Abstract
To elucidate the biological significance of dead bacterial cells in soil to the intra- and interspecies transfer of gene fragments by natural transformation, we have exposed the kanamycin-sensitive recipient Acinetobacter sp. strain BD413(pFG4) to lysates of the kanamycin-resistant donor bacteria Acinetobacter spp., Pseudomonas fluorescens, and Burkholderia cepacia. Detection of gene transfer was facilitated by the recombinational repair of a partially (317 bp) deleted kanamycin resistance gene in the recipient bacterium. The investigation revealed a significant potential of these DNA sources to transform Acinetobacter spp. residing both in sterile and in nonsterile silt loam soil. Heat-treated (80°C, 15 min) cell lysates were capable of transforming strain BD413 after 4 days of incubation in sterile soil and for up to 8 h in nonsterile soil. Transformation efficiencies obtained in vitro and in situ with the various lysates were similar to or exceeded those obtained with conventionally purified DNA. The presence of cell debris did not inhibit transformation in soil, and the debris may protect DNA from rapid biological inactivation. Natural transformation thus provides Acinetobacter spp. with an efficient mechanism to access genetic information from different bacterial species in soil. The relatively short-term biological activity (e.g., transforming activity) of chromosomal DNA in soil contrasts the earlier reported long-term physical stability of DNA, where fractions have been found to persist for several weeks in soil. Thus, there seems to be a clear difference between the physical and the functional significance of chromosomal DNA in soil.
Horizontal gene transfer can be an important mechanism for bacterial adaptation to changing environments (14, 23). Molecular studies have shown that many chromosomal genes from bacteria have a mosaic pattern (2, 5, 15, 19) presumably as a result of recombination with DNA from heterologous sources (10, 30, 31). The mechanisms causing these patterns are, however, seldom known. Natural transformation provides a mechanism of gene transfer that enables competent bacteria to generate genetic variability by “sampling” of DNA present in their surroundings. However, only a few cases of interspecies transfer of chromosomal genes between environmental isolates have been shown to occur by natural transformation. For instance, Majewski and Cohan (28) investigated barriers to transfer of chromosomally encoded antibiotic resistance in Bacillus species in vitro, and Juni (18) reported reclassification of 265 different isolates into the Acinetobacter genus after studies of their relatedness based on natural transformation of an auxotrophic Acinetobacter sp. recipient strain in vitro. Moreover, data on transfer of genetic material between bacterial genera by natural transformation are scarce. Juni (18) reported that strains from genera such as Alcaligenes, Bacillus, Escherichia, Haemophilus, Pseudomonas, Rhizobium, Serratia, and Streptococcus were not able to transform the auxotrophic recipient Acinetobacter strain to prototrophy in vitro. In addition to the barrier generated by increasing sequence divergence, limitations to interspecies gene transfer are active at many stages in soil (33, 36). There may be differences in the cellular protection of DNA such as the presence of a bacterial cell wall. Moreover, interactions of DNA with proteins or other cellular substances and variable methylation patterns may limit its accessibility as a source of transforming DNA for Acinetobacter cells. It is, therefore, unclear to what extent soil bacteria like Acinetobacter spp. actively access and take up DNA from divergent species in their natural habitats such as soil (27).
Studies on the transfer of chromosomal DNA in soil by natural transformation have focused mainly on transfer events with purified DNA in nutrient- or mineral-amended sterile soil (1, 11, 24). Only recently has natural transformation of chromosomal genes been shown to occur in unamended and nonsterile soil (34, 35). Here, we extend these studies by exposing a kanamycin-sensitive recipient bacterium, Acinetobacter sp. strain BD413(pFG4) (12), to lysed cells of the nptII-containing (kanamycin-resistant [Kmr]) donor strains, Acinetobacter spp., Pseudomonas fluorescens R2f, and Burkholderia cepacia P2, in order to demonstrate that there is no efficient barrier in nonsterile soil that inhibits intra- or interspecies natural transformation of the Acinetobacter sp. with homologous DNA released from lysed bacterial cells. Detection of gene transfer was based on the restoration of a partially deleted kanamycin resistance gene (nptII) in the recipient bacterium and established models for monitoring of gene transfer in soil microcosms (12, 34, 35).
MATERIALS AND METHODS
Bacterial strains.
All bacteria used in this study were originally isolated from soil and were spontaneous rifampin-resistant mutants. The strains were stored in 20% glycerol at −70°C and cultured in Luria-Bertani (LB) broth (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, 1 liter of H2O [pH 7.2]). Liquid cultures were grown overnight at 27°C with shaking (225 rpm). Before use, 1-ml aliquots of cultures of the recipient strain, prepared as described by Nielsen et al. (34) and stored at −70°C (in 0.85% NaCl–20% glycerol), were thawed, centrifuged, and resuspended in water. Portions of 100 μl were used immediately for the various transformation studies (12). Final concentration (mean ± standard deviation [SD]) of the inoculum was (6.4 ± 0.9) × 108 CFU/ml of water. For plating and enumeration of CFU, 1.5% agar (Oxoid, Basingstoke, England) was included and the LB agar (LBA) plates were incubated at 30°C for 3 days before counting. The antibiotics (Sigma, St. Louis, Mo.) rifampin, ampicillin, and kanamycin were added to the growth media at 50 μg ml−1.
As the recipient bacterium in all transformations, Acinetobacter sp. strain BD413(pFG4), which carried a 317-bp deletion in the central part of the nptII gene (located on an IncQ plasmid), was used (12). Recombinational repair of this 317-bp deletion was facilitated due to presence of a functional nptII gene in the KTG cassette harbored by the donor bacteria. As donor DNA, either purified chromosomal DNA or cell lysates obtained from the gamma proteobacteria Acinetobacter sp. strain BD413 (18) (chr::KTG [34]) and P. fluorescens R2f (52) (chr::KTG [46]), and the beta proteobacterium B. cepacia P2 (37) (chr::KTG [J. D. van Elsas unpublished data]), with or without a chromosomally inserted (chr::) KTG gene cassette, were used. Bacterial strains without the inserted KTG cassette were included as controls for spontaneous mutations. The KTG cassette (mini-Tn5 [16] inserted into the bacterial chromosomes) consisted of a functional nptII gene (conferring kanamycin resistance to all hosts), the aadb gene, and a truncated Bacillus thuringiensis cryIVB gene (45).
Isolation of donor DNA.
DNA was isolated from the different donor bacteria by using a scaled-up version of the cetyltrimethylammonium bromide protocol of Wilson (54). In addition, the crude DNA solution was reextracted with phenol-chloroform, and chloroform, precipitated with isopropanol, and washed twice in ethanol before quantification on a UV spectrophotometer at 260 and 280 nm (44).
Preparation of cell lysates.
For the preparation of heat-treated cell lysates, overnight cultures of the bacteria were centrifuged at 5,000 × g for 5 min, resuspended in water, recentrifuged, and finally taken up in the initial volume of distilled water. Samples were taken for enumeration of CFU, and 1-ml aliquots were then heat treated at 80°C for 15 min (13). Final concentrations (means ± SD) of the cell lysates (determined in samples withdrawn for CFU counts immediately before heat treatment) were, per milliliter, (2.5 ± 0.2) × 109 for Acinetobacter sp. strain BD413, (2.3 ± 0.2) × 109 for Acinetobacter sp. strain BD413 (chr::KTG), (1.3 ± 0.3) × 109 for P. fluorescens R2f, (2.0 ± 0.1) × 109 for P. fluorescens R2f (chr::KTG), (1.0 ± 0.3) × 109 for B. cepacia P2, and (3.2 ± 0.3) × 109 for B. cepacia P2 (chr::KTG). From the CFU counts, the estimated amount of chromosomal DNA in the lysates, assuming 6 × 10−15 g of DNA per bacterial cell (49), ranged from 7.8 to 19.2 μg/ml. Subsequent plating of the lysates on LBA plates showed that none of the cells could be resuscitated. The lysates were then stored at −20°C and thawed at room temperature before use. Tenfold-concentrated cell lysates were made by heat treating cell suspensions dissolved in 1/10 of the initial volume with water. Sterility of the heat-treated cells was determined by plating 100 μl of undiluted lysate on antibiotic-free LBA plates or 1/10-strength tryptic soy agar plates (Oxoid). For preparation of sterile-filtered cell suspensions, dense cultures of the different bacteria were passed through a 0.22-μm-pore-size nitrocellulose filter (Millipore, Bedford, Mass.). These non-heat-treated filtrates were streaked on LBA plates to confirm sterility and used directly or stored at −20°C before use.
Filter transformation.
Filter transformations were done essentially as described by Nielsen et al. (34). Briefly, 100 μl of the bacterial inoculum was mixed with 100 μl of lysed cells or DNA isolated from either Acinetobacter sp. strain BD413 (chr::KTG), P. fluorescens R2f (chr::KTG), or B. cepacia (chr::KTG) in an Eppendorf tube and then spread onto a nitrocellulose filter (Millipore) put on top of an LBA plate supplemented with ampicillin and rifampin. The DNA used was either purified bacterial DNA at concentrations of 0.1, 1, 10, and 50 μg per 100 μl of solution or cell lysates at concentrations of 1, 10, and 100 μl per 100 μl of solution) After incubation at 30°C overnight, the overgrown filter was transferred to a 50-ml Falcon tube and vortexed with 2 ml of a solution containing 0.85% NaCl and 100 μl of DNase I (5 mg ml−1; Boehringer Mannheim). Tenfold dilutions were plated onto LBA plates supplemented with ampicillin and rifampin (recipient counts) and ampicillin, rifampin, and kanamycin (transformant counts), and CFU counts were determined after incubation of the plates at 30°C for 72 h. Controls consisted of plates obtained from filters containing inoculum and 100 μl of water (to check for the occurrence of spontaneous Kmr mutants and bacterial contamination) and only DNA (10 μl) or 100 μl of lysate (to check for sterility).
Transformation frequencies are given as the number of Acinetobacter sp. CFU growing on transformant-selective LBA plates divided by the number of CFU on recipient-selective plates after the filter or soil transformations. The data from both the filter and soil microcosm experiments are presented as mean values for experiments done in triplicate ± SD (41). Each triplicate experiment was plated undiluted or in 10-fold dilutions on three plates and also repeated at least once in time. The mean variability of the data (CFU counts), given as the coefficient of variation (SD/mean), ranged from 0.2 to 0.4. The limit of detection is given as the reciprocal of the recipient cells (CFU). A Student t test was used to compare the data with regard to significance (P < 0.05 was considered significant).
Transformation in soil microcosms.
A Flevo silt loam soil (FSL) obtained from microplots in Wageningen, The Netherlands, was used in all microcosms. The FSL soil has previously been characterized (45, 51). After sampling, the nonsterile soil was sieved (4-mm mesh) and used directly or stored in plastic bags at 4°C. Sterile soil was obtained after gamma irradiation (4 Mrad) with a 60Co source (Gammaster BV, Ede, The Netherlands). Microcosms consisted of autoclaved polypropylene cylinders of 1-cm3 volume to which 1.2 g of soil was added, as described before (34, 35). The 7-mm-tall cylinders, made of 15-ml polypropylene centrifuge tubes (34, 35), containing the inoculated soil portions were placed on sterile agarose (1.5% [wt/vol] in water) in petri dishes. All transformations in soil were done at 20°C.
For transformation in sterile and nonsterile soil with cell lysates of Acinetobacter sp. strain BD413 (chr::KTG), P. fluorescens R2f (chr::KTG), and B. cepacia P2 (chr::KTG), the recipient inoculum was added to the soil by distributing a 100-μl aliquot onto the surface of each microcosm. The soil microcosms were incubated for 24 h before addition of 100 μl of nutrient solution (5M9L25P; 5-times-concentrated M9 salts [44], 25 times the standard P concentration, 2% lactic acid [35]) or 100 μl of water as a control. After 1 h, 100 μl of cell lysates was added, and the soil microcosms were incubated for a further 24 h before sampling as described above. Microcosms containing added wild-type lysates and inoculum, microcosms without added cell lysates or cells, or those with only cell lysates added were used as controls. The final moisture content was 38%. Due to the short incubation time and use of the agarose support, the soil did not dry substantially over the course of the experiments.
After the various transformation studies in soil, the microcosms were sampled, in triplicate, as follows. The cores of the soil microcosms (approximately 25% of the total dry weight of the soil) were collected by using the wide end of a 1-ml Eppendorf tip, transferred to a 1.5-ml Eppendorf tube with 0.9 ml of 0.1% sodium pyrophosphate (tetrasodium diphosphate decahydrate; Merck) supplemented with 50 μl of DNase I (5 mg ml−1) and Vortex mixed with 0.2 g of sterile gravel (2- to 4-mm diameter). Aliquots of the solution were either plated directly on three LBA plates, with antibiotics as described above for the filter transformation, or serially diluted before plating. The LBA plates used for sampling of nonsterile soil were supplemented with cycloheximide at 100 μg ml−1 to inhibit fungal growth. CFU were enumerated after a 72-h incubation period at 30°C. CFU counts refer to the soil portions, i.e., samples of 0.3 g (dry weight of soil). Colonies obtained from nonsterile soil were identified as Acinetobacter based on colony morphology (34, 35), growth rate, Biolog pattern (see below), and PCR amplification of the restored pFG4 plasmid or the original one carrying the 317-bp deletion (see below).
Stability of cell lysates in vitro and in soil.
The stability of cell lysates of Acinetobacter sp. strain BD413 (chr::KTG), P. fluorescens R2f (chr::KTG), and B. cepacia P2 (chr::KTG), incubated in vitro (in water or 5% humic acid solution) and in sterile and nonsterile soil for up to 8 days, was measured by the ability to transform freshly added Acinetobacter sp. strain BD413(pFG4). For the in vitro studies, 100 μl of each cell lysate was incubated in separate Eppendorf tubes at 20°C with either 100 μl of MilliQ water or 100 μl of a 10% (wt/vol in water) humic acid (Fluka catalog no. 53680) solution. After 0, 1, 2, 4, and 8 days, the solution was mixed with 100 μl of the recipient bacteria and processed further as described for the filter transformations. Sterility of the lysates over the 8-day period was checked by plating on nonselective LBA plates.
For studies in sterile soil, 100 μl of the cell lysates was added to the soil microcosms and incubated for 0, 1, 2, 3, and 4 days at 20°C before the bacterial inoculum (100 μl suspended in 5M9L25P) was added. After 24 h of incubation, the microcosms were sampled and plated, and the CFU count was determined as described above. Sterility of the lysates and nutrient solution was shown by sampling soil microcosms that received water instead of the bacterial inoculum. The absence of spontaneous mutants was confirmed by sampling microcosms receiving only the recipient inoculum and nutrients.
Stability studies using nonsterile soil were performed as described for the sterile soil experiments except that lysates were incubated for 0, 1, 2, 4, 6, 8, and 24 h before addition of the recipient inoculum. In addition, lysates of Acinetobacter sp. strain BD413 (chr::KTG) were added to nonsterile soil 10-fold diluted (adjusted to 100 μl with water) and incubated for up to 8 h before addition of the bacterial recipient.
As a control to determine the possible occurrence of transformants on the selective agar plates, the various cell lysates were added to 24-h nutrient-stimulated (with 5M9L25P) soil microcosms with Acinetobacter spp. and immediately plated as described for the soil transformations. No transformants were found in these assays, indicating that any DNA released from the soil during the suspension and plating procedure did not generate transformants. Furthermore, our experimental procedures were unlikely to yield plate transformants, since sampling of bacterial cells was done at least 24 h after DNA addition (and inoculum addition) to the soil microcosms; previous studies have shown that chromosomal DNA incubated in sterile soil for more than 6 h is not available to Acinetobacter sp. as transforming DNA (34).
Identification of putative transformants.
Putative restoration of the nptII gene in transformants of Acinetobacter sp.(pFG4) was assessed by restreaking colonies and amplifying colony material with primer set P1-P2 (12), using the method described by Hofmann and Brian (17). The PCR mix consisted of 27 μl of MilliQ water, 5 μl of 10× PCR buffer II (Perkin-Elmer), 5 μl of 25 mM MgCl2, 10 μl of 1 mM each deoxynucleoside triphosphate (Perkin-Elmer), 5 μl (2 μM) of each primer, 0.05 μl of T4 gene 32 protein (Pharmacia), and 0.25 μl (10 U/μl) of Stoffel fragment Taq DNA polymerase (Perkin-Elmer). The sequences of primers P1 and P2 were (1236) 5′ TGC TAA AGG AAG CGG AAC ′3 and (2929) 5 ′AGG TCA ACA GGC GGT AAC ′3, respectively. The primers were designed to amplify the Tn5 region from position 1236 to 2929, which includes the nptII promoter, the complete nptII gene, and the bleomycin resistance gene (12). The amplification conditions were 7 min at 95°C, then 1 min at 94°C, 1 min at 62°C, and 2 min at 72°C for 35 cycles, and finally 10 min at 72°C. DNA from a Kmr Acinetobacter sp. strain BD413(pFG4) strain was used as a positive control (the expected band size of the restored gene is 1,693 bp). Colonies obtained from recipient-selective plates (the expected band size of a gene with deletion is 1,376 bp) and wild-type Acinetobacter sp. strain BD413 were used as negative controls.
To confirm the identity of Kmr bacterial cells obtained from transformation studies done in nonsterile soil, colonies were routinely restreaked on transformant-selective plates. Randomly selected colonies were confirmed to have metabolic patterns in Biolog GN plates identical to that of the recipient strain BD413(pFG4). The Biolog plates were used as specified by the manufacturer and quantified on a Biolog MicroLog station (Biolog Microlog3; Biolog, Inc., Hayward, Calif.).
RESULTS
Filter transformations.
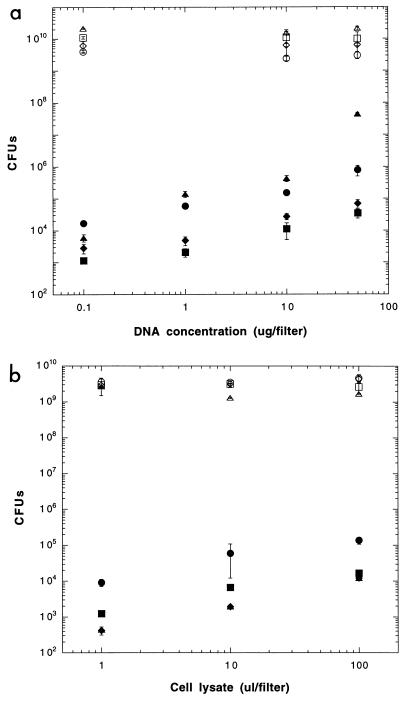
To clarify the effects of increasing concentrations of cell lysates on natural transformation of the recipient bacterium Acinetobacter sp. strain BD413(pFG4) under optimized conditions in vitro, we exposed this recipient to 1, 10, and 100 μl of lysate (per filter) obtained from the Kmr donor bacteria, Acinetobacter sp., P. fluorescens, and B. cepacia. The cell lysates proved to be highly efficient sources of DNA, since lysates from all three strains gave rise to high numbers of transformants (Fig. 1b). The maximum transformation frequencies obtained under optimized in vitro conditions were 3.0 × 10−5 for Acinetobacter sp., 3.5 × 10−6 for P. fluorescens R2f, and 6.3 × 10−6 for B. cepacia P2. An increase of the lysate concentration from 106 to 108 cells per filter increased the numbers of transformants 12- to 25-fold in all cases (Fig. 1b). To determine if the 100 μl of cell lysate used would be saturating for the recipient cells, transformations were also done with a 10-fold-concentrated cell lysate of Acinetobacter sp. (chr::KTG). This concentrate produced a significant higher number (mean ± SD) of transformants ([3.6 ± 0.3] × 106 transformants; frequency, 5.8 × 10−4) compared to the normally used lysate ([1.4 ± 0.3] × 105 transformants; frequency, 3.0 × 10−5). However, since the concentrated cell lysate would represent a further enrichment of an already dense cell suspension, we found the continued use of this concentrate of little value when estimating lysate availability in natural soils. Cell lysates obtained after longer periods of heat treatment, e.g., autoclaving at 120°C for 15 min, did not give rise to any transformants in our studies, presumably due to fragmentation of the DNA, generation of inactive single-stranded DNA, and complexation of DNA with the denatured cell debris. Shorter periods of heating gave occasional growth of survivors.
FIG. 1.
(a) Natural transformation of Acinetobacter sp. strain BD413(pFG4) on filter with increasing concentrations of chromosomal DNA (chr::nptII) from Acinetobacter sp. (circles), P. fluorescens R2f (squares), and B. cepacia P2 (diamonds). Filled symbols, transformants; open symbols, recipients. Triangles show recipients and transformants of Acinetobacter sp. strain BD413 (wild type) with homologous cell lysates (chr::nptII). T bars, SD. (b) Natural transformation of Acinetobacter sp. BD413(pFG4) on filter with increasing concentrations of cell lysates (chr::nptII) from Acinetobacter sp. (circles), P. fluorescens R2f (squares), and B. cepacia P2 (diamonds). Filled symbols, transformants; open symbols, recipients. Triangles show recipients and transformants (symbols are within symbols for Burkholderia transformants) of Acinetobacter sp. strain BD413 (wild type) with homologous cell lysates (chr::nptII). T-bars, SD.
Filter transformations performed with purified DNA instead of lysate (at 0, 0.1, 1, 10, and 50 μg of purified DNA per filter) isolated from the same bacterial strains gave rise to numbers of transformants higher than those obtained with the lysates (Fig. 1b). However, if the numbers of transformants detected are adjusted by the estimated DNA content in the lysates (see Materials and Methods), the lysates were at least as efficient as transforming DNA; differences in transformation frequency of less than fourfold for Acinetobacter sp. DNA and less than threefold for P. fluorescens DNA were found. For B. cepacia, a higher difference in transformation frequency (8- to 15-fold) was seen; the lysate in this case also gave higher numbers of transformants compared to purified DNA.
Sterile-filtered suspensions of non-heat-treated bacterial cultures were also assayed as a source of DNA in the filter transformations to indicate a potential variability of the amount of free DNA available in the cell supernatant. However, Table 1 shows that lower frequencies were obtained with filtrates than with cell lysates (compare Table 1 to Fig. 1b for 100 μl of lysate), presumably indicating the amount of free DNA available in the supernatant compared to total DNA present in the cell debris. There were no clear differences in the recipient counts obtained after the filter transformations done with the above-described DNA sources, indicating that inhibition of recipient growth was not the cause of the variability in the number of transformants observed.
TABLE 1.
Natural transformation of Acinetobacter sp. strain BD413 in filter assays with sterile-filtered cell suspensions (chr::KTG) of Acinetobacter sp., P. fluorescens, and B. cepacia
| Strain (chr::KTG) | Mean no. of transformants (CFU) ± SD | No. of transformants/recipient |
|---|---|---|
| Acinetobacter sp. strain BD413 | (2.4 ± 0.4) × 104 | 1.2 × 10−5 |
| P. fluorescens R2f | (3.1 ± 0.7) × 103 | 1.3 × 10−7 |
| B. cepacia P2 | (2.0 ± 0.3) × 103 | 6.9 × 10−7 |
To check the transformation efficiency of the plasmid-harboring Acinetobacter sp. strain BD413(pFG4) as a recipient for chromosomal DNA and lysates, we compared the transformation frequencies obtained with this strain to frequencies for the wild-type Acinetobacter sp. strain BD413. In transformation studies with purified homologous DNA, the wild-type strain was significantly more transformable than the plasmid-bearing strain when high concentrations of DNA were used (Fig. 1a). This observation could be due to deleterious crossover events in the pFG4 recipient since a recombination of the restored kanamycin resistance-conferring plasmid into a newly generated chromosomal nptII-bearing recipient would cause inactivation of the selected marker gene. However, such a relationship was not found when lysates were used, and the plasmid-bearing strain proved to be a more efficient recipient for lysates than the wild-type strain (Fig. 1b). The presence, or generation during uptake, of more fragmented DNA in the cell lysates, possibly impeding transformation of the wild-type recipient, which requires larger fragments for integration into the chromosome (40), could account for this observation.
Transformation in soil microcosms.
Given the high numbers of bacterial cells in soil (109 to 1010 bacteria per g of soil), dead bacteria could potentially contribute significantly to the gene pool of chromosomal DNA present in this environment. To elucidate the potential of cell lysates obtained from common soil and rhizosphere bacteria to function as DNA sources for natural transformation, we exposed the recipient bacterium Acinetobacter sp. strain BD413(pFG4) to cell lysates obtained from the Kmr (chr::KTG) donor bacteria, Acinetobacter sp. BD413, P. fluorescens R2f, and B. cepacia P2. For transformation in sterile and nonsterile soil, the recipient was added to the soil and incubated for 24 h before addition of nutrient solution and lysates. As seen from Table 2, freshly added cell lysates obtained from all three different species were able to transform the recipient Acinetobacter sp. strain BD413(pFG4) residing in sterile soil. The homologous cell lysates were, however, 4- to 16-fold more efficient for transformation than lysates produced from the heterologous strains. Recipient Acinetobacter cells residing in nonsterile soil were recalcitrant to transformation with heterologous cell lysates. The homologous lysate, however, transformed the recipient at a frequency of 1.1 × 10−6, corresponding to 1.9 × 10−7 transformants per lysed cell (Table 2).
TABLE 2.
Natural transformation and restoration of a 317-bp deleted nptII gene in Acinetobacter sp. strain BD413(pFG4) residing in sterile and nonsterile soil microcosms for 24 h, with added cell lysates of Acinetobacter sp., P. fluorescens, and B. cepaciaa
| Cell lysate (chr::KTG) of: | Sterile soil
|
Nonsterile soil
|
||||
|---|---|---|---|---|---|---|
| Mean no. of transformants (CFU) ± SDb | No. of transformants/recipient | No. of transformants/lysed cellc | Mean no. of transformants (CFU) ± SD | No. of transformants/recipient | No. of transformants/lysed cell | |
| Acinetobacter sp. strain BD413 | 1,290 ± 146 | 7.4 × 10−6 | 5.6 × 10−6 | 44 ± 15 | 1.1 × 10−6 | 1.9 × 10−7 |
| P. fluorescens R2F | 77 ± 29 | 3.1 × 10−7 | 3.8 × 10−7 | NDd | <1.4 × 10−8 | <5.0 × 10−7 |
| B. cepacia P2 | 334 ± 85 | 1.8 × 10−6 | 1.1 × 10−6 | ND | <1.8 × 10−8 | <3.1 × 10−7 |
Following 24 h of bacterial presence in soil, nutrients (5M9L25P) and lysates (after 1 h) were added, and the microcosms were incubated for 24 h at 20°C before plating and enumeration of CFU.
The mean coefficient of variation (SD/mean) of the CFU of transformants and recipients was between 0.3 and 0.4.
The number of lysed cells is the CFU count of the bacterial suspension used for preparation of the lysates.
ND, not detected.
Microcosms with added inoculum, nutrients, and the wild-type strain lysates, or only lysates and nutrients, were used as controls for spontaneous mutations and contamination. None of these treatments produced any colonies that were identified as Acinetobacter spp. on transformant-selective plates. In addition, selected colonies obtained from transformant-selective plates were restreaked and confirmed to be Acinetobacter sp. strain BD413(pFG4) by the metabolic pattern obtained on Biolog GN plates and by the presence of the restored nptII gene sequence as verified by PCR.
Availability of cell lysates for natural transformation in soil.
To clarify the time period during which cell lysates would be available as a source of transforming DNA in soil, the lysates were incubated for increasing amounts of time in sterile (0 to 4 days) and nonsterile (0 to 24 h) soil before addition of the bacterial recipient. As can be seen in Table 3, cell lysates (chr::KTG) incubated in sterile soil were available as transforming DNA for the recipient cells for 3 days for P. fluorescens and B. cepacia lysates or 4 days with Acinetobacter sp. lysates. The highest transformation frequencies were always produced with freshly added lysate to soil. Lysates incubated for 1 day in sterile soil had an average remaining transforming capability of 40%. In nonsterile soil (Table 4), the lysates became rapidly inactivated, and restoration of the nptII gene was seen only with lysates of P. fluorescens and B. cepacia incubated for up to 4 h in soil and with lysates from Acinetobacter sp. incubated for up to 8 h in soil. Incubation of the lysates for 1 h in nonsterile soil reduced their transforming ability by 31% on average. For the Acinetobacter sp. cell lysates, a 10-fold-diluted lysate was also assayed in nonsterile soil. Transformation with this lysate was detectable only for 2 h in soil. After 1 h in soil, the diluted lysate had a remaining transforming activity of 36%.
TABLE 3.
Natural transformation and restoration of a 317-bp deleted nptII gene in Acinetobacter sp. strain BD413(pFG4) with cell lysates (chr::KTG) of Acinetobacter sp., P. fluorescens, and B. cepacia, incubated for up to 4 days in sterile soil before recipient inoculation
| Incubation time (days)a |
Acinetobacter sp. strain BD413
|
P. fluorescens R2f
|
B. cepacia P2
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of transformants (CFU)b | No. of transformants/recipient | No. of transformants/lysed cellc | No. of transformants (CFU) | No. of transformants/recipient | No. of transformants/ lysed cell | No. of transformants (CFU) | No. of transformants/recipient | No. of transformants/lysed cell | |
| 0 | 1,073 | 9.9 × 10−6 | 4.8 × 10−6 | 254 | 2.2 × 10−6 | 1.3 × 10−6 | 66 | 5.3 × 10−7 | 2.1 × 10−7 |
| 1 | 73 | 5.6 × 10−7 | 3.2 × 10−7 | 60 | 3.1 × 10−7 | 2.9 × 10−7 | 59 | 3.6 × 10−7 | 1.9 × 10−7 |
| 2 | 21 | 1.2 × 10−7 | 9.3 × 10−8 | 6 | 3.5 × 10−8 | 2.9 × 10−8 | 45 | 2.5 × 10−7 | 1.4 × 10−7 |
| 3 | 18 | 8.8 × 10−8 | 8.0 × 10−8 | 1 | 5.2 × 10−9 | 4.9 × 10−8 | 12 | 4.8 × 10−8 | 3.8 × 10−8 |
| 4 | 10 | 4.5 × 10−8 | 4.4 × 10−8 | NDd | <5.6 × 10−9 | <5.0 × 10−9 | ND | <5.6 × 10−9 | <3.1 × 10−9 |
Incubation time of lysates in sterile soil before addition of the Acinetobacter sp.(pFG4 ΔnptII), suspended in 5M9L25P. The microcosms with added bacteria were incubated further for 24 h at 20°C before plating and enumeration of CFU.
The variability of the CFU of transformants and recipients, as determined by the coefficient of variation (SD/mean), was between 0.3 and 0.4 (mean values).
The number of lysed cells is given as the CFU count of the bacterial suspension used for preparation of the lysates.
ND, not detected.
TABLE 4.
Natural transformation and restoration of a 317-bp deleted nptII gene in Acinetobacter sp. strain BD413(pFG4) with cell lysates (chr::KTG) of the Acinetobacter sp., P. fluorescens, and B. cepacia, incubated for up to 24 h in nonsterile soil before recipient inoculation
| Incubation time (h)a |
Acinetobacter sp. BD413
|
P. fluorescens R2f
|
B. cepacia P2
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of transformants (CFU)b | No. of transformants/recipient | No. of transformants/lysed cellc | No. of transformants (CFU) | No. of transformants/recipient | No. of transformants/lysed cell | No. of transformants (CFU) | No. of transformants/recipient | No. of transformants/lysed cell | |
| 0 | 276 | 3.0 × 10−6 | 1.2 × 10−6 | 15 | 2.0 × 10−7 | 7.5 × 10−8 | 33 | 4.1 × 10−7 | 1.0 × 10−7 |
| 1 | 148 | 1.7 × 10−6 | 6.4 × 10−7 | 10 | 1.6 × 10−7 | 5.0 × 10−8 | 29 | 3.9 × 10−7 | 9.1 × 10−8 |
| 2 | 55 | 6.2 × 10−7 | 2.4 × 10−7 | 6 | 9.3 × 10−8 | 2.9 × 10−8 | 11 | 2.2 × 10−7 | 3.4 × 10−8 |
| 4 | 18 | 2.1 × 10−7 | 7.8 × 10−8 | 4 | 4.9 × 10−8 | 2.0 × 10−8 | 9 | 1.3 × 10−7 | 2.8 × 10−8 |
| 8 | 14 | 2.1 × 10−7 | 6.1 × 10−8 | NDd | <4.3 × 10−8 | <5.0 × 10−9 | ND | <1.5 × 10−8 | <3.1 × 10−9 |
| 24 | ND | <2.2 × 10−8 | <4.3 × 10−9 | ND | ND | ||||
Incubation time of lysates in nonsterile soil before addition of Acinetobacter sp.(pFG4 ΔnptII) (suspended in 5M9L25P). The microcosms with added bacteria were incubated further for 24 h at 20°C before plating and enumeration of CFU.
The variability of the CFU of transformants and recipients, as determined by the coefficient of variation (SD/mean), was between 0.3 and 0.4 (mean values).
The number of lysed cells is given as the CFU count of the bacterial suspension used for the preparation of the lysates.
ND, not detected.
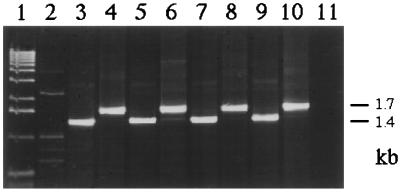
Microcosms containing lysates of wild-type strains of the three donor bacteria, nutrients and the inoculant, or microcosms with lysates and nutrients were used as controls. These microcosms did not give rise to any Acinetobacter transformant colonies (see above). Colonies restreaked from transformant and recipient-selective plates were PCR amplified to confirm recombinational repair of the nptII gene. Figure 2 shows the PCR amplification of DNA from colonies growing on transformant-selective plates transformed with either of the three lysates and colonies growing on the corresponding recipient-selective LBA plates.
FIG. 2.
PCR amplification of bacterial colonies obtained after natural transformation of the Acinetobacter sp. with cell lysates (chr::nptII) of Acinetobacter sp., P. fluorescens, and B. cepacia incubated in nonsterile soil. Lane 1, 1-kb ladder (Gibco/BRL); lane 2, wild-type Acinetobacter sp.; lane 3, Acinetobacter sp. strain BD413(pFG4) inoculant; lane 4, Acinetobacter sp. transformant with restored nptII gene (positive control); lane 5, Acinetobacter sp. recipient; lane 6, Acinetobacter sp. transformant; lane 7, B. cepacia lysate recipient; lane 8, B. cepacia lysate transformant; lane 9, P. fluorescens lysate recipient; lane 10, P. fluorescens lysate transformant; lane 11, PCR mix without added DNA.
Stability of cell lysates in vitro.
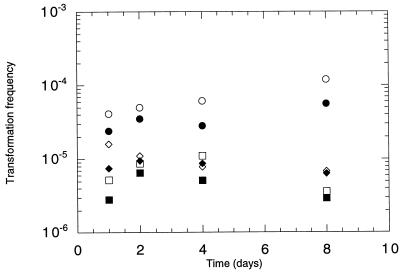
To clarify the stability of transforming DNA in the cell lysates over time, portions of the lysates of the three bacteria were stored in Eppendorf tubes with added water or 5% humic acid. Transformation assays with the water-suspended lysates sampled at day 1, 2, 4, and 8 did not reveal any clear changes in the transforming ability of the lysates (Fig. 3). The addition of 5% humic acid to the lysates affected their transforming activity, giving consistently lower transformation frequencies, and a 32% average reduction was seen. This reduction might be explained by a physical inhibition of the filter transformation by the dark brown humic acid solution. Thus, clear effects of humic acids on protection of DNA (7) or inhibition of transformation (48) were not found.
FIG. 3.
Transformation frequencies of Acinetobacter sp. in vitro with cell lysates (chr::nptII) of Acinetobacter sp. (circles), P. fluorescens R2f (squares), and B. cepacia P2 (diamonds) incubated for 1 to 8 days in water (open symbols) or in 5% humic acid solution (filled symbols).
DISCUSSION
In this study, we used lysates obtained from common soil bacteria (3, 4, 22) to demonstrate that Acinetobacter sp. cells can efficiently access genetic information in cell debris of various bacterial genera present in soil. Based on homologous recombination-mediated restoration of a kanamycin resistance gene (nptII) in the recipient Acinetobacter sp.(pFG4), the uptake of chromosomal gene fragments present in lysates (chr::KTG) of Acinetobacter sp., P. fluorescens R2f, and B. cepacia P2 was shown to occur at frequencies between 10−5 and 10−6 in vitro. A 10-fold drop in the frequencies was seen in sterile soil, and a further reduction from 5- to more than 100-fold was noted in nonsterile soil. The homologous lysate transformed the recipient at a frequency of 1.1 × 10−6 in nonsterile soil. This frequency is similar (when judged determined per microgram of DNA used) to that obtained in studies using purified DNA in nonsterile soil (34; K. M. Nielsen, T. B. Løkken, and A. M. Bones, unpublished data). On average, a less than 10-fold difference in the capability of restoration of the antibiotic resistance gene (via homologous recombination) was seen between isogenic and heterogenic lysates. The reduced transformation frequencies obtained for the heterologous lysates can be caused by differences in the degree of homology to the recipient genome or reduced accessibility of the nptII gene due to cellular debris. Homology to the heterologous strains is found only in the pFG4 plasmid in the recipient Acinetobacter sp., whereas homology to the isogenic lysate is also displayed by the recipient chromosome. Indeed, filter transformations with plasmid-harboring recipients gave, on average, 10-fold-higher numbers of transformants with isogenic lysates than the wild-type recipients; these numbers of transformants were comparable to those obtained with the heterogenic lysates. Up to 1 μg of free DNA (per g of dry soil) has been estimated to be present in soil (of a total of approximately 90 μg of DNA/g of dry soil [49]). Torsvik et al. (50) estimated that at least 4,000 different bacterial types composed the majority of the bacterial diversity seen in soil. Our 14-fold-higher inoculum concentration (compared to the estimated average population size) could be successfully transformed in nonsterile soil with DNA corresponding to the amount found in the lysates of fewer than five clones. Acinetobacter sp. has also been transformed in vitro with the nptII marker gene present in transgenic plants (8, 12), and its DNA uptake is regarded as nonspecific (40, 41). Due to the heterogeneity of the DNA donors used here, it was unclear if DNA escaping their cells would be exploited efficiently as a source of genetic information by Acinetobacter spp. populations in soil. The results obtained both in vitro and in situ indicate that the lack of DNA purity and the variable cellular background did not reduce their transforming ability, which for the isogenic strain was similar to that obtained with purified DNA (Fig. 1a). Kloos et al. (20) also observed equally efficient transformation of Acinetobacter spp. on filters when inducing lysis of donor cells. DNA may be associated with the bacterial slime layer and thereby become stabilized and still be a source of extracellular DNA. Catlin (6) reported transformation of Neisseria meningitidis by DNA from cells and from culture slime, and Juni (18) prepared crude extracts of cells for natural transformation of Acinetobacter spp. by heating sodium dodecyl sulfate-treated cell suspensions at 60°C for 1 h. The apparently enhanced transformation efficiency of the cell lysates compared to the purified DNA on filters might be due to underestimation of the amount of free chromosomal DNA present in the cell supernatant. The sterile-filtered cultures of the different donors all contained high amounts of free DNA, as shown in the transformation assay (Table 1), indicating that none of the bacteria secreted high amounts of DNase during in vitro growth. Thus, the inactivation of the transforming activity of the cell lysates seen in soil seems to be due not to any introduced DNase activity but rather to indigenous activity in soil or other abiotic factors and mechanisms of DNA inactivation present in soil.
Production of extracellular DNA in Acinetobacter spp., Burkholderia spp., and Pseudomonas spp. is known to occur (25, 26, 39, 42). From the differences seen between the transforming activity of the purified DNA, sterile-filtered cell supernatants, and cell lysates of B. cepacia P2 and P. fluorescens R2f, it can be suggested that B. cepacia liberates more free chromosomal DNA than P. fluorescens during in vitro growth since the CFU counts of their lysates were comparable.
Fragments of chromosomal bacterial DNA have been shown to persist in soil for weeks (9, 43). However, this physical stability has not been reflected in similar data demonstrating the long-term biological activity (e.g., transforming activity) of chromosomal DNA in soil. Bacterial DNA introduced into nonsterile soil has been found to be active as transforming DNA for only a few hours (34). Thus, there is a clear discrepancy between the detected physical and the functional significance of DNA in soil (20). Pure DNA is initially hydrolyzed at substantial rates when introduced to soil. The half-life of purified DNA added to soil has been estimated to be 9 to 28 h, depending on the soil's mineral composition (26). The presence of cell debris may be important for the protection of crude DNA against enzymatic hydrolysis and its interaction with the soil matrix. The half-life of DNA associated with dead bacterial cells may therefore differ substantially from estimates obtained with purified DNA. For instance, the half-life of DNA present in bacterial cells in deep-sea sediments was found to be several days (38).
Our data demonstrate that cell lysates are available as a source of transforming DNA for Acinetobacter spp. populations in sterile or nonsterile soil for considerable time periods: P. fluorescens and B. cepacia lysates were available for up to 3 days/4 h and isogenic lysates of the Acinetobacter sp. were available for up to 4 days/8 h. Lysates incubated for 1 day in soil had an average transforming capability of 30 to 40% of the initial value. The relative availability of the DNA present in the lysates was enhanced over time compared to the availability of naked chromosomal DNA for natural transformation of Acinetobacter spp. in the same sterile and nonsterile FSL soil (34; Nielsen et al., unpublished data), indicating that the presence of cell debris may protect DNA from inactivation in soil.
The decreasing transformation frequencies over time were likely to be caused by DNA fragmentation, degradation by nucleases, and possible inactivation of DNA by binding to soil substances. Shorter DNA fragments are known to be less efficient for natural transformation than high-molecular-weight DNA (40).
Thus, we conclude that cell lysates in which DNA may be adhering to polysaccharides, proteins, and/or membranes are generally not inhibitory to natural transformation of Acinetobacter spp. in soil and may, in addition, protect DNA from rapid inactivation. Furthermore, if DNA homology is present (21, 29, 32, 47, 53), gene transfer by natural transformation might provide populations of Acinetobacter cells with a mechanism for generating genetic variability (e.g., mosaic genes) by enabling them to take up chromosomal DNA released from various bacterial donors in their surroundings.
ACKNOWLEDGMENTS
We thank L. Lankwarden and E. Torsetnes for excellent technical assistance and F. Gebhard for providing the Acinetobacter sp. BD413(pFG4) strain.
This work was supported by grant MU:121733 from the Norwegian Research Council to K.M.N.
REFERENCES
- 1.Alvarez A J, Khanna M, Toranzos G A, Stotzky G. Amplification of DNA bound on clay minerals. Mol Ecol. 1998;7:775–778. [Google Scholar]
- 2.Barcus V A, Titheradge A J B, Murray N E. The diversity of alleles at the hsd locus in natural populations of Escherichia coli. Genetics. 1995;140:1187–1197. doi: 10.1093/genetics/140.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann P. Isolation of Acinetobacter from soil and water. J Bacteriol. 1968;96:39–42. doi: 10.1128/jb.96.1.39-42.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevivino A, Sarrocco S, Dalmastri C, Tabacchioni S, Cantale C, Chiarini L. Characterization of a free-living maize-rhizosphere population of Burkholderia cepacia: effect of seed treatment on disease suppression and growth promotion of maize. FEMS Microbiol Ecol. 1998;27:225–237. [Google Scholar]
- 5.Bowler L D, Zhang Q-Y, Riou J-Y, Spratt B G. Interspecies recombination between the penA genes of Neisseria meningitis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitis: natural events and laboratory simulations. J Bacteriol. 1994;176:333–337. doi: 10.1128/jb.176.2.333-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catlin B W. Transformation of Neisseria meningitidis by deoxyribonucleates from cells and from culture slime. J Bacteriol. 1960;79:579–590. doi: 10.1128/jb.79.4.579-590.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crecchio C, Stotzky G. Binding of DNA on humic acids: effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol Biochem. 1998;30:1061–1067. [Google Scholar]
- 8.De Vries J, Wackernagel W. Detection of nptII (kanamycin resistance) genes in genomes of transgenic plants by marker-rescue transformation. Mol Gen Genet. 1998;257:606–613. doi: 10.1007/s004380050688. [DOI] [PubMed] [Google Scholar]
- 9.England L S, Lee H, Trevors J. Persistence of Pseudomonas aureofaciens strains and DNA in soil. Soil Biol Biochem. 1997;29:1521–1527. [Google Scholar]
- 10.Feil E, Zhou J, Maynard Smith J, Spratt B G. A comparison of the nucleotide sequences of the adk and recA genes of pathogenic and commensal Neisseria species: evidence for extensive interspecies recombination within adk. J Mol Evol. 1996;43:631–640. doi: 10.1007/BF02202111. [DOI] [PubMed] [Google Scholar]
- 11.Gallori E, Bazzicalupo M, Dal Canto L, Nannipieri P, Vettori C, Stotzky G. Transformation of Bacillus subtilis by DNA bound on clay in non-sterile soil. FEMS Microbiol Ecol. 1994;15:119–126. [Google Scholar]
- 12.Gebhard F, Smalla K. Transformation of Acinetobacter sp. strain BD413 by transgenic sugar beet DNA. Appl Environ Microbiol. 1998;64:1550–1554. doi: 10.1128/aem.64.4.1550-1554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham J B, Istock C A. Genetic exchange in Bacillus subtilis in soil. Mol Gen Genet. 1978;166:287–290. doi: 10.1007/BF00267620. [DOI] [PubMed] [Google Scholar]
- 14.Graham J B, Istock C A. Gene exchange and natural selection cause Bacillus subtilis to evolve in soil culture. Science. 1979;204:637–639. doi: 10.1126/science.107592. [DOI] [PubMed] [Google Scholar]
- 15.Guttman D S, Dykhuizen D E. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 16.Herrero M, De Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. Appl Environ Microbiol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann D A, Brian D A. Sequencing PCR DNA amplified directly from a bacterial colony. BioTechniques. 1991;11:30–31. [PubMed] [Google Scholar]
- 18.Juni E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapur V, Kanjilal S, Hamrick M R, Li L-L, Whittam T S, Sawyer S A, Musser J M. Molecular population genetic analysis of the streptokinase gene of Streptococcus pyogenes: mosaic alleles generated by recombination. Mol Microbiol. 1995;16:509–519. doi: 10.1111/j.1365-2958.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 20.Kloos D-U, Stratz M, Guttler A, Steffan R J, Timmis K N. Inducible cell lysis system for the study of natural transformation and environmental fate of DNA released by cell death. J Bacteriol. 1994;176:7352–7361. doi: 10.1128/jb.176.23.7352-7361.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kok R G, D'Argeno D A, Ornston L N. Combining localized PCR mutagenesis and natural transformation in direct genetic analysis of a transcriptional regulator gene, pobR. J Bacteriol. 1997;179:4270–4276. doi: 10.1128/jb.179.13.4270-4276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert B, Leyns F, van Rooyen L, Gossele F, Papon Y, Swings J. Rhizobacteria of maize and their antifungal activities. Appl Environ Microbiol. 1987;53:1866–1871. doi: 10.1128/aem.53.8.1866-1871.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence J G, Ochman H. Molecular archeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee G-H, Stotzky G. Transformation is a mechanism of gene transfer in soil. Korean J Microbiol. 1990;28:210–218. [Google Scholar]
- 25.Lorenz M G, Gerjets D, Wackernagel W. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch Microbiol. 1991;156:319–326. doi: 10.1007/BF00263005. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz M G, Wackernagel W. Mechanisms and consequences of horizontal gene transfer in natural bacterial populations. In: Tomiuk J, Wöhrmann K, Sentker A, editors. Transgenic organisms—biological and social implications. Basel, Switzerland: Birkhauser Verlag; 1996. pp. 45–57. [Google Scholar]
- 28.Majewski J, Cohan F M. The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics. 1998;148:13–18. doi: 10.1093/genetics/148.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 30.Maynard Smith J, Dowson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 31.Maynard Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melnikov A, Youngman P J. Random mutagenesis by recombinatorial capture of PCR products in Bacillus subtilis and Acinetobacter calcoaceticus. Nucleic Acids Res. 1999;27:1056–1062. doi: 10.1093/nar/27.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen K M. Barriers to horizontal gene transfer by natural transformation in soil bacteria. APMIS. 1998;106(Suppl. 84):77–84. doi: 10.1111/j.1600-0463.1998.tb05653.x. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen K M, van Weerelt M D M, Berg T N, Bones A M, Hagler A N, van Elsas J D. Natural transformation and availability of transforming DNA to Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol. 1997;63:1945–1952. doi: 10.1128/aem.63.5.1945-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen K M, Bones A M, van Elsas J D. Induced natural transformation of Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol. 1997;63:3972–3977. doi: 10.1128/aem.63.10.3972-3977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen K M, Bones A, Smalla K, van Elsas J D. Horizontal gene transfer from transgenic plants to terrestrial bacteria—a rare event? FEMS Microbiol Rev. 1998;22:79–103. doi: 10.1111/j.1574-6976.1998.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 37.Nijhuis E H, Maat M J, Zeegers I W E, Waalwijk C, van Veen J A. Selection of bacteria suitable for introduction into the rhizosphere of grass. Soil Biol Biochem. 1993;25:885–895. [Google Scholar]
- 38.Novitzky J A. Degradation of dead microbial biomass in a marine sediment. Appl Environ Microbiol. 1986;52:504–509. doi: 10.1128/aem.52.3.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmen R, Hellingwerf K J. Acinetobacter calcoaceticus liberates chromosomal DNA during induction of competence by cell lysis. Curr Microbiol. 1995;30:7–10. doi: 10.1007/BF00294516. [DOI] [PubMed] [Google Scholar]
- 40.Palmen R, Hellingwerf K J. Uptake and processing of DNA by Acinetobacter calcoaceticus. Gene. 1997;192:179–190. doi: 10.1016/s0378-1119(97)00042-5. [DOI] [PubMed] [Google Scholar]
- 41.Palmen R, Vosman B, Buijman P, Breek C K, Hellingwerf K J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 42.Paul J H, David A W. Production of extracellular nucleic acids by genetically altered bacteria in aquatic-environment microcosms. Appl Environ Microbiol. 1989;55:1865–1869. doi: 10.1128/aem.55.8.1865-1869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Recorbet G, Picard C, Normand P, Simonet P. Kinetics of persistence of chromosomal DNA from genetically engineered Escherichia coli introduced into soil. Appl Environ Microbiol. 1993;59:4289–4294. doi: 10.1128/aem.59.12.4289-4294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Smit E, van Elsas J D. Conjugal gene transfer in the soil environment: new approaches and developments. In: Gauthier M, editor. Gene transfers and environment. Berlin, Germany: Springer-Verlag; 1992. pp. 79–94. [Google Scholar]
- 46.Smit E, Wolters A, van Elsas J D. Genetic stability, conjugal transfer and expression of heterologous DNA inserted into different plasmids and the genome of Pseudomonas fluorescens in soil. Rev Microbiol. 1995;26:169–179. [Google Scholar]
- 47.Strätz M, Mau M, Timmis K N. System to study horizontal gene exchange among microorganisms without cultivation of recipients. Mol Microbiol. 1996;22:207–215. doi: 10.1046/j.1365-2958.1996.00099.x. [DOI] [PubMed] [Google Scholar]
- 48.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torsvik V L, Goksøyr J. Determination of bacterial DNA in soil. Soil Biol Biochem. 1978;10:7–12. [Google Scholar]
- 50.Torsvik V L, Goksøyr J, Daae F L. High diversity of DNA in soil. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Elsas J D, Dijkstra A F, Govaert J M, van Veen J A. Survival of Pseudomonas fluorescens and Bacillus subtilis introduced into two soils of different texture in field microplots. FEMS Microbiol Ecol. 1986;38:151–160. [Google Scholar]
- 52.Van Elsas J D, Trevors J, Starodub M E. Bacterial conjugation between pseudomonads in the rhizosphere of wheat. FEMS Microbiol Ecol. 1988;53:299–306. [Google Scholar]
- 53.Vulic M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smit J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 2.4.1–2.4.4. [DOI] [PubMed] [Google Scholar]