed with cell monolayers.
The numbers of viruses recovered from different aqueous solutions by the method described here were significantly different (P < 0.05, as determined by ANOVA) from the numbers determined by the standard plaque assay method (Table 1).
When the samples were either tap water or seawater samples, the numbers of viruses recovered by the method described here were significantly higher (P < 0.05, as determined by ANOVA) than the numbers determined by the standard plaque assay. Neither the volume filtered nor amendment of the sample nor the pore size of the membrane filter affected the efficiency of recovery significantly (P > 0.05, as determined by ANOVA).
However, the recovery of viruses added in a 0.25 M glycine solution depended significantly, (P < 0.05, as determined by ANOVA) on the volume filtered and amendment of the sample. Filtering 100 ml of the solution amended with MgCl2 resulted in levels of recovery that were significantly higher (P < 0.05, as determined by ANOVA) than the levels of recovery obtained with the standard plaque assay method.
Plaque hybridization.
Hybridization with specific probes occurred either directly on a nitrate cellulose membrane filter or after viruses and viral nucleic acids were transferred from a nitrate cellulose membrane filter to a Hybond N+ membrane. Hybridization spots were present on the membrane at sites corresponding to the sites where plaques were observed after the monolayers in the petri dishes were stained (Fig. 2). In contrast, no hybridization occurred with the GA phage-specific probe used as a negative control. However, hybridization after transfer to a Hybond N+ membrane was better than hybridization on the cellulose nitrate membrane filter both in terms of definition of the spots and in terms of sensitivity. The exposure times necessary to produce clear spots (2.5 ± 0.5 h versus 36 ± 12 h) indicate that the method based on transfer to Hybond N+ membranes is much more sensitive than direct hybridization on nitrate cellulose membrane filters. Moreover, the procedure for hybridizing DNA on cellulose nitrate membrane filters is cumbersome, since the cellulose nitrate membrane filters become very fragile after they are heated at 80°C.
Detection of viruses adsorbed on membrane filters by a presence-absence test.
Cells added as described above settled both on the membrane filters and on the rings between the filters and the edges of the plates, where they formed monolayers that could be observed with an inverted microscope. The presence of growing cells at the periphery of a petri dish indicated that viruses were not present in the medium. Extensive cytopathic effects on a cell monolayer indicated that one or more viruses were present in a sample. As in the standard presence-absence tests regularly used for detecting viruses, the suspected positive cultures had to be confirmed by reinoculation of the supernatant into growing cells.
The efficiency of the presence-absence approach was assessed by using the MPNCU method. The numbers of viruses recovered were significantly higher, (P < 0.05, as determined by Student's t test) than the numbers determined by the standard plaque assay (Table 2).
TABLE 2.
Recovery of viruses added to tap water as determined by the most probable number approach (one 50-ml preparation, five 10-ml preparations, and five 1-ml preparations) when the method described here was useda
| Sample | No. of PFU addedb | Most probable no. (100 ml) | % Recoveryc |
|---|---|---|---|
| 1 | 40 | 91 | 227 |
| 2 | 22 | 17 | 77 |
| 3 | 30 | 53 | 177 |
| 4 | 34 | 91 | 268 |
| 5 | 32 | 53 | 166 |
| 6 | 28 | 53 | 189 |
| 7 | 10 | 4 | 40 |
| 8 | 26 | 53 | 204 |
| Mean | 27.7 | 51.8 | 168.5 |
| SD | 9.0 | 30.6 | 77.0 |
The pH values of virus suspensions in water were decreased to 3.5.
The number of viruses added per 100 ml was determined by performing a plaque assay with confluent cell monolayers.
Percentage of viruses recovered after filtration compared to the number of viruses determined by the plaque assay method performed with confluent cell monolayers.
DISCUSSION
In this paper we describe detection of poliovirus 1 adsorbed on cellulose nitrate membrane filters by using BGM cells on the filters. Since it has been shown that other cells grow on cellulose nitrate membrane filters (17) and that viruses other than poliovirus adsorb to such filters (8), we expect that the methodological approach described here may be applicable to a wide range of cells and viruses. Moreover, since most nonenveloped viruses suspended in aqueous solutions adsorb to cellulose nitrate filters (8), it may be possible to use the methodological approach described here to detect viruses in low-titer viral suspensions.
Because we thought that our new method could be used with samples that were not decontaminated, we added ceftazidime to the infected cultures along with penicillin, streptomycin, gentamicin, and nystatin. Using ceftazidime to control bacterial contamination in cell cultures has not been described previously. The use of ceftazidime in conjunction with the antibiotics typically used when viruses in environmental samples are assessed kept the bacteria and fungi under control and had no detrimental effects on virus recovery. The possible use of the method for samples that are not decontaminated suggests that it has additional potential.
The viruses adsorbed on the cellulose nitrate membrane filters were also counted either by the plaque assay method or by the MPNCU method. When any of the quantification methods was used, the numbers of viruses counted after the viruses were adsorbed to the membrane filters were significantly higher (approximately 50% higher) than the numbers of suspended viruses determined by standard plaque assay performed with a cell monolayer. Similar differences were reported when numbers of viruses were determined by the plaque assay performed with a monolayer and by the usual quantal assays (13). We concluded that the methodological approach described here is very useful for concentrating and enumerating viruses in low-titer viral suspensions.
Adjusting the pH to 3.5 and adding 0.05 M MgCl2, techniques which reportedly favor virus adsorption on membrane filters (19, 20), were examined, and we observed no differences when we concentrated and enumerated viruses suspended in water. However, adding MgCl2 is more practical than adjusting the pH to 3.5 since it does not require obtaining measurements during the concentration process.
When tap water and seawater were used, our method allowed us to concentrate and enumerate the viruses in 100- to 500-ml samples with similar efficiencies. Hypothetically, the upper limit for the volume that can be filtered depends on filter clogging, which depends among other factors on the size of the pores in the membranes. We used 3- and 5-μm-pore-size membrane filters. The size of the pores did not affect the efficiency of virus recovery, and presumably the 5-μm-pore-size membranes allow filtration of larger volumes. However, as described above, very often we had problems with 5-μm-pore-size membranes, and therefore, 3-μm-pore-size membrane filters are recommended.
One of the possible applications of the new method is reconcentration of the eluates obtained with most methods based on filter adsorption and elution, which are typically used to concentrate viruses from large volumes of water (8). However, some of the eluants (e.g., glycine and beef extract) used to release the viruses linked to the adsorbents used to concentrate viruses (8, 11) may interfere with virus adsorption on the cellulose nitrate membranes. We tested one of these eluants, 0.25 M glycine buffer (11), which is commonly used to elute virus adsorbed to electropositive filters (18). In contrast to what occurred with water, in this experiment the efficiency of recovery depended both on the content of the sample and on the volume filtered. Amendment with MgCl2 resulted in higher percentages of recovery, and filtering 100 ml resulted in much higher levels of recovery than filtering 500 ml. Glycine probably competed with viruses for adsorption when 500 ml was filtered. When the method described here was used with 100 ml of 0.25 M glycine buffer amended with MgCl2, the levels of recovery were more than 150%; these values were much higher than the values obtained with any of the methods described previously (10, 11, 18).
Another advantage of our method is its straightforwardness for identifying plaque-forming viruses. Indeed, plaque hybridization can be performed either directly on the nitrate cellulose membrane filter or after a preparation is transferred from the nitrate cellulose membrane filter to a Hybond N+ membrane. For the reasons explained above, hybridization after transfer to a Hybond N+ membrane is preferable. Although in the experiments described here radioactively labeled probes were used, successful transfer to Hybond N+ membranes may facilitate hybridization with nonradioactively labeled probes (for example, probes labeled with dioxigenin), which can be detected either colorimetrically or by chemiluminiscence. Hybridization of plaques on membrane filters provides a simple way to study the diversity of mixed populations of viruses growing in a given cell line or to identify a given virus. In addition, quantal assay methods based on detection of cytopathic effects on monolayers covered with liquid medium may allow application of the cell culture-PCR—reverse transcription-PCR approach that combines the advantages of cell culture methods and the advantages of molecular methods (15). This approach may be useful for detecting nonculturable viruses (for example, human rotavirus and hepatitis A viruses).
Thus, the method described here has great potential for quantifying and identifying viruses in low-concentration viral suspensions in aqueous solutions. The potential applications include performing virus inactivation experiments with either disinfectants or antiseptics, counting viruses in medium-range volumes of polluted waters, and counting viruses in concentrates of higher volumes of water. For example, the method could be used to reconcentrate and count viruses concentrated from large volumes of water (8). Despite extensive research on reconcentration procedures that can be used for viruses, only tentative methods are outlined in the Standard Methods for the Examination of Water and Wastewater (1); these methods include organic flocculation of beef extract, aluminium hydroxide precipitation, and polyethylene glycol hydroextraction. The limitations of these reconcentration methods include the relatively low, variable efficiencies of recovery (10, 11, 18) and the high volumes tested (about 100 ml) (11), which result in using a large number of tissue culture flasks or plates. The results described in this paper show that the efficiency of recovery of viruses in 0.25 M glycine buffer was quite good. Both in terms of efficiency of recovery and in terms of reproducibility of results, the method described here performs at least as well for 500 ml and much better for 100 ml than other methods (10, 11, 18). Moreover, it solves the problem of having to assay for the presence of viruses in large volumes, a problem encountered with most methods (11). Thus, reconcentrating by organic flocculation the viruses present in 500 ml of 0.25 M glycine buffer obtained after the viruses adsorbed to a electropositive filter are eluted and enumeration should cost approximately $45 in materials and media, whereas the method described here should cost about $15. The time ratio is also favorable for the method described here and should be around 1:3. Moreover, except for a CO2 incubator, the method described here does not require sophisticated equipment and can be carried out with equipment that is present in routine microbiology laboratories. Very similar reasoning can be applied to the quantal assay for cytopathic effects, such as the MPNCU and 50% tissue culture infective dose methods.
ACKNOWLEDGMENTS
Expertise gained through LIFE project 95/CY/B2/CT/868 MED of the European Commission contributed to this work. We thank the Serveis Cientifico-Tècnics of the University of Barcelona for help with the electron microscope observations and GlaxoWellcome for providing ceftazidime for this work.
This work was supported in part by grant 1997 SGR 00069 from the Generalitat de Catalunya. Laura Mocé-Llivina is a fellow of the Generalitat de Catalunya.
REFERENCES
- 1.Anonymous. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 2.Anonymous. Statistic program. SAS user's guide, version 6.04. Cary, N.C: SAS Institute, Inc.; 1990. [Google Scholar]
- 3.Bosch A, Lucena F, Gironés R, Jofre J. Survey of viral pollution in Besos River. J Water Pollut Control Fed. 1986;58:87–91. [Google Scholar]
- 4.De Man J C. The probability of the most probable numbers. Eur J Appl Microbiol. 1975;1:72–77. [Google Scholar]
- 5.Dykstra M J. Biological electron microscopy. Theory, techniques and troubleshooting. New York, N.Y: Plenum Press; 1992. [Google Scholar]
- 6.Echlin P. Low temperature microscopy and analysis. New York, N.Y: Plenum Press; 1992. [Google Scholar]
- 7.Farrah S R, Gerba C P, Wallis C, Melnick J L. Concentration of viruses from large volumes of tap water using pleated membrane filters. Appl Environ Microbiol. 1976;31:221–226. doi: 10.1128/aem.31.2.221-226.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerba C P. Recovery of viruses from sewage, effluents, and water. In: Berg G, editor. Methods for recovering viruses from the environment. Boca Raton, Fla: CRC Press; 1987. pp. 1–51. [Google Scholar]
- 9.Inokuchi Y, Takahashi R, Hirose T, Inayama S, Jacobson A B, Hirashima A. The complete nucleotide sequence of group II RNA coliphage GA. J Biochem. 1986;99:1169–1180. doi: 10.1093/oxfordjournals.jbchem.a135580. [DOI] [PubMed] [Google Scholar]
- 10.Ju-Fang M, Naranjo J, Gerba C P. Evaluation of MK filters for recovery of enteroviruses from tap water. Appl Environ Microbiol. 1994;60:1974–1977. doi: 10.1128/aem.60.6.1974-1977.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melnick J L, Safferman R, Rao V C, Goyal S, Berg G, Dahling D R, Wright B A, Akin E, Stetler R, Sorber C, Moore B, Sobsey M D, Moore R, Lewis A L, Wellings F M. Round robin investigation on methods for the recovery of poliovirus from drinking water. Appl Environ Microbiol. 1984;47:144–150. doi: 10.1128/aem.47.1.144-150.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris R, Waite W M. Evaluation of procedures for recovery of viruses from water. I. Concentration systems. Water Res. 1980;14:791–793. [Google Scholar]
- 13.Morris R, Waite W M. Evaluation of procedures for recovery of viruses from water. I. Detection systems. Water Res. 1980;14:795–798. [Google Scholar]
- 14.Puig M, Jofre J, Lucena F, Allard A, Wadell G, Gironés R. Detection of adenoviruses and enteroviruses in polluted water by nested PCR amplification. Appl Environ Microbiol. 1994;60:2963–2970. doi: 10.1128/aem.60.8.2963-2970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds K A, Gerba C P, Pepper I L. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Appl Environ Microbiol. 1996;62:1424–1427. doi: 10.1128/aem.62.4.1424-1427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Savage C R, Jr, Bonney J. Extended expression of differentiated function in primary cultures of adult liver parenchymal cells maintained on nitrocellulose filters. Exp Cell Res. 1978;114:307–315. doi: 10.1016/0014-4827(78)90488-3. [DOI] [PubMed] [Google Scholar]
- 18.Sobsey M D, Glass J S. Poliovirus concentration from tap water with electropositive adsorbent filters. Appl Environ Microbiol. 1980;40:201–210. doi: 10.1128/aem.40.2.201-210.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobsey M D. Quality of currently available methodology for monitoring viruses in the environment. Environ Int. 1982;7:39–51. [Google Scholar]
- 20.Wallis C, Henderson M, Melnick J L. Enterovirus concentration on cellulose membranes. Appl Microbiol. 1972;23:476–480. doi: 10.1128/am.23.3.476-480.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]