Abstract
“Norwalk-like viruses” (NLVs) and hepatitis A virus (HAV) are the most common causes of virus-mediated food-borne illness. Epidemiological investigations of outbreaks associated with these viruses have been hindered by the lack of available methods for the detection of NLVs and HAV in foodstuffs. Although reverse transcription (RT)-PCR methods have been useful in detecting NLVs and HAV in bivalve mollusks implicated in outbreaks, to date such methods have not been available for other foods. To address this need, we developed a method to detect NLVs and HAV recovered from food samples. The method involves washing of food samples with a guanidinium-phenol-based reagent, extraction with chloroform, and precipitation in isopropanol. Recovered viral RNA is amplified with HAV- or NLV-specific primers in RT-PCRs, using a viral RNA internal standard control to identify potential sample inhibition. By this method, 10 to 100 PCR units (estimated to be equivalent to 102 to 103 viral genome copies) of HAV and Norwalk virus seeded onto ham, turkey, and roast beef were detected. The method was applied to food samples implicated in an NLV-associated outbreak at a university cafeteria. Sliced deli ham was positive for a genogroup II NLV as determined by using both polymerase- and capsid-specific primers and probes. Sequence analysis of the PCR-amplified capsid region of the genome indicated that the sequence was identical to the sequence from virus detected in the stools of ill students. The developed method is rapid, simple, and efficient.
“Norwalk-like viruses” (NLVs), previously known as small round-structured viruses, are a group of human caliciviruses (HuCVs) belonging to a newly proposed genus in the family Caliciviridae (15). NLVs are the most widely recognized agents of outbreaks of food-borne and waterborne viral gastroenteritis. Recently, the Centers for Disease Control and Prevention determined that 96% of reported outbreaks of nonbacterial gastroenteritis in the United States are caused by NLVs (11). Nausea, vomiting, diarrhea, and an illness lasting 1 to 3 days characterized these outbreaks. Consumption of contaminated food was the most commonly identified mode of transmission (11). Similarly, HuCVs were detected in up to 91% of all outbreaks of gastroenteritis reported in The Netherlands in 1996, confirming the etiologic significance of these viruses (46). Outbreaks of food-borne disease have been associated with consumption of uncooked and cooked shellfish, ice, water, bakery products (frosting), various types of salads (potato, chicken, fruit, and tossed), and cold foods (celery, melon, vermicelli consommé, sandwiches, and cold cooked ham) (18, 33, 42, 47). Consumption of raw or cooked shellfish has resulted in numerous documented outbreaks of HuCV-associated disease (1, 6, 10, 25, 27, 30, 32, 34). HuCVs also have been implicated in outbreaks of gastroenteritis caused by ill or asymptomatic infected food handlers who contaminated food while preparing salads, cold food items, and frosted confectionery items (20, 23, 26, 31, 37–38, 40, 48; A. Curry, T. Riordan, J. Craske, and E. O. Caul, Letter, Lancet ii:864–865, 1987).
Although NLVs are currently recognized as the cause of the majority of outbreaks of viral gastroenteritis, hepatitis A virus (HAV) historically has been the most common virus associated with food-borne and waterborne outbreaks, and there continue to be reports of HAV outbreaks (9, 19, 39). Epidemiological investigations of outbreaks associated with these viruses have been hindered by a lack of available methods for the detection of NLVs and HAV in foodstuffs other than bivalve mollusks. Although reverse transcription-PCR (RT-PCR) methods have been useful in detecting NLVs and HAV in bivalve mollusks implicated in outbreaks (16, 27, 28, 30), to date such methods have not been useful with other foods. The goal of this study was to develop a method to recover HAV and NLVs from food, followed by detection of viral nucleic acid by RT-PCR and subsequent PCR product confirmation by internal oligoprobing or sequencing.
MATERIALS AND METHODS
Virus preparations.
Stools containing Norwalk virus (NV) collected from human volunteers following challenge with NV 8fIIa (13, 22) were diluted to 10% suspensions in phosphate-buffered saline (PBS) (pH 7.4). Titers of NV suspensions were determined by RT-PCR unit (PCRU) endpoint dilution by using heat to release the viral genome from the capsid protein (43).
The cell culture-adapted HM-175 strain of HAV was propagated in FRhK4 cells as previously described (45). A dispersed virus preparation was prepared by adjustment of an FRhK4-grown virus suspension to pH 9.5, followed by passage sequentially through low protein binding 5.0-, 0.45-, and 0.22-μm-pore-size filters (Millipore). The pH of virus-containing filtrates was adjusted aseptically to pH 7, and 50-μl to 1-ml aliquots were stored at −80°C. The titers of HAV stocks were determined by plaque assay (8) and heat release PCRU endpoint dilution.
Food samples used in seeding studies.
Food samples were obtained from the Baylor College of Medicine cafeteria, maintained at 4°C, and used within 24 h. Deli meats, including ham, turkey, and roast beef, were sliced at the cafeteria on the day of sample collection.
Viral seeding of food samples.
Twenty to 40 g of food was placed in sterile 250-ml centrifuge bottles. A total volume of 20 μl of serially diluted virus stocks, corresponding to 10 to 104 PCRU, was pipetted onto the surfaces of individual food samples. Up to six samples were processed per experiment. Each experiment included a food sample that was spiked with sterile water to serve as a negative control during the entire sample processing and viral detection procedures.
Outbreak food samples.
Food items were collected from a university cafeteria implicated in an outbreak of NLV-associated gastroenteritis during the outbreak investigation (N. A. Daniels, D. A. Bergmire-Sweat, K. J. Schwab, K. A. Hendricks, S. Reddy, S. M. Rowe, R. L. Fankhauser, S. S. Monroe, R. L. Atmar, R. I. Glass, and P. Mead, submitted for publication). Samples were shipped to the Baylor College of Medicine and stored at 4°C until processed, as described below.
Sample processing for recovery of viruses from food.
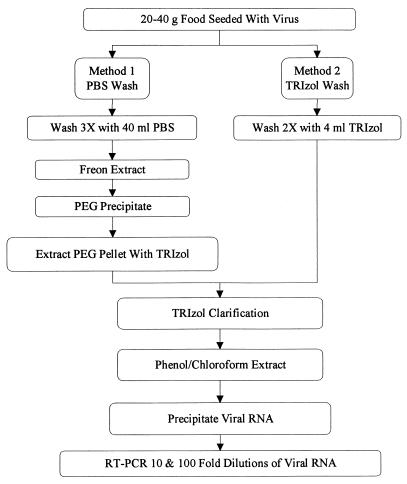
Initially, two methods for recovery of viruses from foodstuffs were examined. Figure 1 is a flow diagram of sample processing by both methods. For the first method (method 1), 40 ml of PBS was added to virus-seeded food samples and mixed for 5 min. The resulting supernatant was collected, and the sample was washed with an additional 40 ml of PBS two additional times. Following each wash, the aqueous solution was decanted into sterile 250-ml centrifuge bottles. The entire wash solution (approximately 120 ml) was mixed with 70 ml of Freon for 5 min and centrifuged at 5,000 × g for 10 min at 4°C, and the aqueous phase was removed and retained. The interface was re-extracted by addition of 30 ml of PBS to the Freon phase, mixing for an additional 5 min, centrifugation at 5,000 × g for 10 min at 4°C, and collection of the aqueous phase. The aqueous phases were then pooled. Virus was precipitated from the aqueous phase by adding polyethylene glycol 6000 (Gallard-Schlesinger Industries, Carle Place, N.Y.) and NaCl to final concentrations of 10% and 0.3 M, respectively, and incubating the mixture for 2 h at 4°C. The samples were centrifuged at 7,000 × g for 30 min at 4°C, the supernatant was discarded, and the pellet was suspended in 8 ml of TRIzol (Gibco BRL, Gaithersburg, Md.) and transferred to Falcon 2059 tubes. The remainder of the method 1 recovery procedure was identical to method 2 following virus recovery from the food, as described below.
FIG. 1.
Flow chart for detection of viruses in foods.
For method 2, approximately 30 g of each food sample was placed in a sterile container and washed with 4 ml of TRIzol for 5 min. The resulting supernatant was collected and the sample was washed with an additional 4 ml of TRIzol for 5 min. The wash retentates were pooled and placed into Falcon 2059 disposable tubes. All subsequent steps were the same for both methods. The TRIzol sample was clarified by centrifugation for 20 min at 8,000 × g and 4°C, and the upper lipid layer and residual food pellet were discarded. The viral RNA-containing aqueous layer was extracted by addition of 1.6 ml of chloroform, mixing for 15 s, incubation for 3 min at room temperature, and centrifugation for 20 min at 8,000 × g and 4°C. The aqueous phase was precipitated in 4 ml of isopropanol by mixing for 30 s, incubation for 10 min at room temperature, centrifugation for 20 min at 8,000 × g and 4°C, and discarding of the supernatant. The resulting RNA pellet was washed with 8 ml of 70% ethanol and centrifuged for 5 min at 7,000 × g and 4°C, and the supernatant was discarded. The RNA pellet was air dried, suspended in 100 μl of RNase-free water, divided into 20-μl portions, and stored at −80°C prior to RT-PCR amplification.
RT-PCR.
RT-PCRs for extracts of seeded samples were performed in 0.2-ml thin-walled tubes with an MJR 200 thermocycler (MJ Research, Watertown, Mass.) with NV-specific primers NVp35 and NVp36 and HAV-specific primers HAVp3 and HAVp4 as described previously (3). NLV genogroup-specific polymerase and capsid primers were used on food extracts from the NLV outbreak investigation (Daniels et al., submitted). Briefly, 20 μl of sample was reverse transcribed in a total volume of 30 μl. The RT mixture contained 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 3.3 μM downstream primer, 667 μM deoxynucleoside triphosphates, 20 U of RNasin (Promega), and 5 U of avian myeloblastosis virus reverse transcriptase (Life Sciences, Inc., St. Petersburg, Fla.). In addition, in seeding studies, the NLV RT mixture contained approximately 50 copies of NV RNA internal standard control (43). The RT reaction mixture was incubated for 1 h at 43°C, heated for 5 min at 95°C, and placed on ice. Seventy microliters of PCR mixture was added to the RT mixture to yield a mixture containing 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, a 1 μM concentration of each primer, 200 μM deoxynucleoside triphosphates, and 5 U of Taq polymerase (Perkin-Elmer, Foster City, Calif.). PCR amplification consisted of an initial 2-min denaturation at 94°C followed by 40 cycles of denaturation at 92°C for 15 s, annealing at 55°C (NVp35-NVp36 and HAVp3-HAVp4) or 50°C (SR33-SR46, Mon381-Mon383, and Mon441-Mon443) for 30 s, and extension for 30 s at 72°C, with a final 5-min extension at 72°C.
The expected product sizes were as follows: 470 bp for NV and 347 bp for the internal standard control with primers NVp35 and NVp36, 248 bp with primers HAVp3 and HAVp4, 123 bp with outbreak investigation polymerase primers SR33 and SR46, 322 bp with capsid primers Mon381 and Mon383, and 268 bp with capsid primers Mon441 and Mon443.
RT-PCR internal standard control.
The NV RNA internal standard control, consisting of NV RNA with a 123-bp deletion, was generated by transcription from an NV plasmid constructed as previously described (43). The internal control transcript was added to NV-specific amplification reactions with primers NVp35 and NVp36. For reactions involving the university outbreak, separate reactions using the internal control transcript and primers NVp35 and NVp36 were set up in order to detect the presence of inhibitors of amplification.
Oligoprobe hybridization and detection.
Virus-specific oligonucleotides NVp69 (29) and HAVp8 (5′ GTGATAGCTCCCACAGGTGC 3′ [negative sense; HAV nucleotides 2346 to 2365]) (7) were used for the detection of NV and HAV, respectively. SR47 (2) and an NLV GII capsid-specific oligonucleotide (35) were used for the outbreak investigation samples. All oligonucleotides were 5′ end labeled with digoxigenin by using terminal transferase according to the manufacturer's instructions (Boehringer Mannheim, Indianapolis, Ind.). Southern blot hybridization assays were performed as described previously (3), with some modifications. Briefly, DNA was transferred to positively charged nylon membranes by vacuum transfer for 2 h and fixed to the membranes by UV cross-linking. The membrane was prehybridized for 30 min at 55°C in prehybridization buffer (Express Hyb; Clontech, Palo Alto, Calif.) and then placed into a hybridization solution (Express Hyb plus a 7 nM concentration of a digoxigenin-labeled probe). After hybridization for 1 h at 55°C, the membrane was washed twice at room temperature for 5 min each in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate and twice at 45°C for 15 min each in 0.5× SSC–0.1% sodium dodecyl sulfate. The hybridized probe was detected by use of the Genius 3 nucleic acid detection kit (Boehringer Mannheim) according to the manufacturer's protocol. Digoxigenin-labeled markers (Boehringer Mannheim) were used for molecular size determination following gel electrophoresis and Southern blot hybridization.
Sequencing of PCR product from outbreak investigation sample.
NLV-specific RT-PCR amplicons from the positive food sample from the outbreak investigation were characterized further by nested PCR amplification of the capsid region followed by direct sequencing of the resulting amplicons. To minimize the risk of carryover contamination, nested PCR amplification was performed in a different wing of the building with entirely different sets of reagents and equipment (enzymes, buffers, tubes, pipettes, and thermal cycler) after all the samples had been processed by the described methods and amplified by standard RT-PCR. Appropriate-sized bands of the nested-PCR product were extracted from agarose gels and purified by Jetsorb purification (Genomed, Research Triangle Park, N.C.). The purified PCR product was sequenced at the Baylor College of Medicine Core Sequencing Facility on an automated sequencer (ABI 373A; Applied Biosystems, Foster City, Calif.).
RESULTS
Comparisons between HAV PFU and PCRU.
To determine the relative detection sensitivities of amplification of viral nucleic acid by RT-PCR and HAV infectivity testing by cell culture, titers of HAV monodispersed virus stocks were determined by PFU cell culture assay (n = 6) and PCRU endpoint dilution (n = 4). Viral titers were estimated to be 2.2 × 107 PFU/ml and 5 × 108 PCRU/ml, respectively (data not shown), giving a ratio of 23 PCRU per PFU.
Recovery of NV seeded onto food by method 1.
Forty-gram samples of ham and turkey seeded with 103 PCRU of NV were processed by method 1. Extractions from both types of meat samples were inhibitory when 20 μl of undiluted RNA was amplified. NV-specific PCR products were detected following amplification of 20-μl 10- and 100-fold dilutions of meat extract (data not shown).
Recovery of NV and HAV seeded onto food by method 2.
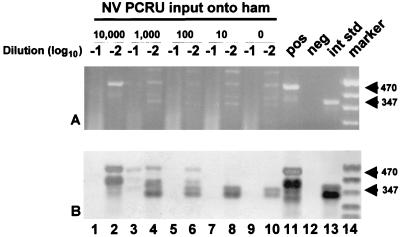
In an effort to streamline the method, a second extraction technique was tested. Ham, turkey, and roast beef (seeded with both NV and HAV at levels decreasing from 104 to 10 PCRU of input virus) were all tested by this method. Recovered RNA was serially diluted 10- and 100-fold in MilliQ RNase-free H2O, and 20 μl of each dilution was amplified by RT-PCR with NV- and HAV-specific primers in separate RT-PCR tubes. A representative test result for ham samples is shown in Fig. 2. NV was detected in hundredfold dilutions of the RNA extracts of ham samples seeded with 104, 103, and 102 PCRU of input virus. Tenfold dilutions of ham samples seeded with 104, 102, 101, and 0 PCRU of virus input samples were negative due to inhibition of amplification, as determined by the absence of both NV-specific (for 104, 102, and 101 PCRU of input virus) and internal standard control-specific amplicons (Fig. 2). Similar levels of detection were obtained for NV and HAV in other meat samples, as shown in Table 1.
FIG. 2.
Detection of NV RNA and internal control RNA by RT-PCR and oligoprobing of 10-fold (−1) and 100-fold (−2) dilutions of RNA extracts from ham samples seeded with decreasing concentrations of NV and processed by method 2. Lanes 9 and 10 are negative sample controls. Other lanes include an NV RT-PCR positive control (pos), a negative reagent control (neg), 50 copies of an internal standard control (int std), and a digoxigenin-labeled molecular size marker (MarkerVIII; Boehringer Mannheim) (marker). (A) Ethidium bromide-stained agarose gel; (B) Southern blot of gel in panel A. Numbers at the right are molecular sizes, in base pairs.
TABLE 1.
Detection of decreasing inputs of NV or HAV seeded onto foodstuffsa
| Foodstuff | No. of samples positive/no. tested at indicated input (PCRU)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 10,000
|
1,000
|
100
|
10
|
|||||
| NV | HAV | NV | HAV | NV | HAV | NV | HAV | |
| Ham | 2/2 | 2/2 | 2/2 | 2/2 | 3/4 | 2/2 | 0/3 | 1/2 |
| Turkey | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 | 1/2 | 2/2 |
| Roast beef | 2/2 | 3/3 | 2/2 | 2/2 | 3/3 | 1/3 | 2/3 | 0/2 |
Twenty to 30 g of food was seeded with decreasing inputs of both NV and HAV. Seeded samples were then processed by method 2, and 10- and 100-fold dilutions of extracted viral RNA were analyzed by RT-PCR and oligoprobing.
The use of the internal standard control in NV RT-PCRs of sample extracts processed by method 2 demonstrated sample inhibition in 62% (ham, 7 of 11; roast beef, 10 of 10, turkey, 1 of 8; total, 18 of 29) of the samples diluted 10-fold prior to amplification. No sample inhibition was seen in any of the samples diluted 100-fold prior to amplification.
Recovery of NLV from food implicated in an outbreak.
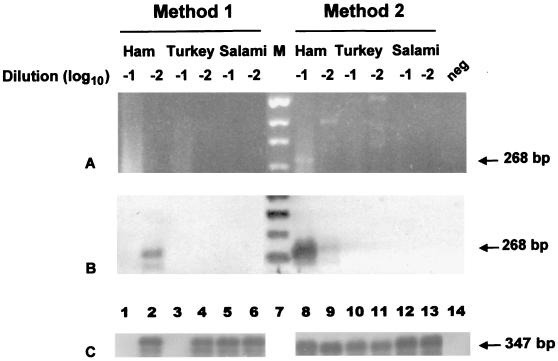
Approximately 20 g each of ham, turkey, and salami obtained during an outbreak of gastroenteritis at a Texas university cafeteria (Daniels et al., submitted) was processed by both of the described methods. NLV-specific products were identified only from the ham samples by using both polymerase- and capsid-specific primers. Figure 3 shows the results of RT-PCR amplification of the capsid region with the primer pair Mon441-Mon443. In a separate reaction with 50 copies of the NV internal standard control, inhibitors were present in 10-fold dilutions of ham and turkey processed by method 1 (Fig. 3C).
FIG. 3.
Detection of NLV RNA and internal control RNA by RT-PCR and oligoprobing of 10-fold (−1) and 100-fold (−2) dilutions of RNA extracts from outbreak food samples processed by both methods. (A) Ethidium bromide-stained agarose gel; (B) Southern blot of gel in panel A. Results were obtained with NLV GII capsid primer pair Mon441-Mon443. (C) Southern blot for each food extract dilution from a separate RT-PCR seeded with 50 copies of NV internal standard RNA, using the primer pair NVp35-NVp36 to identify sample inhibition. Other lanes include a negative reagent control (neg) and a digoxigenin-labeled molecular size marker (M). NLV-specific amplicons (268 bp) are seen in lanes 2, 8, and 9 of panel B. Internal standard control-specific amplicons (347 bp) are seen in lanes 2, 4 to 6, and 8 to 13 of panel C. The internal standard control is absent from lanes 1 and 3 of panel C due to sample inhibition.
Sequencing of NLV PCR products from ham samples.
Virus-specific PCR products from ham samples processed by both methods were sequenced, and the sequences were compared to those obtained from stool samples of ill subjects identified in the outbreak. The capsid sequences from both stool and ham samples were identical over 228 bases. The sequences were determined to be within the NLV GII Lordsdale virus cluster.
DISCUSSION
Outbreaks associated with NLVs are a major health concern worldwide. To date, researchers and epidemiologists have had limited success directly linking foodstuffs associated with NLV outbreaks to the NLV strain identified in stool samples collected from ill individuals during the outbreak investigation. Methods developed to isolate NLV nucleic acid from stools (2, 17, 22, 24, 43) have not been successful for virus detection from environmental samples. Additionally, RT-PCR methods used for detecting NLVs and HAV in bivalve mollusks implicated in outbreaks (16, 27, 28, 30) have not been useful for detection of NLVs in other foods. Gouvea et al. (12) described a method for potential recovery of NV from foods by using guanidinium extraction but did not apply it in outbreak settings. Their method, which includes adsorption of RNA to hydroxyapatite and sequential precipitation with cetyltrimethylammonium bromide and ethanol following guanidinium extraction, is labor-intensive and requires the use of nested PCR for detection of low viral input. Recently, NLV was successfully recovered from well water implicated in an NLV outbreak. The water was filtered and NLVs were eluted from the filter by using basic amino acids as an eluent. The NLVs were then detected by RT-PCR (5).
This is the first study to describe methods for NLV recovery from foods other than bivalve mollusks and the successful application of these methods in an outbreak investigation. We describe two methods for the isolation and purification of NLVs from deli meats (Fig. 1). Method 1 requires washing of the surfaces of the deli meats with PBS, clarification of the wash solution with Freon to remove inhibitory substances (such as lipids), and polyethylene glycol precipitation to further concentrate and purify the sample prior to TRIzol extraction. Method 2 consists of a simpler procedure in which the deli meat is washed directly with the TRIzol solution. Use of both of these methods was successful in recovering NLV from deli ham implicated in a food-borne outbreak of viral gastroenteritis (Fig. 3). Although more time-consuming than method 2, method 1 has the advantage of concentrating and purifying intact viral particles from foodstuffs. Isolation of intact particles could be useful for incorporating the method into an NLV antibody capture system or determining viral infectivity if an NLV cell culture system is ever developed. However, the majority of this research focused on method 2 because it is the simplest and most straightforward of the two methods. By using method 2, an input of as few as 10 PCRU of NV or HAV could be detected and 100 to 1,000 PCRU were consistently detected in every deli meat tested (Table 1). Increasing the volume of TRIzol used to wash the meats as described for method 2 did not improve sample quality. Similar levels of viral recovery and sample inhibition were detected (data not shown).
The RT-PCR assays used in these experiments can detect as few as 10 to 30 RNA transcripts of NV and HAV (3, 43). Based on the observed PCRU/PFU ratio of 23:1 for HAV, a genomic copy (particle)/PFU ratio for HAV was estimated to be 230:1 to 690:1. This is similar to the 200:1 ratio reported for poliovirus (41). These data suggest that the TRIzol wash procedure (method 2) can detect <1 PFU of HAV and consistently detects ∼4 PFU. Although no culture system allowing a calculation of the NV infectious dose exists, the procedure consistently detected 102 to 103 viral genomic copies.
The developed purification and recovery methods are efficient for both HAV and the prototype NLV, NV (a genogroup I virus). During the outbreak investigation, we were able to detect a genogroup II strain of NLV, indicating that the method is efficient for both NLV genogroups. Unfortunately, we did not have a sufficient stock of a genogroup II NLV to use during seeding experiments to directly prove that genogroup II recovery is similar to genogroup I recovery, although we believe this to be the case. Thus, this method appears to have adequate sensitivity for the detection of the most important viruses associated with food-borne outbreaks.
The incorporation of an internal standard RNA control into each RT-PCR tube was important to identify inhibitors and eliminate false negatives (Fig. 2, lanes 1, 5, 7, and 9, and Fig. 3C, lanes 1 and 3). The internal standard contains a 123-nucleotide deletion compared to native NV RNA (43). As a result, the amplification product is easily distinguished from the full-length viral RNA product on an agarose gel. In the constructed internal standard, the sequences complementary to primers NVp35 and NVp110 are adjacent. NVp110 is a degenerate oligonucleotide which can be used in RT-PCR assays to detect a wide range of human caliciviruses (29). Thus, the internal control amplification product is approximately the same size whether generated by NVp35 for NV or by NVp110 for many of the other HuCVs. Finally, the deleted region includes the sequence recognized by probes that recognize NV (probes NVp116 and SR65d) or other related caliciviruses (e.g., probes NVp117, NVp118, and SR47d) (2, 29). Thus, hybridized PCR products from food samples containing products from amplified internal control RNA can be analyzed by size differences following Southern transfer or by using specific probes.
Internal standard controls can also be designed to provide a method for quantitative assessment of the level of virus present in a food sample. Determining the level of viral contamination could be important and useful during outbreak investigations, especially if data are collected over time to give a measure of risk assessment.
Figure 2 shows representative RT-PCR and Southern transfer oligoprobe hybridization results for deli ham. The importance of confirmation of RT-PCR gel results by internal probing is evident from the multiple bands observed in the gel. In fact, the negative control ham sample contains bands very close in size to the positive control (lane 10). Following Southern transfer and oligoprobing, lane 10 does not contain any signal near the size expected for amplified NV (470 bp), clearly indicating that the bands observed in the gel were nonspecific. In a recent study (14) in which the authors assessed the presence of HuCV in shellfish, multiple bands were observed in gels without further confirmation of specificity by probing or sequencing. The results obtained by members of our group in these and prior studies (3, 44) have shown that errors in interpretation will occur when visual interpretation of gels is used alone to analyze results; results must be confirmed by probe hybridization. It is interesting that when amplifying HuCV RNA by RT-PCR, members of our group and others (2, 4, 21, 44) have consistently observed positive bands of different molecular weights following oligoprobing of Southern-transferred HuCV PCR products. The additional bands are present only in virus-positive samples; i.e., they have never been observed in virus-negative samples. Thus, these bands do not change the specificity of the confirmatory test.
NLVs are a major cause of outbreaks of nonbacterial gastroenteritis (11), and many of these outbreaks are caused by transmission via contaminated foods (11). The public health significance of detecting NLVs in food has been addressed only via risk assessment assumptions, with no empirical verification. The lack of available assays for the direct detection of NLVs in food has made it difficult for state and federal agricultural and health agencies to determine which foodstuff is the cause of a food-borne outbreak, and patterns of food-borne transmission of disease cannot be assessed effectively. In addition, regulatory health institutions as well as the public are now requesting more direct information on the potential of viral pathogens being present in the foods that are consumed by the public. The development of a robust, specific, and sensitive method to recover NLVs from food will permit identification of contaminated food and improve our understanding of the modes of food contamination and NLV transmission, thereby contributing useful information to better public health policy and safety.
ACKNOWLEDGMENTS
This work was supported in part by grant NA77FD0080 from the National Oceanic and Atmospheric Administration. K.J.S. is supported by training grant T32 AI07471 from the National Institutes of Health.
We thank Baylor College of Medicine Food Services Manager Terri Chin for supplying pathogen-free deli items for the seeding studies.
REFERENCES
- 1.Ando T, Jin Q, Gentsch J R, Monroe S S, Noel J S, Dowell S F, Cicirello H G, Kohn M A, Glass R I. Epidemiologic applications of novel molecular methods to detect and differentiate small round structured viruses (Norwalk-like viruses) J Med Virol. 1995;47:145–152. doi: 10.1002/jmv.1890470207. [DOI] [PubMed] [Google Scholar]
- 2.Ando T, Monroe S S, Gentsch J R, Jin Q, Lewis D C, Glass R I. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J Clin Microbiol. 1995;33:64–71. doi: 10.1128/jcm.33.1.64-71.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar R L, Neill F H, Romalde J L, Le Guyader F, Woodley C M, Metcalf T G, Estes M K. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl Environ Microbiol. 1995;61:3014–3018. doi: 10.1128/aem.61.8.3014-3018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atmar R L, Neill F H, Woodley C M, Manger R, Fout G S, Burkhardt W, Leja L, McGovern E R, Le Guyader F, Metcalf T G, Estes M K. Collaborative evaluation of a method for the detection of Norwalk virus in shellfish tissues by PCR. Appl Environ Microbiol. 1996;62:254–258. doi: 10.1128/aem.62.1.254-258.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beller M, Ellis A, Lee S H, Drebot M A, Jenkerson S A, Funk E, Sobsey M D, Simmons O D I, Monroe S S, Ando T, Noel J, Petric M, Middaugh J P, Spika J S. Outbreak of viral gastroenteritis due to a contaminated well. International consequences. JAMA. 1997;278:563–568. [PubMed] [Google Scholar]
- 6.Chalmers J W, McMillan J H. An outbreak of viral gastroenteritis associated with adequately prepared oysters. Epidemiol Infect. 1995;115:163–167. doi: 10.1017/s0950268800058222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J I, Ticehurst J R, Purcell R H, Buckler-White A, Baroudy B M. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol. 1987;61:50–59. doi: 10.1128/jvi.61.1.50-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cromeans T, Sobsey M D, Fields H A. Development of a plaque assay for a cytopathic, rapidly replicating isolate of hepatitis A virus. J Med Virol. 1987;22:45–56. doi: 10.1002/jmv.1890220107. [DOI] [PubMed] [Google Scholar]
- 9.De Serres G, Cromeans T L, Levesque B, Brassard N, Barthe C, Dionne M, Prud'homme H, Paradis D, Shapiro C N, Nainan O V, Margolis H S. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J Infect Dis. 1999;179:37–43. doi: 10.1086/314565. [DOI] [PubMed] [Google Scholar]
- 10.Dowell S F, Groves C, Kirkland K B, Cicirello H G, Ando T, Jin Q, Gentsch J R, Monroe S S, Humphrey C D, Slemp C, Dwyer D M, Meriwether R A, Glass R I. A multistate outbreak of oyster-associated gastroenteritis: implications for interstate tracing of contaminated seafood. J Infect Dis. 1995;171:1497–1503. doi: 10.1093/infdis/171.6.1497. [DOI] [PubMed] [Google Scholar]
- 11.Fankhauser R L, Noel J S, Monroe S S, Ando T, Glass R I. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J Infect Dis. 1998;178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- 12.Gouvea V, Santos N, Timenetsky M C, Estes M K. Identification of Norwalk virus in artificially seeded shellfish and selected foods. J Virol Methods. 1994;48:177–187. doi: 10.1016/0166-0934(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 13.Graham D Y, Jiang X, Tanaka T, Opekun A R, Madore H P, Estes M K. Norwalk virus infection of volunteers: new insights based on improved assays. J Infect Dis. 1994;170:34–43. doi: 10.1093/infdis/170.1.34. [DOI] [PubMed] [Google Scholar]
- 14.Green J, Henshilwood K, Gallimore C I, Brown D W, Lees D N. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl Environ Microbiol. 1998;64:858–863. doi: 10.1128/aem.64.3.858-863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, K. Y., T. Ando, M. S. Balayan, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H.-J. Thiel.Caliciviridae. In M. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. Mayo, D. McGeoch, C. R. Pringle, and R. Wickner (ed.), Virus taxonomy: 7th report of the International Committee on Taxonomy of Viruses, in press. Academic Press, Orlando, Fla.
- 16.Hafliger D, Gilgen M, Luthy J, Hubner P. Seminested RT-PCR systems for small round structured viruses and detection of enteric viruses in seafood. Int J Food Microbiol. 1997;37:27–36. doi: 10.1016/s0168-1605(97)00041-x. [DOI] [PubMed] [Google Scholar]
- 17.Hale A D, Green J, Brown D W. Comparison of four RNA extraction methods for the detection of small round structured viruses in faecal specimens. J Virol Methods. 1996;57:195–201. doi: 10.1016/0166-0934(95)01966-9. [DOI] [PubMed] [Google Scholar]
- 18.Hedberg C W, Osterholm M T. Outbreaks of food-borne and waterborne viral gastroenteritis. Clin Microbiol Rev. 1993;6:199–210. doi: 10.1128/cmr.6.3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutin Y J, Pool V, Cramer E H, Nainan O V, Weth J, Williams I T, Goldstein S T, Gensheimer K F, Bell B P, Shapiro C N, Alter M J, Margolis H S. A multistate, foodborne outbreak of hepatitis A. National Hepatitis A Investigation Team. N Engl J Med. 1999;340:595–602. doi: 10.1056/NEJM199902253400802. [DOI] [PubMed] [Google Scholar]
- 20.Iversen A M, Gill M, Bartlett C L, Cubitt W D, McSwiggan D A. Two outbreaks of foodborne gastroenteritis caused by a small round structured virus: evidence of prolonged infectivity in a food handler. Lancet. 1987;ii:556–558. doi: 10.1016/s0140-6736(87)92933-3. [DOI] [PubMed] [Google Scholar]
- 21.Jaykus L A, De Leon R, Sobsey M D. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl Environ Microbiol. 1996;62:2074–2080. doi: 10.1128/aem.62.6.2074-2080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X, Wang J, Graham D Y, Estes M K. Detection of Norwalk virus in stool by polymerase chain reaction. J Clin Microbiol. 1992;30:2529–2534. doi: 10.1128/jcm.30.10.2529-2534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilgore P E, Belay E D, Hamlin D M, Noel J S, Humphrey C D, Gary H E, Jr, Ando T, Monroe S S, Kludt P E, Rosenthal D S, Freeman J, Glass R I. A university outbreak of gastroenteritis due to a small round-structured virus. Application of molecular diagnostics to identify the etiologic agent and patterns of transmission. J Infect Dis. 1996;173:787–793. doi: 10.1093/infdis/173.4.787. [DOI] [PubMed] [Google Scholar]
- 24.Kogawa K, Nakata S, Ukae S, Adachi N, Numata K, Matson D O, Estes M K, Chiba S. Dot blot hybridization with a cDNA probe derived from the human calicivirus Sapporo 1982 strain. Arch Virol. 1996;141:1949–1959. doi: 10.1007/BF01718206. [DOI] [PubMed] [Google Scholar]
- 25.Kohn M A, Farley T A, Ando T, Curtis M, Wilson S A, Jin Q, Monroe S S, Baron R C, McFarland L M, Glass R I. An outbreak of Norwalk virus gastroenteritis associated with eating raw oysters. Implications for maintaining safe oyster beds. JAMA. 1995;273:466–471. doi: 10.1001/jama.1995.03520300040034. [DOI] [PubMed] [Google Scholar]
- 26.Kuritsky J N, Osterholm M T, Greenberg H B, Korlath J A, Godes J R, Hedberg C W, Forfang J C, Kapikian A Z, McCullough J C, White K E. Norwalk gastroenteritis: a community outbreak associated with bakery product consumption. Ann Intern Med. 1984;100:519–521. doi: 10.7326/0003-4819-100-4-519. [DOI] [PubMed] [Google Scholar]
- 27.Lees D N, Henshilwood K, Green J, Gallimore C I, Brown D W G. Detection of small round structured viruses in shellfish by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:4418–4424. doi: 10.1128/aem.61.12.4418-4424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Guyader F, Dubois E, Menard D, Pommepuy M. Detection of hepatitis A virus, rotavirus, and enterovirus in naturally contaminated shellfish and sediment by reverse transcription-seminested PCR. Appl Environ Microbiol. 1994;60:3665–3671. doi: 10.1128/aem.60.10.3665-3671.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Guyader F, Estes M K, Hardy M E, Neill F H, Green J, Brown D, Atmar R L. Evaluation of a degenerate primer for the PCR detection of human caliciviruses. Arch Virol. 1996;141:2225–2235. doi: 10.1007/BF01718228. [DOI] [PubMed] [Google Scholar]
- 30.Le Guyader F, Neill F H, Estes M K, Monroe S S, Ando T, Atmar R L. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl Environ Microbiol. 1996;62:4268–4272. doi: 10.1128/aem.62.11.4268-4272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo S V, Connolly A M, Palmer S R, Wright D, Thomas P D, Joynson D. The role of the pre-symptomatic food handler in a common source outbreak of food-borne SRSV gastroenteritis in a group of hospitals. Epidemiol Infect. 1994;113:513–521. doi: 10.1017/s0950268800068527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonnell S, Kirkland K B, Hlady W G, Aristeguieta C, Hopkins R S, Monroe S S, Glass R I. Failure of cooking to prevent shellfish-associated viral gastroenteritis. Arch Intern Med. 1997;157:111–116. [PubMed] [Google Scholar]
- 33.Metcalf T G, Melnick J L, Estes M K. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu Rev Microbiol. 1995;49:461–487. doi: 10.1146/annurev.mi.49.100195.002333. [DOI] [PubMed] [Google Scholar]
- 34.Morse D L, Guzewich J J, Hanrahan J P, Stricof R, Shayegani M, Deibel R, Grabau J C, Nowak N A, Herrmann J E, Cukor G, et al. Widespread outbreaks of clam- and oyster-associated gastroenteritis. Role of Norwalk virus. N Engl J Med. 1986;314:678–681. doi: 10.1056/NEJM198603133141103. [DOI] [PubMed] [Google Scholar]
- 35.Noel J S, Ando T, Leite J P, Green K Y, Dingle K E, Estes M K, Seto Y, Monroe S S, Glass R I. The correlation of patient immune responses with genetically characterized small round-structured viruses involved in outbreaks of nonbacterial acute gastroenteritis in the United States, 1990 to 1995. J Med Virol. 1997;53:372–383. doi: 10.1002/(sici)1096-9071(199712)53:4<372::aid-jmv10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 36.Parashar U D, Dow L, Fankhauser R L, Humphrey C D, Miller J, Ando T, Williams K S, Eddy C R, Noel J S, Ingram T, Bresee J S, Monroe S S, Glass R I. An outbreak of viral gastroenteritis associated with consumption of sandwiches: implications for the control of transmission by food handlers. Epidemiol Infect. 1998;121:615–621. doi: 10.1017/s0950268898001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson T, Hutchings P, Palmer S. Outbreak of SRSV gastroenteritis at an international conference traced to food handled by a post-symptomatic caterer. Epidemiol Infect. 1993;111:157–162. doi: 10.1017/s0950268800056776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson W, Haswell P, Fryers P T, Green J. Outbreak of small round structured virus gastroenteritis arose after kitchen assistant vomited. Commun Dis Rep CDR Rev. 1997;7:R101–R103. [PubMed] [Google Scholar]
- 39.Pebody R G, Leino T, Ruutu P, Kinnunen L, Davidkin I, Nohynek H, Leinikki P. Foodborne outbreaks of hepatitis A in a low endemic country: an emerging problem? Epidemiol Infect. 1998;120:55–59. doi: 10.1017/s0950268897008340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid J A, Caul E O, White D G, Palmer S R. Role of infected food handler in hotel outbreak of Norwalk-like viral gastroenteritis: implications for control. Lancet. 1988;ii:321–323. doi: 10.1016/s0140-6736(88)92367-7. [DOI] [PubMed] [Google Scholar]
- 41.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 42.Schwab, K. J., M. K. Estes, and R. L. Atmar. Norwalk and other human caliciviruses: molecular characterization, epidemiology and pathogenesis. In J. W. Cary, J. E. Linz, M. A. Stein, and C. Bhatnagar (ed.), Microbial foodborne diseases: mechanisms of pathogenicity and toxin synthesis, in press. Technomic Publishing Company, Inc., Lancaster, Pa.
- 43.Schwab K J, Estes M K, Neill F H, Atmar R L. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J Clin Microbiol. 1997;35:511–514. doi: 10.1128/jcm.35.2.511-514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwab K J, Neill F H, Estes M K, Metcalf T G, Atmar R L. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J Food Prot. 1998;61:1674–1680. doi: 10.4315/0362-028x-61.12.1674. [DOI] [PubMed] [Google Scholar]
- 45.Simmonds R S, Szucs G, Metcalf T G, Melnick J L. Persistently infected cultures as a source of hepatitis A virus. Appl Environ Microbiol. 1985;49:749–755. doi: 10.1128/aem.49.4.749-755.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinje J, Altena S A, Koopmans M. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J Infect Dis. 1997;176:1374–1378. doi: 10.1086/517325. [DOI] [PubMed] [Google Scholar]
- 47.Wanke C A, Guerrant R L. Viral hepatitis and gastroenteritis transmitted by shellfish and water. Infect Dis Clin N Am. 1987;1:649–664. [PubMed] [Google Scholar]
- 48.White K E, Osterholm M T, Mariotti J A, Korlath J A, Lawrence D H, Ristinen T L, Greenberg H B. A foodborne outbreak of Norwalk virus gastroenteritis. Evidence for post-recovery transmission. Am J Epidemiol. 1986;124:120–126. doi: 10.1093/oxfordjournals.aje.a114356. [DOI] [PubMed] [Google Scholar]