Abstract
Induction of cellular senescence or cancer is associated with a reshaping of the nuclear envelope and a broad reorganization of heterochromatin. At the periphery of mammalian nuclei, heterochromatin is stabilized at the nuclear lamina via lamina-associated domains (LADs). Alterations in the composition of the nuclear lamina during senescence lead to a loss of peripheral heterochromatin, repositioning of LADs, and changes in epigenetic states of LADs. Cancer initiation and progression are also accompanied by a massive reprogramming of the epigenome, particularly in domains coinciding with LADs. Here, we review recent knowledge on alterations in chromatin organization and in the epigenome that affect LADs and related genomic domains in senescence and cancer.
Keywords: cancer, EMT, epigenome, heterochromatin, lamin, lamina-associated domain, senescence
1. Introduction
Spatial chromatin organization in the mammalian nucleus is under the influence of extracellular stimuli and is a crucial regulator of gene expression programs. Induction of cellular senescence, a state characterized by an inability to re-enter the cell cycle, and cancer, which requires cell proliferation, interferes with normal cell fate and tissue homeostasis. Senescence and cancer are functionally connected, as senescence is implicated in cancer prevention by causing cell division arrest, and in cancer aggressiveness through the secretion of bioactive compounds [1,2].
Induction of senescence and cancer elicits massive rearrangements in the epigenome and in chromatin architecture. The latter includes a re-wiring of short- and long-range chromosomal interactions and of associations of chromosomes with the nuclear lamina (NL), at the nuclear periphery, via a repositioning of lamina-associated domains (LADs). Cancer cells, in addition, display genomic rearrangements that have consequences on three-dimensional (3D) and radial (center-to-periphery) chromatin organization, which are reinforced by epigenetic alterations of DNA and histones. Genomic and 3D chromatin changes resulting from senescence and cancer initiation have been recently reviewed elsewhere (senescence [3,4,5,6]; cancer [7,8,9]) and are not addressed here.
Increasing evidence indicates that the nuclear periphery is a dynamic compartment and plays a role in the establishment of high-order senescence and cancer chromatin phenotypes. Here, we adopt a nuclear sideline view of chromatin and address current knowledge on chromatin structural changes and epigenetic alterations affecting LADs and related domains in senescence and cancer.
2. Chromatin Organization at the Nuclear Periphery: Consistency and Variation in LADs
The 3D organization of chromatin is able to change in response to external signals during development, differentiation, and tissue homeostasis. This view rests on discoveries of genomic elements and architectural proteins that structurally shape the genome at multiple levels [10,11,12]. At the largest scale, chromosomes are organized in territories broadly divided into lose, euchromatic, and transcriptionally active A compartments, and more compact heterochromatic and largely repressed B compartments enriched in LADs. Within compartments, topological domains of higher chromatin contact frequencies are important for gene expression regulation, and establish long-range contacts with other topological domains, shaping higher-order chromatin architecture [13,14,15].
At the nuclear periphery, chromatin is stabilized by interactions with the nuclear envelope, an outer and inner nuclear membrane system interfacing chromatin via the nuclear lamina (NL). The NL is a meshwork intermediate filaments of A-type lamins (lamin A/C or LMNA/C), splicing products of the LMNA gene, and B-type lamins (lamins B1 and B2), encoded by the LMNB1 and LMNB2 genes [16]. The NL dynamically sequesters signaling molecules, chromatin modifiers, and transcription factors and imposes a spatio-temporal regulation of DNA replication and transcription [17]. Interactions of chromatin with the NL occur through hundreds of LADs unevenly distributed throughout the genome [18,19]. LADs are a conserved feature of genome organization in virtually all cell types; they collectively make up over 30% of the genome and are individually 10 kilobases (kb) to 10 megabases (Mb) in size. LADs are gene poor (<2 genes/Mb, compared to ~8 genes/Mb on average in the human genome) and harbor features of heterochromatin such as di- and tri-methylated histone H3 lysine 9 (H3K9me2, H3K9me3). As a result, LADs constitute a repressive peripheral compartment of the nucleus.
LADs are heterogeneous in their composition, dynamics, and transcriptional status (Figure 1A). Nevertheless, most LADs are well conserved across cell types and during differentiation [20,21,22,23]. Such constitutive LADs (cLADs) display the strongest LMNB1 enrichment, they are AT rich, enriched in long interspersed elements (LINEs), and strongly heterochromatic. Yet, cLADs may also harbor narrow euchromatic areas of lower LMNB1 contact frequency which are detectable by their enrichment in active histone modifications (such as H3K4me1, H3K4me3, and acetylated H3K27 [H3K27ac]) and expressed genes that escape the overall repressive context of the NL [24,25]. Moreover, during TGFβ-induced epithelial-to-mesenchymal transition (EMT), a fraction of LMNB1 has been shown to associate with euchromatic LADs containing active genes pertinent for EMT [26]. These are distinct from the euchromatic areas found in cLADs, but both could be of relevance in senescence and cancer genome architecture.
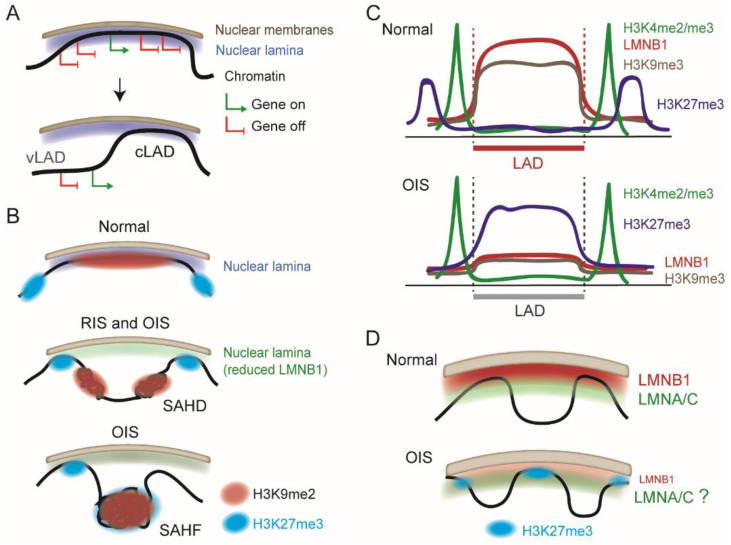
Figure 1.
Lamina-associated domains (LADs) are dynamic features of peripheral chromatin architecture in senescence. (A) Chromatin association with the nuclear lamina via a LAD. LADs mostly harbor repressed genes, although occasional “escaper” regions contain expressed genes. While constitutive LADs (cLADs) are conserved during differentiation and between cell types, variable LADs (vLADs) are gained or lost; vLAD repositioning does not always concur with a change in gene expression. (B) Formation of SAHDs and SAHFs during replication-induced senescence (RIS) and oncogene-induced senescence (OIS). (C) Summary of aggregated profiles of LMNB1 and indicated histone modifications across LADs in normal cells and after OIS. (D) Speculative model of remodeling of nuclear lamina composition in OIS nuclei. Whereas loss of LMNB1 is documented, whether LMNA/C constitutes the main component of the lamina remains to be examined. OIS LADs are rich in H3K27me3 (see also panel B).
Variable (v)LADs arise during differentiation and therefore differ between cell types [19] (Figure 1A). vLADs emerge or disappear as entire domains or as edges of LADs that gain or lose contact with the NL [25], often but not always in line with the activation of cell type-specific genes within them [20,21,25,27,28]. Some vLADs are radially repositioned in the nucleus when losing NL association [29] but this is not always the case [27,30]. vLADs are smaller, more gene dense, and tend to be less heterochromatic than cLADs, with lower H3K9me2/me3 levels. Other subsets of LADs (often vLADs) harboring active histone modifications are bound by a nucleoplasmic pool of LMNA/C in the nuclear interior, where they may play a role in the regulation of chromatin organization, epigenetic marking of regulatory elements, and gene expression [31,32,33].
3. Chromatin Remodeling Elicited by Senescence
Senescence is the result of an irreversible cell proliferation arrest due to exhaustion of cellular replicative potential [2]. Replication-induced senescence (RIS) results from progressively shortening telomeres that are ultimately detected as irreparable DNA damage [3]. Senescence is also a response to various insults such as oxidative stress, DNA damage, chemotherapeutic agents, and stress signals elicited during development, wound healing, and regeneration. Oncogene activation also leads to a variation in the senescence process called oncogene-induced senescence (OIS) [2]. RIS and OIS lead to common characteristics including morphological changes, senescence-associated β-galactosidase accumulation, activation of cell cycle inhibition pathways via upregulation of P16INK4A and P21CIP1, reduced levels of LMNB1, and the senescence-associated secretory phenotype (SASP), a spectrum of damage-associated molecular patterns, bioactive lipids, and secreted pro-inflammatory molecules [34].
Senescent human fibroblasts exhibit broad chromatin re-organization manifested by alterations in 3D chromosomal contacts, DNA hypomethylation, distension and de-repression of centromeric repeats, and epigenetic reactivation of transposable elements that enhances chromatin accessibility [3,35,36]. RIS nuclei also frequently contain compacted chromosome arms, while both RIS and OIS nuclei display increased long-range chromosomal cis-interactions to form senescence-associated heterochromatin domains (SAHDs) rich in H3K9me3, LINEs, and LADs [37] (Figure 1B).
RIS and OIS, however, also differ in the chromatin architectural changes they elicit. A hallmark of OIS, which is rare in RIS, is the formation of senescence-associated heterochromatin foci (SAHFs), large masses of heterochromatin localized away from the NL [38] (Figure 1B). Microscopy and whole-genome chromosome conformation (Hi-C) data show that OIS nuclei display a much larger gain of chromosomal contacts than RIS nuclei, while losing intra-SAHD contacts and gaining long-range inter-SAHD interactions, forming the basis of SAHFs [37]. SAHFs display an atypical heterochromatin architecture, with H3K9me3 and heterochromatin protein 1 alpha HP1α/CBX5 enriched in the center and facultative heterochromatin marked by H3K27me3 localized in the outer layer [38,39,40] (Figure 1B). This geometry results from a spatial rearrangement of H3K9me3 and H3K27me3 domains rather than from epigenomic erasure and re-writing of these marks. The significance of SAHFs in the senescence phenotype in vivo remains unclear, however, as these structures have so far only been observed in cultured cells.
4. Repositioning of LADs during Oncogene-Induced Senescence
The loss of LMNB1 characterizing senescence is probably at the origin of the formation of SAHFs away from the NL [41,42,43,44], effectively repositioning LADs. This LMNB1 loss is not uniform along the NL and preferentially occurs in regions richest in AT content and in H3K9me2/me3 [40,42,45,46]. OIS-induced repositioning of H3K9me2/me3 LADs is not accompanied by significant changes in the expression of protein-coding genes in these domains [40], consistent with the persistence of constitutive heterochromatin after the loss of NL interaction [45]. Of note, whether local areas of gene activity identified in cLADs in normal cycling or differentiated cells [24,25] retain their euchromatic and active states in an OIS context remains to be examined, but since they contain mostly cell type-specific genes [25], they may become repressed within the compact SAHFs, as part of the hijacking of cell fate induced by senescence.
The relationship between loss of NL interaction, SAHF formation, and gene expression changes in senescent cells is complex and may in part depend on the mode of senescence induction, early or late senescence stages, and the nature of genomic elements considered [47]. Repetitive DNA sequences can be transcriptionally deregulated in relation to their repositioning relative to the NL [47]. In OIS nuclei, telomeres preferentially relocalize to the NL [48] where, probably due to reduced lamin levels, they undergo structural rearrangement, loss of chromatin integrity, and dysfunction [49]. Supporting this view, loss of LMNA/C in mouse cells results in loss of telomere maintenance and reactivation of non-coding Terra transcription [50]. Downregulation of LMNB1 in OIS nuclei also leads to detachment of centromeric repeats from the NL, nucleoplasmic relocalization, satellite distension, and activation of these repeats, contributing to genomic instability [51]. These observations are in line with the increased expression level of not only L1 repeats [36,52], but also of a number of long terminal repeat (LTR) and DNA transposons, and several classical satellite and α-satellite repeats in late RIS cells [36,53,54]. Interestingly, SAHF formation may affect gene expression at more distal sites relative to SAHFs themselves. Indeed, protein-coding genes located 500–700 kb away from SAHFs have been found to be significantly downregulated [55]. In contrast, SAHF formation also brings together euchromatic protein-coding loci to enable their activation, including genes involved in cancer and inflammation responses (but not SASP genes) [37]. In fact, the large-scale genomic reorganization characterizing OIS also elicits expression of senescence-associated genes as a result of condensin-mediated transitions from repressive B compartments to active A compartments, and reinforcement of boundaries between these compartments [55].
As AT-rich domains lose LMNB1 interaction during senescence, smaller more GC-rich regions (so-called H-isochores) gain lamin interactions [45]. OIS-induced LADs are H3K27me3-rich [39,40], reflecting a reconfiguration of chromatin states in these domains (Figure 1B,C). In non-senescent cells, LADs are interspersed by H3K27me3 areas often found~50–200 kb away from LAD borders [15,56], or sometimes adjacent to the LAD [57]. Thus, H3K27me3-rich OIS-induced LADs may be positioned by an exchange of H3K9me2/me3 LADs with H3K27me3 domains in cis. One may however not formally exclude the possibility of a de novo H3K27 methylation of domains ending up as LADs in OIS nuclei.
The OIS-induced loss of LMNB1 at the NL expectedly leads to a shift in the ratio of LMNA/C to LMNB1 at the lamina, putatively favoring a proportion of LADs only bound to LMNA/C—a feature reported in hepatocarcinoma cells [30] and in normal human mesenchymal stem cells (our unpublished observations). Whereas LMNB1 preferentially interacts with H3K9me2/me3 domains, LMNA/C has been shown to be intertwined and interact with the polycomb repressor complex 2 containing the H3K27 methyltransferase EZH2 [58,59]. Consequently, a proportion of LMNA/C and H3K27me3 domains are in proximity. So, in line with the idea of an OIS-induced LAD shift, H3K27me3-rich LADs [39,40] could speculatively be predominantly bound to LMNA/C at the NL (Figure 1D). Exploring the fate of LMNA/C LADs during OIS-induced LAD repositioning may provide new mechanistic insights into these radial genome reconfigurations. This view is supported by the causal link between LMNA mutations and progeria, a premature aging syndrome characterized by a loss of heterochromatin at the NL [60,61].
GC content, along with replication timing, has been proposed to predict LAD rearrangement during OIS [37,45]; this is consistent with the higher GC content of H3K27me3 than H3K9me2/me3 domains [62], and with the higher gene density of OIS-induced LADs than bona fide cLADs [48]. Since genes within these H3K27me3-rich LADs overall retain their initial expression level [40], they are likely to be regulated in a lamin-independent manner, or alternatively, the NL of senescent cells loses its repressive influence through mechanisms that remain to be determined.
5. What Do We Know about LADs in Cancer?
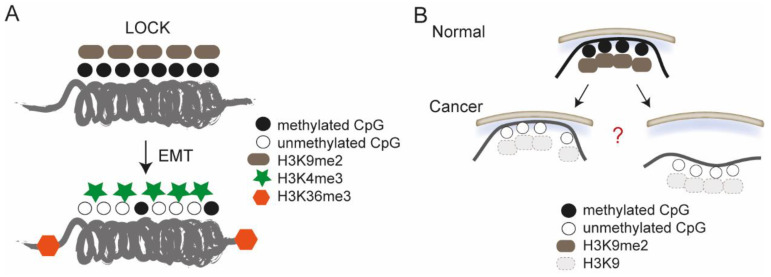
Surprisingly little. Cancer cells are characterized by a reprogramming of their epigenome [63,64] and by spatial chromatin rearrangements exacerbated by variations in DNA content and deformations of the nuclear envelope [65]. These predict impairments in interactions of chromatin with the NL, but LADs have not systematically been examined in tumors. The role of LADs in development and differentiation has however raised the idea that they may also be involved in cancer initiation and progression. Domains the closest to LADs which have been characterized during TGFβ-induced EMT are so-called “large organized chromatin lysine modifications” (LOCKs). LOCKs have been initially identified in embryonic stem cells and in various cell types as domains rich in H3K9me2 and which strikingly overlap with LADs [66]. TGFβ-induced EMT elicits a reduction in H3K9me2 levels in these LOCKs and an increase in euchromatic histone modifications, including H3K4me3 in GC-rich areas along with H3K36me3 in regions flanking LOCKs [67] (Figure 2A). This reprogramming of LOCKs involves the H3K4/K9 demethylase LSD1 and occurs downstream of the remodeling of cell adhesion and cytoskeletal processes elicited by TGFβ [67].
Figure 2.
Epigenetic reprogramming of large organized chromatin K9 modifications (LOCKs)/LADs in cancer cells. (A) Epigenetic alterations targeted to LOCKs upon TGFβ-induced epithelial-to-mesenchymal transition (EMT). (B) LOCKs/LADs, naturally enriched in H3K9me2 and DNA methylated (top), lose their heterochromatic states in cancer (bottom). What is still unclear is whether cancer cell LADs are actually H3K9me2 poor and DNA hypomethylated, and whether LOCKs/LADs losing H3K9me2 detach from the nuclear lamina (bottom right) or are retained at the lamina as euchromatic LADs (bottom left).
A key epigenetic alteration in colorectal tumors is a partial loss of DNA CpG methylation in domains that strikingly coincide with LADs mapped in fibroblasts in a separate study [68] (Figure 2A) (anecdotally, this suggests that LADs from different cell types may be used as a proxy for LADs in tumor cells). In addition, metastatic foci in pancreatic ductal adenocarcinoma display broad reductions in H3K9me2 along with DNA hypomethylation in domains analogous to LOCKs [69], suggesting these effectively take place in LADs. What remains unknown is whether LOCKs/LADs losing H3K9me2 in cancer cells detach from the NL or are retained as LADs that have lost their heterochromatic state (Figure 2B).
LADs emerge as features of genome susceptibility to carcinogens. UV-induced DNA lesions in cultured cells prevail in LADs in fibroblasts and show enrichment at the nuclear periphery in 3D structural genome models [70]. This radial susceptibility mirrors the frequencies of (driver and/or passenger) mutations associated with melanoma, which in majority display the C > T conversion signature of UV-induced DNA lesions [71,72]. Thus, carcinogen susceptibility and mutation frequency go in pair with the association of the genome with the NL [70].
In addition to LADs, heterochromatin is tethered against nucleoli, as nucleolus-associated domains (NADs) [73]. NADs overlap with LADs, suggesting that the nucleolus and the NL are interchangeable scaffolds for heterochromatin. Increases in nucleoli size and number are hallmark features of many tumor types [74] and may enhance the nucleolar surface available for NADs, contributing to the remodeling of high-order chromatin architecture in tumor cells.
6. Re-KODing LOCKs in Tumor Cells?
Nuclear peripheral chromatin does not solely consist of LADs. Inter-LAD domains referred to as “H3K9me2-only domains” (KODs) have been shown in mouse embryonic stem cells to be enriched in cell type-specific enhancers [75]. KODs are also marked by H3K9me3 and are intriguingly also enriched in H3K27me3 (anecdotally questioning their denomination). Along with LOCKs/LADs, these domains could play a role in senescence or cancer chromatin architecture and gene regulation: loss of H3K9me2 in LOCKs [67,68,69] may also affect H3K9me2-only domains and favor transcriptional activity in these regions. Since KODs contain pluripotency genes [75], this would be compatible with the de-differentiation phenotype of tumor cells. Heterochromatin in these domains may also facilitate enhancer interactions with cognate genes in adjacent LADs, contributing to their upregulation, a feature in line with the euchromatinization of LADs/LOCKs in tumor cells. Alternatively, reconfiguration of peripheral chromatin in cancer cell nuclei may guide inter-LAD regulatory elements to tumor-specific LADs and conversely, may reposition normally in-LAD enhancers into inter-LADs.
7. Euchromatic LADs Associated with EMT
Whereas LADs associated with LMNB1 are typically heterochromatic in normal cells, interactions of LMNB1 with euchromatic LADs (eLADs) have been reported in TGFβ-induced EMT of normal mouse mammary epithelial cells [26]. Structurally, EMT is characterized by A/B compartment switches, with eLAD enriched in EMT A compartments. LMNB1 is enriched at the borders of topological domains showing increased border strength (i.e., enhanced stability), as well as at active genes relevant for EMT. Reducing LMNB1 levels alters EMT gene expression and phenotype, suggesting a role for LMNB1 in functionally stabilizing the genome during EMT in this model system [26]. It will be interesting to determine whether LMNB1 localization at active genes also occurs in human cellular models of EMT.
The results of Pascual-Reguant et al. are based on chromatin preparations enriched in euchromatic fragments, which favor enrichment of immunoprecipitated protein (including lamins) in these regions [31]. So, their findings do not exclude a co-existence of heterochromatin LMNB1 LADs, although EMT-associated A/B re-compartmentalization may reposition a fraction of these LADs into euchromatin. Bioinformatically, detection of lamin enrichment at genes vs. domains (LADs) is influenced by the domain or (narrow) peak calling algorithm used [76]. Applying both types of algorithms may unveil new patterns of lamin-genome interactions and provide additional insights into the regulation of transcriptional programs in normal, senescence, and cancer states.
8. Concluding Remarks
Observations highlighted in this review open the door to further investigations of LAD and inter-LAD peripheral chromatin dynamics, and of emerging roles of A- and B-type lamins in cancer initiation and progression [77,78,79,80,81]. Various tumors experience different stiffness constraints in vivo, which will expectedly modulate the ratio of A- to B-type lamins in the NL [82,83]. This is also probably exacerbated by the lower LMNB1 levels in tumors and may be affected by tumor type or stage, or tumor zone, between which lamin levels can vary [84,85]. A- and B-type lamins may also play distinct roles in cancer due to differences in their binding partners [16], the distinct polymers they form and their distinct localizations in the NL [86], the LADs they associate with [30], and lastly, the LMNA/C-specific nucleoplasmic soluble pool [31,87]. Altered LMNA/C levels in cancer may differentially affect these sub-nuclear pools and confer phenotypes not mediated by changes in levels of B-type lamins in the NL. A role for LMNA/C in cancer is receiving increasing attention. Evidence implicates LMNA/C in preserving nuclear integrity in ovarian epithelial cancer subtypes [88] and non-small-cell lung carcinoma cells [79]. Accordingly, lower LMNA/C levels in invasive breast cancer cells confer increased nuclear deformations enabling enhanced cell invasiveness [89]. Recent implications of LMNA/C in the p53/p16INK4A senescence pathway [80] suggest an interplay between chromatin and signaling functions modulated by A-type lamins which will be worth investigating more closely.
It will be informative to characterize LAD dynamics and composition in models of cancer progression or in cancer cells that resist chemotherapeutic agents, as well as the impact of senescence-inducing cancer therapies on LADs. A challenge in investigating lamin–genome interactions in cancer, however, is the expected large variability between tumor type, stage, site, and composition, and between cells within a specific tumor zone. Genomic rearrangements may further enhance LAD variability, which may be greater within a tumor than between different normal cell types. Tumors also harbor several cell types that unless discriminated will contaminate (epi)genomic analyses of the actual cancer cells. Interpretable models of cancer progression of known genome content will be beneficial to mechanically investigate the dynamics of chromatin rearrangements at the nuclear periphery.
Future investigations combining structural and fluorescence imaging [86,90], with rapidly evolving genomics approaches pertaining to single-cell cancer genomics [91], will increase our understanding of how the nuclear envelope and chromatin architecture contribute to regulating cell fate in health and disease. Deep learning methods aimed to recognize nuclear morphologies will in addition enhance cancer diagnosis accuracy [92]. Many genetic, epigenomic, and structural factors influence genome reorganization in cancer cells. Being able to distinguish the contribution of LADs to cancer cell phenotypes amidst these factors remains a major challenge.
Author Contributions
A.B., J.M.-Ø., N.M.G. and P.C. discussed the content of the paper; A.B. and P.C. wrote the manuscript and made figures, and all authors edited the manuscript before submission. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Our work is funded by the Norwegian Cancer Society, South-East Health Norway, the Research Council of Norway and the University of Oslo.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lecot P., Alimirah F., Desprez P.Y., Campisi J., Wiley C. Context-dependent effects of cellular senescence in cancer development. Br. J. Cancer. 2016;114:1180–1184. doi: 10.1038/bjc.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J., Liu M., Hong D., Zeng M., Zhang X. The Paradoxical Role of Cellular Senescence in Cancer. Front. Cell Dev. Biol. 2021;9:722205. doi: 10.3389/fcell.2021.722205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Criscione S.W., Teo Y.V., Neretti N. The Chromatin Landscape of Cellular Senescence. Trends Genet. 2016;32:751–761. doi: 10.1016/j.tig.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crouch J., Shvedova M., Thanapaul R., Botchkarev V., Roh D. Epigenetic Regulation of Cellular Senescence. Cells. 2022;11:672. doi: 10.3390/cells11040672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao X., Wang C., Zhang R. Chromatin basis of the senescence-associated secretory phenotype. Trends Cell Biol. 2022;32:513–526. doi: 10.1016/j.tcb.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangelinck A., Mann C. DNA methylation and histone variants in aging and cancer. Int. Rev. Cell Mol. Biol. 2021;364:1–110. doi: 10.1016/bs.ircmb.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Deng S., Feng Y., Pauklin S. 3D chromatin architecture and transcription regulation in cancer. J. Hematol. Oncol. 2022;15:49. doi: 10.1186/s13045-022-01271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grillo G., Lupien M. Cancer-associated chromatin variants uncover the oncogenic role of transposable elements. Curr. Opin. Genet. Dev. 2022;74:101911. doi: 10.1016/j.gde.2022.101911. [DOI] [PubMed] [Google Scholar]
- 9.Quiroga I.Y., Ahn J.H., Wang G.G., Phanstiel D. Oncogenic fusion proteins and their role in three-dimensional chromatin structure, phase separation, and cancer. Curr. Opin. Genet. Dev. 2022;74:101901. doi: 10.1016/j.gde.2022.101901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowley M.J., Corces V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marti-Renom M.A., Almouzni G., Bickmore W.A., Bystricky K., Cavalli G., Fraser P., Gasser S.M., Giorgetti L., Heard E., Nicodemi M., et al. Challenges and guidelines toward 4D nucleome data and model standards. Nat. Genet. 2018;50:1352–1358. doi: 10.1038/s41588-018-0236-3. [DOI] [PubMed] [Google Scholar]
- 12.Mirny L.A., Imakaev M., Abdennur N. Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 2019;58:142–152. doi: 10.1016/j.ceb.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niskanen H., Tuszynska I., Zaborowski R., Heinaniemi M., Yla-Herttuala S., Wilczynski B., Kaikkonen M.U. Endothelial cell differentiation is encompassed by changes in long range interactions between inactive chromatin regions. Nucleic Acids Res. 2018;46:1724–1740. doi: 10.1093/nar/gkx1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinodoz S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., Lai M.M., Shishkin A.A., Bhat P., Takei Y., et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 2018;174:744–757. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsen J., Liyakat Ali T.M., Nekrasov M., Delbarre E., Baudement M.O., Kurscheid S., Tremethick D., Collas P. Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat. Genet. 2019;51:835–843. doi: 10.1038/s41588-019-0392-0. [DOI] [PubMed] [Google Scholar]
- 16.Burke B., Stewart C.L. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 2013;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- 17.Buchwalter A., Kaneshiro J.M., Hetzer M.W. Coaching from the sidelines: The nuclear periphery in genome regulation. Nat. Rev. Genet. 2019;20:39–50. doi: 10.1038/s41576-018-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Steensel B., Belmont A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell. 2017;169:780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briand N., Collas P. Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020;21:85. doi: 10.1186/s13059-020-02003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meuleman W., Peric-Hupkes D., Kind J., Beaudry J.B., Pagie L., Kellis M., Reinders M., Wessels L., van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keough K.C., Shah P.P., Wickramasinghe N.M., Dundes C.E., Chen A., Salomon R.E.A., Whalen S., Loh K.M., Dubois N., Pollard K.S., et al. An atlas of lamina-associated chromatin across thirteen human cell types reveals cell-type-specific and multiple subtypes of peripheral heterochromatin. bioRxiv. 2020 doi: 10.1101/2020.07.23.218768. [DOI] [Google Scholar]
- 22.Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W., Solovei I., Brugman W., Graf S., Flicek P., Kerkhoven R.M., van Lohuizen M., et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rønningen T., Shah A., Oldenburg A.R., Vekterud K., Delbarre E., Moskaug J.O., Collas P. Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 2015;25:1825–1835. doi: 10.1101/gr.193748.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leemans C., van der Zwalm M.C.H., Brueckner L., Comoglio F., van Schaik T., Pagie L., van Arensbergen J., van Steensel B. Promoter-Intrinsic and Local Chromatin Features Determine Gene Repression in LADs. Cell. 2019;177:852–864. doi: 10.1016/j.cell.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madsen-Østerbye J., Abdelhalim M., Baudement M.O., Collas P. Local euchromatin enrichment in lamina-associated domains anticipates their re-positioning in the adipogenic lineage. Genome Biol. 2022;23:1–19. doi: 10.1186/s13059-022-02662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pascual-Reguant L., Blanco E., Galan S., Le Dily F., Cuartero Y., Serra-Bardenys G., Di Carlo V., Iturbide A., Cebria-Costa J.P., Nonell L., et al. Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun. 2018;9:3420. doi: 10.1038/s41467-018-05912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czapiewski R., Batrakou D.G., de las Heras J., Carter R.N., Sivakumar A., Sliwinska M., Dixon C.R., Webb S., Lattanzi G., Morton N.M., et al. Genomic loci mispositioning in Tmem120a knockout mice yields latent lipodystrophy. Nat. Commun. 2022;13:321. doi: 10.1038/s41467-021-27869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robson M.I., de Las Heras J.I., Czapiewski R., Le Thanh P., Booth D.G., Kelly D.A., Webb S., Kerr A.R.W., Schirmer E.C. Tissue-Specific Gene Repositioning by Muscle Nuclear Membrane Proteins Enhances Repression of Critical Developmental Genes during Myogenesis. Mol. Cell. 2016;62:834–847. doi: 10.1016/j.molcel.2016.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy K.L., Zullo J.M., Bertolino E., Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 30.Forsberg F., Brunet A., Ali T.M.L., Collas P. Interplay of lamin A and lamin B LADs on the radial positioning of chromatin. Nucleus. 2019;10:7–20. doi: 10.1080/19491034.2019.1570810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gesson K., Rescheneder P., Skoruppa M.P., von Haeseler A., Dechat T., Foisner R. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 2016;26:462–473. doi: 10.1101/gr.196220.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldenburg A., Briand N., Sorensen A.L., Cahyani I., Shah A., Moskaug J.O., Collas P. A lipodystrophy-causing lamin A mutant alters conformation and epigenetic regulation of the anti-adipogenic MIR335 locus. J. Cell Biol. 2017;216:2731–2743. doi: 10.1083/jcb.201701043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikegami K., Secchia S., Almakki O., Lieb J.D., Moskowitz I.P. Phosphorylated Lamin A/C in the Nuclear Interior Binds Active Enhancers Associated with Abnormal Transcription in Progeria. Dev. Cell. 2020;52:699–713. doi: 10.1016/j.devcel.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiley C.D., Campisi J. The metabolic roots of senescence: Mechanisms and opportunities for intervention. Nat. Metab. 2021;3:1290–1301. doi: 10.1038/s42255-021-00483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Cecco M., Criscione S.W., Peterson A.L., Neretti N., Sedivy J.M., Kreiling J.A. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. 2013;5:867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Cecco M., Criscione S.W., Peckham E.J., Hillenmeyer S., Hamm E.A., Manivannan J., Peterson A.L., Kreiling J.A., Neretti N., Sedivy J.M. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12:247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sati S., Bonev B., Szabo Q., Jost D., Bensadoun P., Serra F., Loubiere V., Papadopoulos G.L., Rivera-Mulia J.C., Fritsch L., et al. 4D Genome Rewiring during Oncogene-Induced and Replicative Senescence. Mol. Cell. 2020;78:522–538. doi: 10.1016/j.molcel.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narita M., Nunez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 39.Chandra T., Kirschner K., Thuret J.Y., Pope B.D., Ryba T., Newman S., Ahmed K., Samarajiwa S.A., Salama R., Carroll T., et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol. Cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadaie M., Salama R., Carroll T., Tomimatsu K., Chandra T., Young A.R., Narita M., Perez-Mancera P.A., Bennett D.C., Chong H., et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 2013;27:1800–1808. doi: 10.1101/gad.217281.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimi T., Butin-Israeli V., Adam S.A., Hamanaka R.B., Goldman A.E., Lucas C.A., Shumaker D.K., Kosak S.T., Chandel N.S., Goldman R.D. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25:2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah P.P., Donahue G., Otte G.L., Capell B.C., Nelson D.M., Cao K., Aggarwala V., Cruickshanks H.A., Rai T.S., McBryan T., et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dreesen O., Chojnowski A., Ong P.F., Zhao T.Y., Common J.E., Lunny D., Lane E.B., Lee S.J., Vardy L.A., Stewart C.L., et al. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J. Cell Biol. 2013;200:605–617. doi: 10.1083/jcb.201206121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freund A., Laberge R.M., Demaria M., Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23:2066–2075. doi: 10.1091/mbc.e11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandra T., Ewels P.A., Schoenfelder S., Furlan-Magaril M., Wingett S.W., Kirschner K., Thuret J.Y., Andrews S., Fraser P., Reik W. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015;10:471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiang M., Michieletto D., Brackley C.A., Rattanavirotkul N., Mohammed H., Marenduzzo D., Chandra T. Polymer Modeling Predicts Chromosome Reorganization in Senescence. Cell Rep. 2019;28:3212–3223. doi: 10.1016/j.celrep.2019.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rocha A., Dalgarno A., Neretti N. The functional impact of nuclear reorganization in cellular senescence. Brief Funct. Genom. 2022;21:24–34. doi: 10.1093/bfgp/elab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenain C., de Graaf C.A., Pagie L., Visser N.L., de Haas M., de Vries S.S., Peric-Hupkes D., van Steensel B., Peeper D.S. Massive reshaping of genome-nuclear lamina interactions during oncogene-induced senescence. Genome Res. 2017;27:1634–1644. doi: 10.1101/gr.225763.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood A.M., Rendtlew Danielsen J.M., Lucas C.A., Rice E.L., Scalzo D., Shimi T., Goldman R.D., Smith E.D., Le Beau M.M., Kosak S.T. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 2014;5:5467. doi: 10.1038/ncomms6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez-Suarez I., Redwood A.B., Perkins S.M., Vermolen B., Lichtensztejin D., Grotsky D.A., Morgado-Palacin L., Gapud E.J., Sleckman B.P., Sullivan T., et al. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009;28:2414–2427. doi: 10.1038/emboj.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lukasova E., Kovarik A., Kozubek S. Consequences of Lamin B1 and Lamin B Receptor Downregulation in Senescence. Cells. 2018;7:11. doi: 10.3390/cells7020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., Caligiana A., Brocculi G., Adney E.M., Boeke J.D., et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;566:73–78. doi: 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Criscione S.W., De Cecco M., Siranosian B., Zhang Y., Kreiling J.A., Sedivy J.M., Neretti N. Reorganization of chromosome architecture in replicative cellular senescence. Sci. Adv. 2016;2:e1500882. doi: 10.1126/sciadv.1500882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.KarakUlah G., Yandim C. Signature changes in the expressions of protein-coding genes, lncRNAs, and repeat elements in early and late cellular senescence. Turk J. Biol. 2020;44:356–370. doi: 10.3906/biy-2005-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwasaki O., Tanizawa H., Kim K.D., Kossenkov A., Nacarelli T., Tashiro S., Majumdar S., Showe L.C., Zhang R., Noma K.I. Involvement of condensin in cellular senescence through gene regulation and compartmental reorganization. Nat. Commun. 2019;10:5688. doi: 10.1038/s41467-019-13604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harr J.C., Luperchio T.R., Wong X., Cohen E., Wheelan S.J., Reddy K.L. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S., Luperchio T.R., Wong X., Doan E.B., Byrd A.T., Roy Choudhury K., Reddy K.L., Krangel M.S. A Lamina-Associated Domain Border Governs Nuclear Lamina Interactions, Transcription, and Recombination of the Tcrb Locus. Cell Rep. 2018;25:1729–1740. doi: 10.1016/j.celrep.2018.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cesarini E., Mozzetta C., Marullo F., Gregoretti F., Gargiulo A., Columbaro M., Cortesi A., Antonelli L., Di Pelino S., Squarzoni S., et al. Lamin A/C sustains PcG protein architecture, maintaining transcriptional repression at target genes. J. Cell Biol. 2015;211:533–551. doi: 10.1083/jcb.201504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marullo F., Cesarini E., Antonelli L., Gregoretti F., Oliva G., Lanzuolo C. Nucleoplasmic Lamin A/C and Polycomb group of proteins: An evolutionarily conserved interplay. Nucleus. 2016;7:103–111. doi: 10.1080/19491034.2016.1157675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohler F., Bormann F., Raddatz G., Gutekunst J., Corless S., Musch T., Lonsdorf A.S., Erhardt S., Lyko F., Rodriguez-Paredes M. Epigenetic deregulation of lamina-associated domains in Hutchinson-Gilford progeria syndrome. Genome Med. 2020;12:46. doi: 10.1186/s13073-020-00749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Worman H.J., Schirmer E.C. Nuclear membrane diversity: Underlying tissue-specific pathologies in disease? Curr. Opin. Cell Biol. 2015;34:101–112. doi: 10.1016/j.ceb.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zullo J.M., Demarco I.A., Pique-Regi R., Gaffney D.J., Epstein C.B., Spooner C.J., Luperchio T.R., Bernstein B.E., Pritchard J.K., Reddy K.L., et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 63.Timp W., Feinberg A.P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madakashira B.P., Sadler K.C. DNA Methylation, Nuclear Organization, and Cancer. Front. Genet. 2017;8:76. doi: 10.3389/fgene.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddy K.L., Feinberg A.P. Higher order chromatin organization in cancer. Semin. Cancer Biol. 2013;23:109–115. doi: 10.1016/j.semcancer.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wen B., Wu H., Shinkai Y., Irizarry R.A., Feinberg A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald O.G., Wu H., Timp W., Doi A., Feinberg A.P. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat. Struct. Mol. Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berman B.P., Weisenberger D.J., Aman J.F., Hinoue T., Ramjan Z., Liu Y., Noushmehr H., Lange C.P., van Dijk C.M., Tollenaar R.A., et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2011;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDonald O.G., Li X., Saunders T., Tryggvadottir R., Mentch S.J., Warmoes M.O., Word A.E., Carrer A., Salz T.H., Natsume S., et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 2017;49:367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Nieto P.E., Schwartz E.K., King D.A., Paulsen J., Collas P., Herrera R.E., Morrison A.J. Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 2017;36:2829–2843. doi: 10.15252/embj.201796717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.P., Nickerson E., Auclair D., Li L., Place C., et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bersaglieri C., Santoro R. Genome Organization in and around the Nucleolus. Cells. 2019;8:579. doi: 10.3390/cells8060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orsolic I., Jurada D., Pullen N., Oren M., Eliopoulos A.G., Volarevic S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin. Cancer Biol. 2016;37:36–50. doi: 10.1016/j.semcancer.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Smith C.L., Poleshko A., Epstein J.A. The nuclear periphery is a scaffold for tissue-specific enhancers. Nucleic Acids Res. 2021;49:6181–6195. doi: 10.1093/nar/gkab392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lund E.G., Oldenburg A.R., Collas P. Enriched Domain Detector: A program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res. 2014;42:e92. doi: 10.1093/nar/gku324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Irianto J., Pfeifer C.R., Ivanovska I.L., Swift J., Discher D.E. Nuclear lamins in cancer. Cell Mol Bioeng. 2016;9:258–267. doi: 10.1007/s12195-016-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jia Y., Vong J.S., Asafova A., Garvalov B.K., Caputo L., Cordero J., Singh A., Boettger T., Gunther S., Fink L., et al. Lamin B1 loss promotes lung cancer development and metastasis by epigenetic derepression of RET. J. Exp. Med. 2019;216:1377–1395. doi: 10.1084/jem.20181394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Wang J., Huang W., Cai J., Ba J., Wang Y., Ke Q., Huang Y., Liu X., Qiu Y., et al. Nuclear Nestin deficiency drives tumor senescence via lamin A/C-dependent nuclear deformation. Nat. Commun. 2018;9:3613. doi: 10.1038/s41467-018-05808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon M.H., Kang S.M., Lee S.J., Woo T.G., Oh A.Y., Park S., Ha N.C., Park B.J. p53 induces senescence through Lamin A/C stabilization-mediated nuclear deformation. Cell Death Dis. 2019;10:107. doi: 10.1038/s41419-019-1378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang J., Ao Y., Zhang Z., Mo Y., Peng L., Jiang Y., Wang Z., Liu B. Lamin A safeguards the m(6) A methylase METTL14 nuclear speckle reservoir to prevent cellular senescence. Aging Cell. 2020;19:e13215. doi: 10.1111/acel.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swift J., Discher D.E. The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci. 2014;127:3005–3015. doi: 10.1242/jcs.149203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.W., Tewari M., et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hutchison C.J., Worman H.J. A-type lamins: Guardians of the soma? Nat. Cell Biol. 2004;6:1062–1067. doi: 10.1038/ncb1104-1062. [DOI] [PubMed] [Google Scholar]
- 85.Willis N.D., Cox T.R., Rahman-Casans S.F., Smits K., Przyborski S.A., van den Brandt P., van E.M., Weijenberg M., Wilson R.G., de B.A., et al. Lamin A/C is a risk biomarker in colorectal cancer. PLoS ONE. 2008;20:e2988. doi: 10.1371/journal.pone.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turgay Y., Eibauer M., Goldman A.E., Shimi T., Khayat M., Ben-Harush K., Dubrovsky-Gaupp A., Sapra K.T., Goldman R.D., Medalia O. The molecular architecture of lamins in somatic cells. Nature. 2017;543:261–264. doi: 10.1038/nature21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Naetar N., Ferraioli S., Foisner R. Lamins in the nuclear interior-life outside the lamina. J. Cell Sci. 2017;130:2087–2096. doi: 10.1242/jcs.203430. [DOI] [PubMed] [Google Scholar]
- 88.Watabe S., Kobayashi S., Hatori M., Nishijima Y., Inoue N., Ikota H., Iwase A., Yokoo H., Saio M. Role of Lamin A and emerin in maintaining nuclear morphology in different subtypes of ovarian epithelial cancer. Oncol. Lett. 2022;23:9. doi: 10.3892/ol.2021.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bell E.S., Shah P., Zuela-Sopilniak N., Kim D., McGregor A.L., Isermann P., Davidson P.M., Elacqua J.J., Lakins J.N., Vahdat L., et al. Low lamin A levels enhance confined cell migration and metastatic capacity in breast cancer. bioRxiv. 2021 doi: 10.1101/2021.07.12.451842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kittisopikul M., Shimi T., Tatli M., Tran J.R., Zheng Y., Medalia O., Jaqaman K., Adam S.A., Goldman R.D. Computational analyses reveal spatial relationships between nuclear pore complexes and specific lamins. J. Cell Biol. 2021;220:e202007082. doi: 10.1083/jcb.202007082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gohil S.H., Iorgulescu J.B., Braun D.A., Keskin D.B., Livak K.J. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021;18:244–256. doi: 10.1038/s41571-020-00449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sengupta D., Ali S.N., Bhattacharya A., Mustafi J., Mukhopadhyay A., Sengupta K. A deep hybrid learning pipeline for accurate diagnosis of ovarian cancer based on nuclear morphology. PLoS ONE. 2022;17:e0261181. doi: 10.1371/journal.pone.0261181. [DOI] [PMC free article] [PubMed] [Google Scholar]