Abstract
This is the first description, to our knowledge, of the distribution of human polyomavirus and simian virus 40 (SV40) in urban sewage. Using a nested-PCR procedure, we report the detection of human polyomaviruses JC virus (JCV) and BK virus (BKV) but not SV40 in a high percentage of urban sewage samples obtained from widely divergent geographical areas in Europe and Africa. For a total of 28 samples analyzed, JCV was detected in 26, BKV was detected in 22, and none was positive for SV40. All geographical areas showed a high prevalence of these viruses with mean estimated values of JC viral particles per ml on the order of 103 in Barcelona (Spain) and Nancy (France) and 102 in Pretoria (South Africa) and Umeå (Sweden) and mean values of BK viral particles on the order of 102 in Pretoria and Barcelona and 101 in Nancy and Umeå. This compares with estimated mean values of 102 to 103 for human adenovirus that was evaluated as a control. Nucleotide sequence analysis of the amplified DNA from some of the samples is also presented and represents the sequence of the most abundant JC and BK viral strains in these samples. The nucleotide sequence of the JCV detected was also analyzed in a phylogenetic study and for genomic characterization in the regulatory region. This study has shown that human polyomaviruses are spread in high concentrations in the sewage of different geographical areas and are present in contaminated environments. The frequency and concentration of JCV detected in the environment and the absence of described animal hosts suggest that JCV may be useful as a marker for fecal pollution of anthropogenic origin. The results also support the idea previously described that the strains of JCV are closely related to the ethnic origin of the population studied. The procedure applied should also be useful in future studies of population patterns of viral excretion and as a tool in epidemiological studies for the detection of changes in the prevalence of specific viral pathogens.
JC virus (JCV) and BK virus (BKV) are human viruses classified in the genus Polyomavirus of the family Papovaviridae. JCV is etiologically associated with a fatal demyelinating disease known as progressive multifocal leukoencephalopathy (PML) which has emerged as a frequent complication of AIDS in human immunodeficiency virus-infected individuals (9). Infection with BKV has been associated with diseases of the urinary tract including hemorrhagic cystitis and ureteral stenosis (8). Most of the primary infections with JCV and BKV occur early in childhood and are asymptomatic. In this regard, recent data suggest that some BKV infections may be transplacental (30). Human infections with JCV appear to be population associated in that the genotype of JCV excreted by individuals of defined ethnicities is in high proportion determined by the geographical origin of the ethnic group rather than the JCV genotypes that are prevalent in their current location (5). The latent infections established by these viruses persist indefinitely in infected individuals (37).
Simian virus 40 (SV40) is a simian virus that is closely related to JCV and BKV. This polyomavirus establishes latent infections in nonhuman primates and has been associated with a PML-like condition in simian immunodeficiency virus (SIV)-infected monkeys (19). SV40 has been transmitted to humans experimentally, and evidence that SV40 may be circulating in the human population is accumulating (13). Large numbers of humans were exposed to SV40 when inoculated with the polio vaccine prepared in rhesus monkey cells between 1955 and 1961. Several recent studies have described the detection of SV40-like DNA in tumors from children and young adults who were born after SV40 was removed from the polio vaccine (11, 14, 15, 25, 26, 38). These tumors include choroid plexus tumors, ependymomas, and osteosarcomas. SV40-like DNA sequences have also been detected in mesotheliomas in adults who could have been exposed to SV40-contaminated polio vaccines (14).
JCV, BKV, and SV40 are nonenveloped virions containing double-stranded, closed-circular DNA genomes of approximately 5 kb. The genomes of these viruses have a common organizational structure and are homologous over about 75% of their respective nucleotides (37). Each of these viruses has been found to be oncogenic when injected into rodents, and each has the capacity to transform rodent and human cells in tissue culture (37). Their oncogenic capacity, their persistently latent state in infected tissues, and their association with some types of tumors make these viruses potential human tumor viruses (37).
To provide an independent means of assessing the population association of JCV and the regional prevalence of JCV, BKV, and SV40 or any of their genotypes, an analysis of the relative viral output of an entire community or locality could be useful. Since JCV and BKV are excreted in the urine and SV40 has been found in the feces of infected primates and humans, it seemed reasonable to assume that if a significant percentage of the human population were shedding these viruses in their excreta, then all three should be present and possibly detectable in urban sewage. In addition to the potential use of polyomavirus contamination as a marker of human waste in water sources and other environmental locations, being able to detect and study these viruses in sewage would provide a unique opportunity to evaluate and monitor over time those strains that are prevalent in specific geographical areas. Furthermore, the detection of SV40 in sewage would be reasonable evidence that SV40 is circulating in the human population.
In a previous study (32–34), we have developed nested-PCR procedures for the amplification of viral nucleic acids to detect human adenovirus (human Ad), enteroviruses, and hepatitis A and E viruses in sewage, environmental samples, and shellfish. The aim of this study was to apply the experience gained from our previous work to initiate an assessment of the presence of JCV, BKV, and SV40 in sewage of different geographical areas. For this study, we have adapted these methodologies to look for JCV, BKV, and SV40 in urban sewage. Using our previous data (33) on the prevalence of human Ad in sewage as a point of reference, we have detected JCV and BKV but not SV40 in urban sewage samples from widely divergent geographical locations and have developed sequencing data for the most prevalent strains of JCV and BKV that are present in these samples.
MATERIALS AND METHODS
Viruses.
SV40 DNA strain 776 (Gibco BRL) and viral particles of SV40 strain WT 800, kindly donated by Ferran Azorin from the Institute Juan de la Cierva, Consejo Superior de Investigaciones Científicas, Barcelona, Spain, were used in this study as positive controls. A urine sample from a healthy 38-week-pregnant woman was tested for the presence of JCV and BKV and found to be positive for both viruses. The viruses present in 12 ml of urine were concentrated by ultracentrifugation, suspended in 100 μl of phosphate-buffered saline (PBS), and used as a positive control. Clinical samples of cerebrospinal fluid (CSF) from PML patients were kindly donated by José Luis Pérez from the Microbiology Department of the Hospital de Bellvitge, Barcelona, were tested for the presence of JCV by nested PCR, and were used as a positive JCV control and for comparative sequence analysis between viral strains of clinical and environmental origin. Ad type 2 (Ad2) (prototype) was grown on A549 cells, and then viruses were partially purified and stored at −80°C.
Sewage samples.
A total of 28 raw sewage samples from different geographical areas were analyzed. Sixteen samples were collected in the sewers of Barcelona (Spain) and were found to contain a mean of 1.7 × 106 fecal coliform bacteria/100 ml with a variance of 5.6 × 105. These samples were collected from September 1997 to February 1998. Each sample was collected in sterile 500-ml polyethylene containers, kept at 4°C for less than 8 h until the viral particles were concentrated in PBS, and stored at −80°C.
Four samples were collected in Umeå, Sweden, during September and October 1997. Four samples were collected in Nancy (France) during March 1998, and four samples were collected in Pretoria (South Africa) in October 1997. The sewage samples collected in Umeå and Nancy were stored at −80°C and shipped to Spain, where they were analyzed. From each sewage sample collected in Pretoria, 30-ml aliquots were centrifuged (229,600 × g for 1 h at 4°C) and the 2-ml sediments were shipped to Spain at room temperature immediately (within 24 h) after being processed.
Concentration of viral particles and nucleic acid extraction.
The method applied for the recovery of viral particles and nucleic acid extraction was based on previous studies (18, 33). Briefly, 40 ml of sewage sample was ultracentrifuged (229,600 × g for 1 h at 4°C) to pellet all the viral particles together with any suspended material. From concentrated Pretoria samples, 2 ml was resuspended in 6 ml of 2× PBS and ultracentrifuged (229,600 × g for 1 h at 4°C). The next step in the treatment of all the sewage samples was the elution of the sediment by mixing it with 4 ml of 0.25 N glycine buffer (pH 9.5) on ice for 30 min, and the suspended solids were separated by centrifugation at 12,000 × g for 15 min after the addition of 5 ml of 2× PBS. The viruses were finally pelleted by ultracentrifugation (229,600 × g for 1 h at 4°C), resuspended in 0.1 ml of 1× PBS, and stored at −80°C. Since the DNA extracted from 10 μl of the viral concentrate corresponded to a sample volume of 4 ml and was used as inoculum for the PCR test, the results will be reported in reference to this volume. According to previous studies, the yield after adding poliovirus type 1 LSc 2ab in sewage samples with similar conditions was 70% PFU.
Viral nucleic acids were extracted by a procedure that provides clean nucleic acids for molecular studies. This procedure uses guanidinium thiocyanate and adsorption of the nucleic acids to silica particles (12).
Sensitivity of the method.
Raw urban sewage samples were spiked with WT 800 SV40 viral particles (105/ml) in order to evaluate the sensitivity of virus extraction and the presence of inhibitors for the PCR. The polyomavirus-like particles of a urine sample positive by PCR for JCV and BKV were quantified by transmission electron microscopy (10, 31). Briefly, 10 ml of urine was harvested directly onto electron microscope grids (400-mesh Ni grids) supported with carbon-coated Formvar film by ultracentrifugation (197,500 × g for 90 min) with a swing-out rotor. Afterwards, the supernatant was removed, the grids were air dried, and the sample was stained with 3% uranyl acetate for 20 s. Polyomavirus-like particles were counted in 200 randomly selected fields of view, by using a Hitachi-600AB transmission electron microscope at 75 kV and 80,000× magnification. Serial dilutions were used to estimate the sensitivity of the detection method.
Enzymatic amplification.
For the detection of the specific viral genomes in raw sewage, urine, or CSF, 10-μl aliquots of the extracted nucleic acids were used in each test, corresponding to 4 ml of sewage sample, 10 μl of urine, or 10 μl of CSF.
Amplification was carried out in a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate, 2 U of Ampli Taq DNA polymerase (Perkin-Elmer Cetus), and the corresponding primers at their corresponding concentrations (0.08 μM external primers and 0.07 μM internal primers for Ad amplification and 0.5 μM external and internal primers for all polyomavirus amplifications). Thermal cycling of the amplification mixture was performed in a programmable heat block (Gene Amp PCR System 2400; Perkin-Elmer). In all PCR assays for human Ad detection, the first cycle of denaturation was carried out for 4 min at 94°C. The conditions for the 29-cycle amplification were as follows: denaturing at 92°C for 60 s, annealing at 55°C for 60 s, and extension at 72°C for 75 s. All amplifications were completed with a 4-min, 72°C extension period. The PCR amplifications of Ad genomes were carried out with external primers HR and HL and internal primers NHR and NHL, described and tested in previous studies (6, 7).
In all PCR assays for polyomavirus detection, the first cycle of denaturation was carried out for 4 min at 94°C. The conditions for the 29-cycle amplification were as follows: denaturing at 92°C for 60 s, annealing for 60 s, and extension at 72°C for 75 s. Amplifications were completed with a 4-min, 72°C extension period. Primers and annealing temperatures used in this study are described in Tables 1 and 2.
TABLE 1.
Oligonucleotide primers used for PCR amplification and sequencing of human Ad and BKVa
| Virus type (region) | Position (nt) | Amplification reaction | Primer | Product size (bp) | Annealing temp (°C) | Sequence |
|---|---|---|---|---|---|---|
| Ad2 (hexon) | ||||||
| Ad40 (hexon) | 18858–18883 | First-left | HL | 301 | 55 | 5′-GCCGCAGTGGTCTTACATGCACATC-3′ |
| Ad41 (hexon) | 19136–19158 | First-right | HR | 5′-CAGCACGCCGCGGATGTCCAAAGT-3′ | ||
| Ad2 (hexon) | 18937–18960 | Nested-left | NHL | 143 | 55 | 5′-GCCACCGAGACGTACTTCAGCCTG-3′ |
| Ad2 (hexon) | 19051–19079 | Nested-right | NHR | 5′-TGTACGAGTACGCGGTATCCTCGCGGTC-3′ | ||
| BKV, JCV (VP2/VP3) | 1352–1367 | First-left | BJ1 | 765 | 46 | 5′-TATTGCMCCAGGAGGT-3′ |
| BKV, JCV (VP1) | 2132–2148 | First-right | BJ2 | 5′-AACATTTTCYCCTCCTG-3′ | ||
| BKV (VP1) | 1762–178 | Nested-left | BK4 | 290 | 50 | 5′-AGTAGATTTCCACAGGTTAG-3′ |
| BKV (VP1) | 1486–1506 | Nested-right | BK6 | 5′-CCAGGGGCAGCTCCCAAAAAG-3′ | ||
| BKV (VP1) | 2132–2148 | Seminested-left | BK2 | 388 | 46 | 5′-AACATTTTCCCCTCCTG-3′ |
| BKV (VP1) | 1758–1776 | Seminested-right | BK5 | 5′-GGACCTAACCTGTGGAAAT-3′ |
M = A + C; Y = C + T. For Ad, the sequence positions are referred to the Ad2 hexon region; for BKV and JCV, the sequence positions are referred to the BKV-Dunlop sequence.
TABLE 2.
Oligonucleotide primers used for PCR amplification and sequencing of JCV and SV40
| Virus type (region)a | Position (nt)b | Amplification reaction | Primer | Product size (bp) | Annealing temp (°C) | Sequence |
|---|---|---|---|---|---|---|
| JCV (VP1 region) | 1710–1734 | Nested-left | JLP15 | 215 | 63 | 5′-ACAGTGTGGCCAGAATTCCACTACC-3′c |
| JCV (VP1 region) | 1902–1924 | Nested-right | JLP16 | 5′-TAAAGCCTCCCCCCCAACAGAAA-3′c | ||
| JCV (IG region) | 2062–2087 | First-left | EP1A | 737 | 59 | 5′-TGAATGTTGGGTTCCTGATCCCACC-3′ |
| JCV (IG region) | 2774–2798 | First-right | EP2A | 5′-ACCCATTCTTGACTTTCCTAGAGAG-3′ | ||
| JCV (IG region) | 2099–2124 | Nested-left | P1A | 668 | 59 | 5′-CAAGATATTTTGGGACACTAACAGG-3′d |
| JCV (IG region) | 2742–2766 | Nested-right | P2A | 5′-CCATGTCCAGAGTCTTCTGCTTCAG-3′d | ||
| JCV (IG region) | 2511–2536 | Nested-right | JCSR | 478 | 55 | 5′-TGATTACAGCATTTTTGTCTGCAAC-3′ |
| JCV (IG region) | 2364–2388 | Nested-left | JCSL | 5′-GGAAGTCCTTCTGTTAATTAAATCAG-3′ | ||
| JCV (R region) | 4992–5011 | First-left | JR1 | 586 | 55 | 5′-CCCTATTCAGCACTTTGTCC-3′e |
| JCV (R region) | 428–447 | First-right | JR2 | 5′-CAAACCACTGTGTCTCTGTC-3′e | ||
| JCV (R region) | 5060–5079 | Nested-left | JR3 | 382 | 55 | 5′-GGGAATTTCCCTGGCCTCCT-3′e |
| JCV (R region) | 297–317 | Nested-right | JR4 | 5′-ACTTTCACAGAAGCCTTACG-3′e | ||
| SV40 (TAg) | 2614–2633 | First-left | SV1 | 336 | 46 | 5′-AATTTGTGATGCTATTGCTT-3′ |
| SV40 (TAg) | 2932–2950 | First-right | SV2 | 5′-TTGGAGTTTTAGAAAGGCT-3′ | ||
| SV40 (TAg) | 2746–2766 | Nested-left | SV3 | 157 | 50 | 5′-CTACAAATGTGGTATGGCTGA-3′ |
| SV40 (TAg) | 2883–2903 | Nested-right | SV4 | 5′-AGCCAGGAAAATGCTGATAA-3′ | ||
| SV40 (TAg) | 2609–2631 | First-left | SV1A | 336 | 55 | 5′-TGTGAAATTTGTGATGCTATTGC-3′ |
| SV40 (TAg) | 2922–2945 | First-right | SV2A | 5′-GTTTTAGATTGGCTAAGAAACAGT-3′ | ||
| SV40 (TAg) | 2746–2766 | Nested-left | SV5 | 119 | 50 | 5′-CTACARATGTGRTATGGCTGA-3′fg |
| SV40 (TAg) | 2844–2864 | Nested-right | SV6 | 5′-AAGAACATGGAAGACTCAGGG-3′f |
VP1, virion protein 1; TAg, T antigen.
For JCV, the sequence positions are relative to the JCV-Mad-1 sequence; for SV40, the sequence positions are relative to a standard SV40 sequence.
From the work of Agostini et al. (5).
Modified from the work of Kunitake et al. (23).
From the work of Monaco et al. (27).
From the work of Lednicky et al. (24).
R = A + G.
The PCR amplifications of SV40 genomes were carried out with external primers SV1 and SV2 and internal primers SV3 and SV4. External primers SV1A and SV2A and internal primers SV5 and SV6 (Table 2), which detect the SV40 variants that were described for primates by Lednicky et al. (24), were used for analyzing all the samples from Pretoria, Nancy, and Umeå and four samples from Barcelona (BCN11, -12, -14, and -15). JCV genomes were amplified with BJ1 and BJ2 as external primers and JLP15 and JLP16 as internal primers (5). All the samples were also analyzed for JCV with EP1A and EP2A as external primers and P1A and P2A as internal primers (23). This second set showed higher sensitivity for the amplification of JCV genomes.
BKV genomes were amplified with BJ1 and BJ2 external primers and BK4 and BK6 internal primers. All the primers used in this study are represented in Tables 1 and 2.
The results were analyzed by agarose gel electrophoresis with ethidium bromide as a stain.
Quality control of the amplification method.
To reduce the probability of sample contamination by amplified DNA molecules, standard precautions were applied in all the manipulations. Separate areas of the laboratory were used for reagents, treatment of samples, and manipulation of amplified samples. All the samples were analyzed twice in independent experiments, and a negative control was added every two samples (a negative control is an amplification reaction mixture with the same reagents as in the test tubes of the samples but without the inoculum of viral nucleic acids). Treatment with uracil-DNA-glycosylase for the degradation of amplified material that could contaminate the samples was performed in previous studies (34), but they were not considered necessary for the routine analysis. Direct extract and a 10-fold dilution of the nucleic acid extracts were analyzed routinely on highly polluted samples in order to avoid false negatives because of inhibition of the reactions. This could occur only in a minority of the samples, according to the positive results observed previously with sewage samples supplemented with viruses.
Analysis of the viral genomes.
The amplicons of 12 JCV-positive samples were sequenced with the primers for amplification of the intergenic (IG) region, P1A, P2A, JCSR, and JCSL (23). Nine of these 12 JCV-positive samples were also sequenced with primers for amplifying the regulatory (R) region, JR1, JR2, JR3, and JR4 (27). The amplicons of five BKV-positive samples were also sequenced with BK2 and BK5 primers.
Nested-PCR products were purified with the QUIAquick PCR purification kit (Qiagen, Inc.). Both strands of the purified DNA amplicons were sequenced with the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with Ampli Taq DNA polymerase FS (Perkin-Elmer, Applied Biosystems) following the manufacturer's instructions. The results were checked with the ABI PRISM 377 automated sequencer (Perkin-Elmer, Applied Biosystems). The sequences were compared with the GenBank and EMBL databases by using the BLAST program of the National Center for Biotechnology Information. Sequences were aligned by using the BOXSHADE 3.21 program of the EMBNET.CH. GenBank accession numbers of the JCV sequences used for phylogenetic studies are shown in Table 3.
TABLE 3.
List of the JCV types used in the phylogenetic analysisa
| Genotype | Strain no. | GenBank accession no. | Reference |
|---|---|---|---|
| Type 1 (Europe) | |||
| 1A | Mad-1 | JO227 | 17 |
| 1A | 124 | AFO15526 | 3 |
| 1B | 123 | AFO15227 | 4 |
| Type 2 (Asia) | |||
| 2A | Tokyo-1 | AFO30085 | 4 |
| 2A | 224 | AFO15529 | 4 |
| 2A | 225 | AFO15530 | 4 |
| 2A | 226 | AFO15531 | 4 |
| 2B | 223 | AFO15532 | 4 |
| 2B | 227 | AFO15533 | 4 |
| 2C | 228 | AFO15534 | 4 |
| 2C | 229 | AFO15535 | 4 |
| 2D | 230 | AFO15536 | 4 |
| Type 3 (Africa) | |||
| 3A | 308 | U73500 | 1 |
| 3A | 312 | U73502 | 1 |
| 3B | 311 | U73501 | 1 |
| Type 4 (European and African American) | 402 | AFO15528 | 2 |
| Type 5 (minor European) | 501 | AFO15684 | 4 |
| Type 6 (West Africa) | 601 | AFO15537 | Unpublished data |
| Type 7 (Southeast Asia) | Tai-3 | U61771 | 28 |
From the work of Jobes et al. (20).
Phylogenetic analysis of the JCV sequenced regions was performed with the nucleic acid maximum likelihood method (dnaml), version 3.572c, included in the PHYLIP software package (16). The phylogenetic tree was visualized with the TREEVIEW 1.5 program (29). The JCV genotypes included in the phylogenetic analysis are described in Table 3.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under accession no. AF119345 to AF119356 (JCV IG region sequences), AF120240 to AF120242 (JCV R region sequences), and AF120243 to AF120247 (BKV VP1 region sequences).
RESULTS
Sewage analysis.
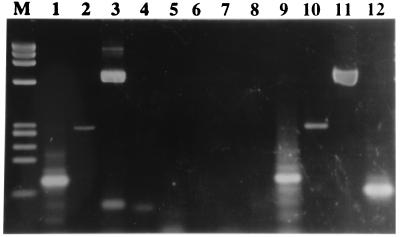
The results obtained are shown in Tables 4 and 5 and reflect a very high level of excretion of human Ad, JCV, and BKV and the absence of positive results for SV40. About 93% of the samples collected in Barcelona were positive for human Ad and JCV, and 10 (62.5%) were positive for BKV. Figure 1 shows the agarose gel electrophoresis of one sewage sample positive for human Ads, BKV, and JCV and negative for SV40; small-size bands, nonspecific or made by amplification of combined primers, can be seen in the gel, as they appear frequently in amplified samples. Samples collected in Pretoria (four samples) and Nancy (four samples) showed positive results for all viruses with the exception of SV40. All four samples collected in Umeå were also positive for human Ads and JCV, and three of these four samples were positive for BKV. From a total of 28 sewage samples analyzed, 96% were positive for human Ads and JCV and 77.8% were positive for BKV. Results of the nested-PCR tests and serial dilution experiments are shown in Tables 4 and 5. Very high numbers of JCV particles are observed, with the concentration estimated as between 102 and 104 viral particles per 4 ml of the sewage samples analyzed. On the basis of the results obtained in the serial dilution experiments, the concentrations estimated for human Ad were 101 to 104 viral particles and for BKV were 101 to 103 BK viral particles in the 4 ml of the sewage samples analyzed.
TABLE 4.
Nested-PCR results in urban sewage samples collected in Barcelonaa (site 1)
| Sample and date of collection (day-mo-yr) | HAd | BKV | JCV | SV40 |
|---|---|---|---|---|
| BCN1 (29-09-97) | + (−1) | + (−1) | + (−2) | − |
| BCN2 (07-10-97) | + (−1) | + (−1) | + (−2) | − |
| BCN3 (21-10-97) | + (−1) | + (−2) | + (−3) | − |
| BCN4 (28-10-97) | + (0) | + (−1) | NT | − |
| BCN5 (07-11-97) | + (−1) | + (0) | + (−1) | − |
| BCN6 (11-11-97) | + (−1) | + (−1) | + (−1) | − |
| BCN7 (19-11-97) | + (−2) | + (−1) | + (−2) | − |
| BCN8 (05-12-97) | + (−2) | + (−1) | + (−2) | − |
| BCN9 (17-12-97) | − | − | − | − |
| BCN10 (30-12-97) | + (−2) | + (−1) | + (−3) | − |
| BCN11 (09-01-98) | + (0) | − | + (−1) | − |
| BCN12 (12-01-98) | + (−2) | + (0) | + (−3) | − |
| BCN13 (16-01-98) | + (−1) | + (0) | + (−3) | − |
| BCN14 (23-01-98) | + (−2) | − | + (−2) | − |
| BCN15 (02-02-98) | + (−2) | − | + (−1) | − |
| BCN16 (11-02-98) | + (−2) | − | + (−3) | − |
Results obtained after nested-PCR analysis of 4 ml of sewage. HAd, human Ad; +, positive; −, negative; NT, not tested. Numbers in parentheses after results are the highest decimal dilutions positive by nested PCR.
TABLE 5.
Nested-PCR results in urban sewage samples from different countriesa
| Sample | HAd | BKV | JCV | SV40 |
|---|---|---|---|---|
| Site 2 (Umeå) | ||||
| UMEA1 | + (−2) | + (0) | + (−2) | − |
| UMEA2 | + (0) | − | + (−1) | − |
| UMEA3 | + (0) | + (0) | + (−2) | − |
| UMEA4 | + (−1) | + (0) | + (−1) | − |
| Site 3 (Nancy) | ||||
| NANCY1 | + (−2) | + (−1) | + (−3) | − |
| NANCY2 | + (−2) | + (−1) | + (−3) | − |
| NANCY3 | + (−1) | + (−1) | + (−3) | − |
| NANCY4 | + (−2) | + (0) | + (−2) | − |
| Site 4 (Pretoria) | ||||
| PRETORIA1 | + (−1) | + (−2) | + (−2) | − |
| PRETORIA2 | + (−3) | + (−1) | + (−1) | − |
| PRETORIA3 | + (−3) | + (−1) | + (−1) | − |
| PRETORIA4 | + (−2) | + (−1) | + (−1) | − |
Results obtained after nested-PCR analysis of 4 ml of sewage for sites 2 and 3 and 1.2 ml of sewage for site 4. HAd, human Ad; +, positive; −, negative. The numbers in parentheses are the highest decimal dilutions positive by nested PCR.
FIG. 1.
Agarose gel electrophoresis showing amplified DNA after nested PCR of one sewage sample that was positive for human Ad, BKV, and JCV and negative for SV40 (lanes 1, 2, 3, and 4, respectively). Lanes 5, 6, 7, and 8 are the corresponding negative controls. Lanes 9, 10, 11, and 12 are positive controls. Lane M, molecular weight standard, φX174 HaeIII digest.
Sensitivity of the method.
In the sewage samples supplemented with SV40 viral particles, the viral genomes were detected by nested PCR with an estimated sensitivity of five genomes compared with the stock suspensions that showed a nested-PCR detection unit five times higher (one genome). The applied nested-PCR procedure with SV40 DNA detected one SV40 genome. This is the level of sensitivity of detection of three different strains of human Ad as described in previous studies (34). The results observed with SV40 DNA agree with the sensitivity described previously for the detection of human Ads and enteroviruses in sewage samples (between 10 and 100 viral particles per 4-ml sample).
The number of polyomavirus-like particles counted in a urine sample by electronic microscopy was 8.7 × 105/ml. According to this, the sensitivity observed after nested PCR in the urine sample is at least 5.2 viral particles/4 ml of JCV and at least 520 BKV particles per 4 ml.
According to these results, we estimate the minimum concentration of viruses producing a positive result in the nested-PCR test for sewage samples to be on the order of 10 viral particles.
Molecular characterization of the viruses detected.
We sequenced 461 nucleotides of the IG region of 12 JCV-positive samples randomly selected: five samples collected in Barcelona (one from a urine sample), two from Pretoria, one from Nancy, and one from Umeå. Three CSF samples from PML patients from different regions in Spain were also sequenced as positive controls.
The sequences analyzed confirmed the specificity of the nested-PCR amplification, since all viral sequences were identified as the expected polyomavirus.
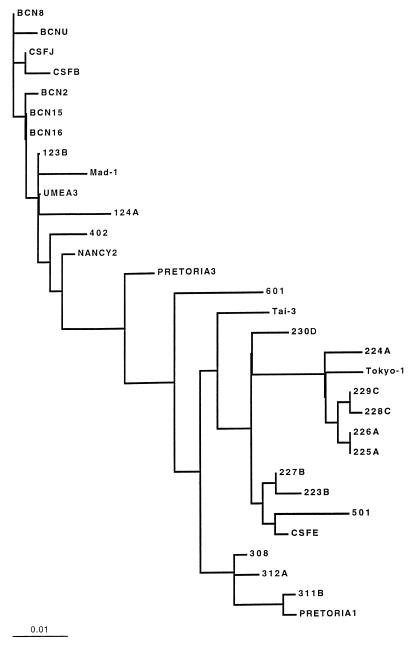
The alignment of the sequences of the JCV obtained is shown in Fig. 2, and the relationship of the detected strains with the nucleotide sequences of the EMBL and GenBank sequence data banks is shown in Table 6. The results of the maximum likelihood method applied to the phylogenetic analysis are shown in Fig. 3. The analysis of the R region of the JCV shows that all strains detected in sewage and urine samples and sequenced are archetype strains; the JCV obtained from the clinical samples presented specific rearrangements in the regulatory region (Fig. 4).
FIG. 2.
Sequence alignment of the IG regions of 12 JCV-positive samples. Dots indicate sequence identities. R = A + G; W = A + T.
TABLE 6.
Comparison of the sequence of the JCV detected with the IG primers with previously described isolates
| Sample | Most closely related isolate(s)a
|
Nucleotide difference from the most closely related strain (no. of nucleotides different/total no. of nucleotides) | |
|---|---|---|---|
| Strain | City and country | ||
| BCNU | UK-2 | London, United Kingdom | 4/461 |
| IT-1 | Rome, Italy | 4/461 | |
| BCN2 | UK-2 | London, United Kingdom | 2/461 |
| BCN8 | UK-2 | London, United Kingdom | 1/461 |
| BCN15 | IT-1 | Rome, Italy | 1/461 |
| UK-2 | London, United Kingdom | 1/461 | |
| BCN16 | UK-2 | London, United Kingdom | 2/461 |
| NANCY2 | RS-3 | Novosibirsk, Russia | 4/461 |
| G-5 | Illertisen, Germany | 4/461 | |
| HU-5 | Budapest, Hungary | 4/461 | |
| UMEA3 | CR-5 | Prague, Czech Republic | 0/461 |
| PRETORIA1 | SO-3 | Welkom, South Africa | 1/461 |
| SO-1 | Welkom, South Africa | 1/461 | |
| PRETORIA3 | MA-4 | Nouakchott, Mauritania | 6/461 |
| MA-5 | Nouakchott, Mauritania | 6/461 | |
| MA-1 | Nouakchott, Mauritania | 6/461 | |
| MR-2 | Fez-frane, Morocco | 6/461 | |
| CSFB | UK-2 | London, United Kingdom | 2/461 |
| CSFE | N-4 | Deventer, The Netherlands | 3/461 |
| CSFJ | UK-2 | London, United Kingdom | 2/461 |
From the work of Kunitake et al. (23).
FIG. 3.
Phylogenetic analysis of the IG regions of 12 sequenced JCV strains compared with previously described JCV subtypes (Table 3) by the maximum likelihood method.
FIG. 4.
Sequences of the R regions of JCV from nine analyzed samples. CSFE and CSFB are clinical samples of CSF from two patients. Archetypal sequences are observed in samples BCNU, BCN15, BCN16, BCN8, BCN2, NANCY2, and UMEA3. Underlined nucleotides are identical to the archetypal sequence. Nucleotides in boldface and in italics represent duplications present in these sequences.
The nucleotide sequences of positive BKV amplicons from one sample collected in Pretoria, one from Nancy, and three samples from Barcelona were compared. These samples showed between two and four nucleotide differences in a fragment of 248 nucleotides (Fig. 5).
FIG. 5.
Sequence alignment of the VP1 regions of five BKV-positive samples. Dots indicate sequence identities.
DISCUSSION
We are not aware of previously reported data on the presence of human polyomavirus in sewage. In this regard, we have detected high concentrations of JCV and BKV in the sewage of different cities of widely divergent geographical origins and have studied the most prevalent strains of JCV excreted.
For this purpose, we have amplified and sequenced JCV DNA in the region designated the IG region, which encompasses the 3′-terminal sequences of both T-antigen and VP1 (major capsid protein) genes, since it contains abundant nucleotide variation compared with other regions. These variations have been described as a means of tracing human migrations (5, 39). The strong relationship of the sequenced JCV strains with previously described viral isolates from related populations is remarkable. The sequenced strains described for sewage samples from European countries are strongly related to European subtypes, and the two strains from the Pretoria area are related to African isolates. PRETORIA1 is related to strains isolated from sub-Saharan populations, and PRETORIA3 is more closely related to isolates from Mauritania and North Africa which appear more similar to the European strains, correlating with the relationship of these populations. We also show in this study that the most common JCV strains that are excreted in the areas studied show archetypal R regions, as described for urine samples. In agreement also with previous studies, the three PML-derived strains detected from CSF samples presented individual rearrangements of these genomic regions (40).
The fact that immunocompetent hosts frequently excrete JCV in urine (21, 36) indicates that renal JCV is not latent under these conditions but replicates to generate progeny that are excreted in urine. Thus, JCV persistence in the kidney may be characterized by continuous viral replication and virus shedding. This would be required for JCV to be transmitted among humans, given that JCV infection has been described as very inefficient (22).
It has also been reported that JCV in urine is infectious, and urine is the most likely source of JCV infection in humans. Although the cells that support JCV replication at portals of entry remain to be identified, some data suggest that tonsil tissue could be a possible site of the initial viral infection (27). BKV seems to circulate independently of JCV in the population. As with JCV, BKV has been detected in kidney but also in tonsils and other tissues as well (35).
The nested amplicons of BKV obtained from four sewage samples and one urine sample were sequenced in order to confirm their identity as BKV. The differences observed in the 248 sequenced nucleotides were around 0.8 to 2% as described in the literature (36). Some indeterminations were observed in nucleotides that showed variability when sequences from the data banks were compared. This is the case for the sample PRETORIA1, suggesting the presence of a mixture of related strains in this sewage sample. Further sequencing studies are required for the identification of the specific BKV genomic subtypes.
In previous studies, we have isolated hepatitis E virus (32), human Ad, and enterovirus (33) from similar sewage samples, and these viruses are infectious. Those polyomaviruses that remain intact as viral particles in sewage may be infectious and could be transmitted to other humans in a fecally polluted environment.
The semiquantitative results for the detection of the polyomavirus indicate the concentration of the viral particles detected. Definitive numbers could be obtained only by quantitative PCR test. The finding of lower titers of BKV than of JCV could be attributable to a lower sensitivity of the PCR amplification and/or to a lower frequency of BKV shedding in immunocompetent persons as described by Shah et al. (36).
We did not detect SV40 in any of these sewage samples. The results indicate that, if SV40 is excreted in human urine or feces, its concentration in the analyzed sewage samples is lower than 10 viral particles in 4 ml of raw sewage.
The high prevalence of human Ad detected in all of these geographical areas is consistent with previous studies carried out in Barcelona, where it was suggested that this parameter could be a good viral indicator of the fecal contamination of human origin in the environment and shellfish (33).
The detection by PCR of polyomaviruses in sewage allows the identification and analysis of the genomes of viral strains that are infecting the population and gives information on the spread, frequency, and distribution of these viruses. These data are also needed for the study of the epidemiology of the related diseases. However, the molecular techniques applied do not give information on the level of infectivity of the strains detected in the sewage samples, and further studies will be needed in order to establish the stability of human polyomavirus in the environment. It is also clear that this approach is likely to provide a method with a higher level of specificity, sensitivity, and speed than isolation of viruses in cell culture. In these experiments, the differences between the nucleotide sequences of the detected genomes and those of the viruses of our positive controls argue against the possibility of laboratory contamination.
The human polyomavirus has been shown to be present in high concentrations in the sewage of the different geographical areas studied, especially JCV, and its specificity as a human virus may be useful as a marker for fecal pollution of anthropogenic origin. The high level of excretion detected also supports the idea previously described that fecal-oral transmission (including contamination from urine) will probably happen soon in vivo, inside the family or from closely related people and less frequently later in life from other polluted sources.
The procedure that we used to study the presence of viruses in the sewage of a community may be useful as a tool for studying changes in epidemiological patterns of some viral infections and in future studies for the analysis of environmental dynamics.
ACKNOWLEDGMENTS
This work was supported by the Center for Biologics Evaluation and Research, Food and Drug Administration, project FDA 122987 00 97 GF 00. Sílvia Bofill-Mas and Sonia Pina are fellows of the Generalitat de Catalunya.
We thank Serveis Cientifico-Tècnics of the University of Barcelona for their help in the sequencing of the PCR products. We also thank Willie Grabow from the University of Pretoria, South Africa, Louis Schwartzbrod from Nancy University, France, and Göran Wadell, from Umeå University, Sweden, for their kind collaboration in obtaining the samples in their areas. We thank Jaume Bertranpetit and Francesc Calafell from the Universitat Pompeu Fabra from Barcelona for their collaboration in the phylogenetic analysis of the samples. We also thank Andrew M. Lewis from the Office of Vaccine Research and Review, CBER, FDA, for useful consultation.
REFERENCES
- 1.Agostini H T, Ryschkewitsch C F, Brubaker G R, Shao J, Stoner G L. Five complete genomes of JC virus type 3 from Africans and African Americans. Arch Virol. 1997;142:637–655. doi: 10.1007/s007050050108. [DOI] [PubMed] [Google Scholar]
- 2.Agostini H T, Ryschkewitsch C F, Stoner G L. Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. J Clin Microbiol. 1996;34:159–164. doi: 10.1128/jcm.34.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agostini H T, Ryschkewitsch C F, Stoner G L. JC virus type 1 has multiple subtypes; three new complete genomes. J Gen Virol. 1998;79:801–805. doi: 10.1099/0022-1317-79-4-801. [DOI] [PubMed] [Google Scholar]
- 4.Agostini H T, Shishido-Hara Y, Baumhefner R W, Singer E J, Ryschkewitsch C F, Stoner G L. JC virus type 2: definition of subtypes based on DNA sequence analysis of ten complete genomes. J Virol. 1998;79:1143–1151. doi: 10.1099/0022-1317-79-5-1143. [DOI] [PubMed] [Google Scholar]
- 5.Agostini H T, Yanagihara R, Davis V, Ryschkewitsch C F, Stoner G L. Asian genotypes of JC virus in Native Americans and in a Pacific Island population: markers of viral evolution and human migration. Proc Natl Acad Sci USA. 1997;94:14542–14546. doi: 10.1073/pnas.94.26.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allard A, Albinsson B, Wadell G. Detection of adenovirus in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J Med Virol. 1992;37:149–157. doi: 10.1002/jmv.1890370214. [DOI] [PubMed] [Google Scholar]
- 7.Allard A, Girones R, Juto P, Wadell G. Polymerase chain reaction for detection of adenoviruses in stool samples. J Clin Microbiol. 1990;28:2659–2667. doi: 10.1128/jcm.28.12.2659-2667.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthur R R, Shah K V. Occurrence and significance of papoviridae BK and JC in the urine. Prog Med Virol. 1989;36:42–61. [PubMed] [Google Scholar]
- 9.Berger J R, Kaszovita B, Donovan P J, Dickinson G. Progressive multifocal leukoencephalopathy associated with human immunodeficiency virus infection. Ann Intern Med. 1987;107:78–87. doi: 10.7326/0003-4819-107-1-78. [DOI] [PubMed] [Google Scholar]
- 10.Bergh O, Børsheim K Y, Bratbak G, Hendal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 11.Bergsagel D J, Finegold M J, Butel J S, Kupsky W J, Garcea R I. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumors of childhood. N Engl J Med. 1992;326:988–993. doi: 10.1056/NEJM199204093261504. [DOI] [PubMed] [Google Scholar]
- 12.Boom R, Sol C J A, Salimans M M M, Jansen C J, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butel J S, Lednicky J A. Cell and molecular biology of simian virus 40: implications for human infectious and disease. J Natl Cancer Inst. 1999;91:119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- 14.Carbone M, Pass H I, Rizzo P, Marianetti M, Di Muzio M, Mew D J, Levine A S, Procopio A. Simian virus 40-like DNA sequences in human pleural mesothelioma. Oncogene. 1994;9:1781–1790. [PubMed] [Google Scholar]
- 15.Carbone M, Rizzo P, Procopio A, Giuliano T, Pass H I, Gebhardt M C, Mangham C, Hansen M, Malkin D F, Bushart G, Pompetti F, Picci P, Levine A S, Bergsagel J D, Garcea R L. SV40-like sequences in human bone tumors. Oncogene. 1996;13:527–533. [PubMed] [Google Scholar]
- 16.Felsenstein J. PHYLIP. Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 17.Frisque R J, Bream G L, Canella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girones R, Puig M, Allard A, Lucena F, Wadell G, Jofre J. Detection of adenovirus and enterovirus by PCR amplification in polluted waters. Water Sci Technol. 1995;31:351–357. [Google Scholar]
- 19.Ilyinskii P O, Daniel M E, Horvath C J, Desrosiers R C. Genetic analysis of simian virus 40 from brains and kidneys of macaque monkeys. J Virol. 1992;66:6353–6360. doi: 10.1128/jvi.66.11.6353-6360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jobes D V, Sylvester C C, Ryschkewitsch C F, Stoner G L. Phylogenetic analysis of 22 complete genomes of the human polyomavirus JC virus. J Gen Virol. 1998;79:2491–2498. doi: 10.1099/0022-1317-79-10-2491. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura T, Kunitake T, Guo J, Tominaga T, Kawabe K, Yogo Y. Transmission of the human polyomavirus JC virus occurs both within the family and outside the family. J Clin Microbiol. 1995;32:2359–2363. doi: 10.1128/jcm.32.10.2359-2363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura T, Sugimoto C, Kato A, Ebihara H, Suzuki M, Taguchi F, Kawabe K, Yogo Y. Persistent JC virus (JCV) infection is demonstrated by continuous shedding of the same JCV strains. J Clin Microbiol. 1997;35:1255–1257. doi: 10.1128/jcm.35.5.1255-1257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunitake T, Kitamura T, Guo J, Taguchi F, Kawabe K, Yogo Y. Parent-to-child transmission is relatively common in the spread of the human polyomavirus JC virus. J Clin Microbiol. 1995;33:1448–1451. doi: 10.1128/jcm.33.6.1448-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lednicky J A, Arrington A S, Stewart A R, Dai X M, Wong C, Jafar S, Murphey-Corb M, Butel J S. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J Virol. 1998;72:3980–3990. doi: 10.1128/jvi.72.5.3980-3990.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lednicky J A, Garcea R L, Bergsagel D J, Butel J S. Natural simian virus 40 strains are present in human choroid plexus and ependymoma tumors. Virology. 1995;212:710–717. doi: 10.1006/viro.1995.1529. [DOI] [PubMed] [Google Scholar]
- 26.Martini F, Iaccheri L, Lazzarin L, Carinci P, Corallini A, Gerosa M, Iuzzolino P, Barbanti-Brodano G, Tognon M. SV40 early region and large T antigen in human brain tumors, peripheral blood cells, and sperm fluids from healthy individuals. Cancer Res. 1996;56:4820–4825. [PubMed] [Google Scholar]
- 27.Monaco M C G, Jensen P N, Hou J, Durham L C, Major E O. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J Virol. 1998;72:9918–9923. doi: 10.1128/jvi.72.12.9918-9923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou W C, Tsai R T, Wang M, Fung C Y, Hseu T H, Chang D. Genomic cloning and sequence analysis of Taiwan-3 human polyomavirus JC virus. J Formos Med Assoc. 1997;96:511–516. [PubMed] [Google Scholar]
- 29.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 30.Pietropaolo V, DiTaranto C, Degener A M, Orsi N, Jin L, Sinibaldi L, Baiochini A, Mellis M. Transplacental transmission of human polyomavirus BK. J Med Virol. 1998;56:372–376. doi: 10.1002/(sici)1096-9071(199812)56:4<372::aid-jmv14>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Pina S, Creus A, González N, Girones R, Felip M, Sommaruga R. Abundance, morphology and distribution of planktonic virus-like particles in two high mountain lakes. J Plankton Res. 1998;20:2413–2421. [Google Scholar]
- 32.Pina S, Jofre J, Emerson S U, Purcell R H, Girones R. Characterization of a strain of infectious hepatitis E virus isolated from sewage in an area where hepatitis E is not endemic. Appl Environ Microbiol. 1998;64:4485–4488. doi: 10.1128/aem.64.11.4485-4488.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pina S, Puig M, Lucena F, Jofre J, Girones R. Viral pollution in the environment and shellfish: human adenovirus detection by PCR as an index of human viruses. Appl Environ Microbiol. 1998;64:3376–3382. doi: 10.1128/aem.64.9.3376-3382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puig M, Jofre J, Lucena F, Allard A, Wadell G, Girones R. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl Environ Microbiol. 1994;60:2963–2970. doi: 10.1128/aem.60.8.2963-2970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosen S, Harmon W, Krensky A M, et al. Tubuinterstitial nephritis associated with polyomavirus (BK type) infection. N Engl J Med. 1983;308:1192–1196. doi: 10.1056/NEJM198305193082004. [DOI] [PubMed] [Google Scholar]
- 36.Shah K V, Daniel R W, Strickler H D, Goedert J J. Investigation of human urine for genomic sequences of the primate polyomaviruses simian virus 40, BK, and JC virus. J Infect Dis. 1997;176:1618–1621. doi: 10.1086/517340. [DOI] [PubMed] [Google Scholar]
- 37.Shah K V. Polyomaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Raven Publishers; 1995. pp. 1997–2025. [Google Scholar]
- 38.Stewart A R, Lednicky J A, Butel J S. Sequence analyses of human tumor-associated SV40 DNAs and SV40 viral isolates from monkeys and humans. J Neurovirol. 1998;4:182–193. doi: 10.3109/13550289809114518. [DOI] [PubMed] [Google Scholar]
- 39.Sugimoto C, Kitamura T, Guo J, Al-Ahdal M N, Shchelkunov S N, Otova B, Ondrejka P, Chollet J Y, El-Safi S, Ettayebi M, Grésenguet G, Kocagöz T, Chaiyarasamee S, Thant K Z, Thein S, Moe K, Kobayashi N, Taguchi F, Yogo Y. Typing of urinary JCV DNA offers a novel means of tracing human migrations. Proc Natl Acad Sci USA. 1997;94:9191–9196. doi: 10.1073/pnas.94.17.9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso X, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]