Abstract
Morbidity and mortality in response to SARS-CoV-2 infection are significantly elevated in people of advanced age. To understand the underlying biology of this phenotype, we utilize the golden hamster model to compare how the innate and adaptive immune responses to SARS-CoV-2 infection differed between younger and older animals. We find that while both hamster cohorts showed similar virus kinetics in the lungs, the host response in older animals was dampened, with diminished tissue repair in the respiratory tract post-infection. Characterization of the adaptive immune response also revealed age-related differences, including fewer germinal center B cells in older hamsters, resulting in reduced potency of neutralizing antibodies. Moreover, older animals demonstrate elevated suppressor T cells and neutrophils in the respiratory tract, correlating with an increase in TGF-β and IL-17 induction. Together, these data support that diminished immunity is one of the underlying causes of age-related morbidity.
Keywords: neutrophils, aging, COVID-19, suppressor T cells, Ki67, Bcl6, Ccr6, MPO, FoxP3, comorbidity
Graphical abstract
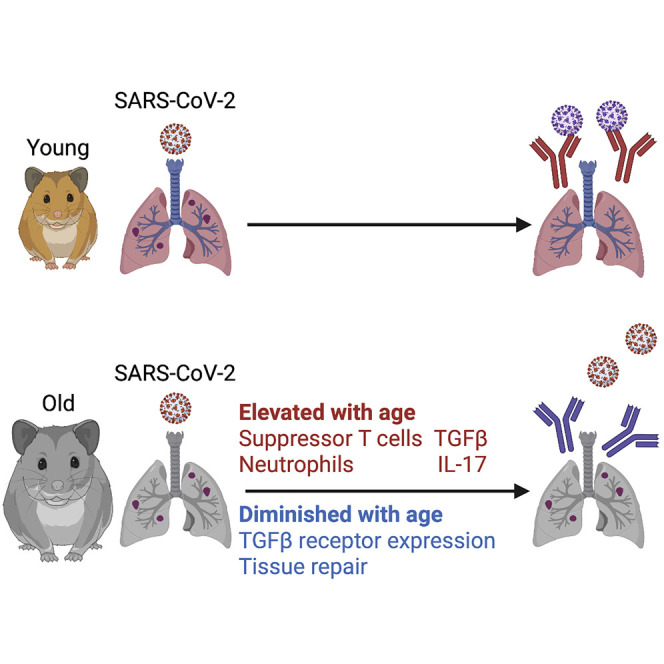
Using the hamster model, Oishi et al. report on how age influences the host response to SARS-CoV-2. Older hamsters have a diminished antiviral response and are slower to repair damage induced by infection. These age-related differences correlate with changes in the adaptive immune cell repertoire.
Introduction
The host response to virus infection often relies on the initial recognition of pathogen-associated molecular patterns that then culminate in transcriptional engagement of antiviral host factors (Iwasaki, 2012; Koyama et al., 2008). Despite the evolutionary success of this antiviral system, many viruses respond to this selective pressure by generating potent antagonistic strategies to subvert it, resulting in morbidity and mortality (Banerjee et al., 2020; Basler et al., 2000; Wang et al., 1999). In general, viruses capable of circumventing our defenses invoke the most disease in the old, correlating with the maturity of their immune response. Interestingly, while this trend holds true for many viruses of pandemic potential, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) appears to selectively target the aged population (O'Driscoll et al., 2021; Verity et al., 2020). Here we set out to better define the biology underlying this phenotype.
The host response to virus infection begins with the recognition of non-self-materials and induces the activation of the type I and III interferon family of genes (IFN-I/-III) as well as additional cytokines responsible for recruiting and activating the adaptive immune response (Kato and Fujita, 2014; Mesev et al., 2019; Park and Iwasaki, 2020; Sokol and Luster, 2015). Infected cells that detect the presence of virus replication respond by inducing an IFN-I/-III-mediated call to arms, which signals to surrounding cells—inducing them to upregulate hundreds of so-called IFN-stimulated genes (ISGs), which work in concert to slow the rate of virus replication (Der et al., 1998; Munnur et al., 2021; Sanyal et al., 2013; Schoggins et al., 2011). In addition, virus infection also results in nuclear factor κB (NF-κB) signaling and the subsequent production of chemokines and other cytokines—representing a call for reinforcements, to ensure the infection can be controlled (Allen et al., 2009; Bonizzi and Karin, 2004; Khalil et al., 2021; Poole et al., 2008; Song and Li, 2021; Velazquez-Salinas et al., 2019).
The call for reinforcements induces the migration of innate immune cells, such as natural killer (NK) cells, neutrophils, monocytes, and dendritic cells, to the site of infection, providing new defensive resources (Chen et al., 2018; Tay et al., 2020). For example, neutrophils can engulf foreign material and induce additional proinflammatory stimuli, whereas monocytes and dendritic cells (DCs) can capture and present protein components of the pathogen to lymphocytes (antigen presentation). Despite the fact that neutrophils participate in virus clearance in the early stage of infection, they also create an atmosphere that leads to tissue injury (Chiang et al., 2020; Narasaraju et al., 2020). Because of this feature, the number of neutrophils is considered as a clinical marker related to acute respiratory distress syndrome in coronavirus disease 2019 (COVID-19) patients. After antigen presentation, specific T and B cells migrate to the site of infection where they target antigen-containing material (Chen et al., 2018). Virus infection indues CD4+ T cells to differentiate to specific cytokine-producing effector helper subsets. B cells undergo affinity maturation in germinal centers (GCs) to produce high-affinity antibodies. In addition to these active processes, the adaptive immune response also requires extensive regulation, which predominantly comes in the form of suppressor T cells, such as FoxP3+CD8+, or regulatory T cells (Tregs) (Veiga-Parga et al., 2013). These lineages suppress the further activation of T and B cells to prevent over-activation of the immune response (Veiga-Parga et al., 2013). Following the recruitment of immune cells to the site of infection, cells signal to each other to often promote further inflammation, resulting in an environment sometimes referred to as cytokine storm. This inflammatory environment often includes proinflammatory interleukins (ILs), such as IL-1, IL-6, IL-12, and IL-17, which aid in the activation of the aforementioned cells and the subsequent establishment of adaptive immunity (Couper et al., 2008; Guo et al., 2019; Kaneko et al., 2019; Lu et al., 2021; Velazquez-Salinas et al., 2019). This process returns to baseline levels, when the source of the response has been cleared or immune cell exhaustion is reached.
Interestingly, as we age, so too does our immune response. The innate immune system has been reported to be stronger in youth and subsequently decreases at a steady state thereafter. Moreover, studies have found that aging affects immune cell biology in the content of virus infection (Hernandez-Vargas et al., 2014; Palacios-Pedrero et al., 2021; Shaw et al., 2011). For example, Tregs, which generate transforming growth factor β (TGF-β) and suppress the function of B and T cells, have been reported to accumulate with age (Rocamora-Reverte et al., 2020). Similarly, IL-17, a signature cytokine of T helper 17 (Th17) cells, has also been found to increase with age and has been suggested to influence the pathology of a wider array of diseases through its ability to activate both neutrophils and glial cells (Allen et al., 2015; Stout-Delgado et al., 2009). These age-related changes to the immune response result in a slower activation of antiviral defenses and delays in recovery.
As the golden hamster model has proven to phenocopy COVID-19 biology, age-dependent differences of COVID-19 were examined using young (6–10 weeks old) and older (27 weeks old) hamster cohorts (Imai et al., 2020; Ohno et al., 2021; Osterrieder et al., 2020; Rosenke et al., 2020; Selvaraj et al., 2021). These age-defined cohort studies were used to assess how aging influences morbidity. These data found that the severity of disease corresponded with lower levels of virus replication and significant tissue damage (Osterrieder et al., 2020). Here, we corroborate these results and expand on them further through molecular characterization of virus-host interactions and the immune response.
Results
Older hamsters demonstrate diminished innate and repair responses to SARS-CoV-2 infection
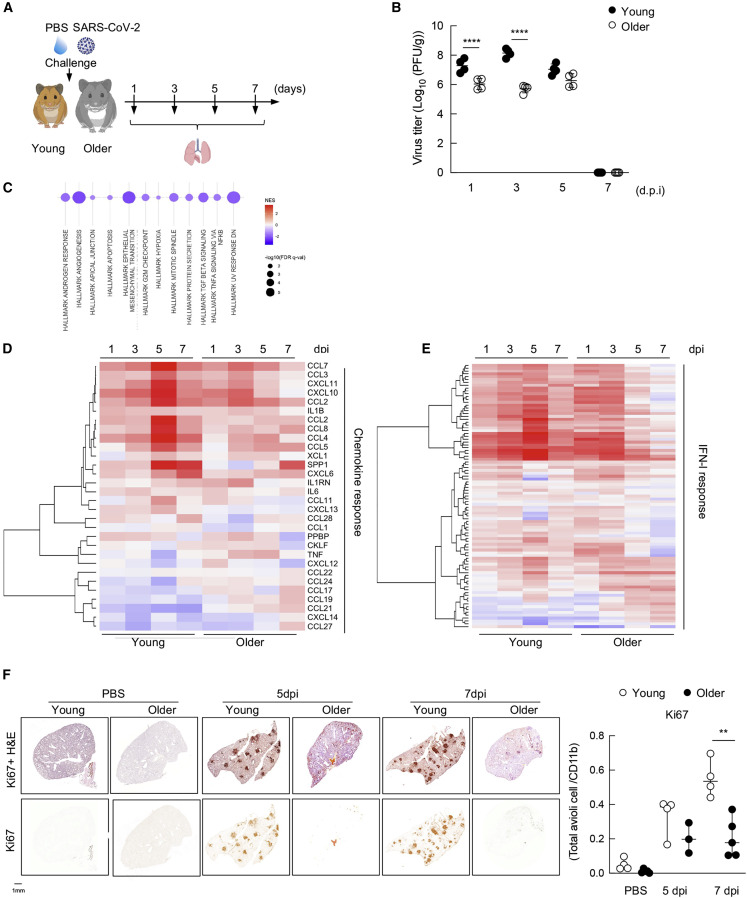
To elucidate the skewed pathogenesis of SARS-CoV-2 as it relates to aging, we generated two golden hamster populations and compared how they responded to SARS-CoV-2 infection. To this end, we followed the reported definitions of aging in the hamster, including cohorts of 6–9 weeks (young) or 40+ weeks (older) of age (Figure 1A) (Imai et al., 2020; Ohno et al., 2021; Osterrieder et al., 2020; Rosenke et al., 2020; Selvaraj et al., 2021). Intranasal inoculation of 1,000 plaque-forming units (PFUs) of SARS-CoV-2 were examined at 1, 3, 5, and 7 days post-infection (dpi) to determine viral load. To this end, lung tissue was obtained for each time point, homogenized, and used to assess SARS-CoV-2 replication by plaque assay (Figure 1B). These data demonstrated elevated levels of virus during the initial phase of infection within the young hamster cohort, showing significant increases on days 1 and 3 post-infection as compared with older hamsters. However, virus levels are comparable between older groups at 5 and 7 dpi (Figure 1B).
Figure 1.
Older hamsters demonstrate diminished innate and repair responses to SARS-CoV-2 infection
(A) Experimental protocol of young and older hamster infection. Syrian golden hamsters (6–9 weeks aged as young and 40 weeks aged as older) were infected intra nasally with either PBS-mock or 103 PFUs of SARS-CoV-2 (n = 4, at each time point). Lung organs were collected at 1, 3, 5, and 7 days post-infection (dpi) for virus titer and gene expression analysis.
(B) Virus titer in the lung from young and older hamster at 1, 3, 5, and 7 dpi. Lungs were collected at each time point and homogenized with PBS. Plaque assay was performed using Vero E6 cells. ∗∗∗∗ p < 0.0001.
(C) Gene set enrichment analysis using the MSigDB Hallmark gene sets was performed on differential expression datasets generated from comparison of naive young versus older hamsters. Dot plot visualizes analysis output, with dot size scaled to significance [−log10(false discovery rate [FDR] q value [q-val])] and dot color correlated with normalized enrichment score (NES). Only significant enrichments (FDR q-val < 0.05) are visualized.
(D) Heatmap depicting the expression levels of chemokines differentially expressed in lung samples of hamsters at 1, 3, 5, and 7 dpi.
(E) Same as (D) for IFN-I-related genes, as defined by the Hallmark IFN-I annotation.
(F) Immunohistochemical (IHC) staining of anti-Ki67 (lower) and IHC with H&E staining (upper) on the lung from PBS-treated or infected young and older hamsters at 5 and 7 dpi (left). Quantification of the ratio of Ki67+ cells from 3 to 5 hamsters (right). For IHC protein expression quantification, four sections of each analyzed slide were quantified by Qupath software. ∗∗ p < 0.01.
To gain a better understanding for the increase in virus replication in young versus older hamsters, we performed RNA sequencing (RNA-seq) on the lungs from uninfected animals representing each population (Figure 1C). These data demonstrated that at baseline, three well-annotated pathways were significantly diminished in the older animals, including TGF-β and NF-κB signaling as well as cells undergoing division (Table S1). While these data collectively suggest that aged animals have an altered immune response, the significant decrease in NF-κB signaling is particularly noteworthy, because this biology has been shown to be essential for virus replication and thus may explain the initial decrease observed in virus levels (Nilsson-Payant et al., 2021).
To further characterize the host response in young versus older hamsters, we performed RNA-seq on total lung samples from all infected time points. In agreement with the annotation data from naive animals, we find that cytokine induction, which relies heavily on NF-κB biology (Karin and Greten, 2005; Taniguchi and Karin, 2018), is significantly diminished in the older animals (Figure 1D). We find lower levels of Ccl2, Ccl4, Ccl8, and Ccl28 as well as the proinflammatory cytokine Il1β in older hamsters (Bonizzi and Karin, 2004; Karin and Greten, 2005). These analyses demonstrated that virus-induced cytokines do rebound in the older animals on day 3 but fail to reach the same peak levels observed at 5 and 7 dpi in young animals (Figure 1D). Analyzing these same datasets against the IFN-I response showed a comparable ISG profile following the initial 3 dpi (Figure 1E). However, despite comparable virus levels on day 5, the ISG response in older animals rapidly diminishes for reasons that remain unclear (Figures 1B and 1E). These altered expression patterns of virus-induced cell signaling suggest that aging influences the duration of the host response. These data can be further visualized by volcano plot to demonstrate how each cohort responds to virus on days 1, 3, 5, and 7 (Figures S1A–S1D and Table S2. Differentially expressed genes between young and older at 1 dpi, related to Figure 1, Table S3. Differentially expressed genes between young and older at 3 dpi, related to Figure 1, Table S4. Differentially expressed genes between young and older at 5 dpi, related to Figure 1, Table S5. Differentially expressed genes between young and older at 7 dpi, related to Figure 1).
To gain a better understanding with regard to the physiological consequences of the aged immune response, we next focused on the differentially expressed genes (DEGs) from days 5 and 7 post-infection (Figures S1C and S1D and Tables S4 and S5), where virus levels were comparable between cohorts (Figure 1B). In addition to the aforementioned ISGs, these data found significant differences relating to immune cell infiltration in the respiratory tract, perhaps influenced by the chemokine profiles observed at 1 dpi. For example, in older animals, we observe a significant increase in neutrophil-related gene expression, as denoted by a log 2-fold change (L2FC) greater than 5 for the S100a8 and S100a9 transcripts that have been shown to regulate CD11b expression and neutrophil recruitment (Figure S1C and Table S4) (Mellett and Khader, 2021; Scott et al., 2020). In contrast, macrophage/DC markers, such as Cd163, Cc5, Clec4a, Mafb, and Slamf8, show a significant downregulation in response to virus in the older cohort (Table S4). In contrast, the host response on 7 dpi showed a dramatic loss of histone-modification-associated transcripts (e.g., Tet2, Tet3, Ogt, and Suz12) in addition to other markers associated with activated T cells (e.g., Nfat5) in older hamsters (Table S5). These data provide further support for the idea that there is diminished immune cell recruitment to the respiratory tract in older animals, which likely also results in the observed loss of sustained ISGs from total lung RNA (Figure 1E).
In addition to immune markers, RNA-seq data also implicated a significant downregulation (L2FC of −2) in Ki67 (Table S3). Because Ki67 is a prominent proliferation marker that can be used to denote repair (Major et al., 2020; Zhao et al., 2020), we performed immunohistochemistry to ascertain whether our RNA-seq data could be validated independently. To this end, mock and infected lungs from our aged hamster cohorts were fixed and analyzed by H&E staining and probed for Ki67 expression (Figure 1F). In agreement with the transcriptional data, Ki67 staining showed a dramatic difference between cohorts, implicating defects in the kinetics and/or biology of lung repair in older animals.
Aging induces a diminished adaptive immune response following SARS-CoV-2 infection
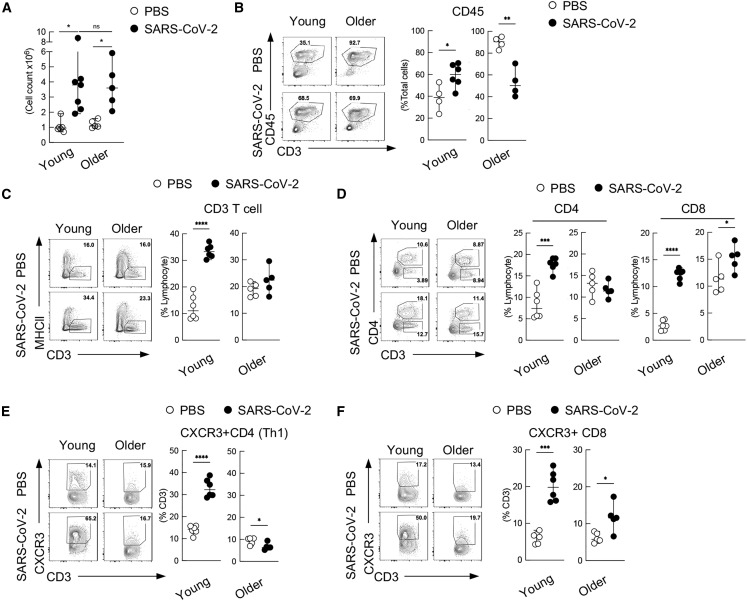
Given the marked differences in chemokines induced in response to SARS-CoV-2, coupled to mRNA-seq data, we next characterized the cellular and humoral response using more traditional assays recently developed for the hamster model (Horiuchi et al., 2021). To this end, young versus older cohorts of hamsters were infected with 1,000 PFUs of SARS-CoV-2 and total number of lung cells separated by Ficoll (immune-cell-enriched fraction) were analyzed at 7 dpi (Figure 2A). These analyses demonstrated that the overall recruitment of immune-enriched cells to the lung were comparable, regardless of age, despite the finding that hematopoietic lineages were dramatically skewed in the older animals (Table S4). To further define the cell population recruited to the lung in response to SARS-CoV-2, we repeated the experiment using our developed flow cytometry methods (Figures S2A and S2B) (Horiuchi et al., 2021). These data demonstrated that older animals had a significant elevation of CD45+ hematopoietic cells at baseline, a trend that inversed by 7 dpi (Figure 2B). Further resolution of the CD45+ cell population found that young animals expand CD3+ T cells, where there was no increase in the population in older animals (Figure 2C). We further defined the component of CD3+ T cells as in CD4+ T cells and CD8+ T cells. In contrast to young animals, older animals showed a plateau of CD4+ T cells in response to SARS-CoV-2 (Figure 2D), although in agreement with our CD45+ count, a high baseline level of these cells was also observed in this cohort. Examining Th1 cell populations (as in CXCR3+CD4+ T cells) corroborated the trends observed for CD4+ cells in both cohorts, showing a significant upregulation in the young, with a downregulation in the older animals (Figure 2E). In contrast, profiling of CXCR3+CD8+ cells was in agreement with the trends observed for total CD8+ cell counts, with significant expansion in both young and older animals in response to SARS-CoV-2 (Figure 2F). These data confirm earlier reports about the suppressed expansion of T cells in response to virus in aged cohort while additionally demonstrating the impacts of aging on the ability to mobilize the adaptive immune response (Jagger et al., 2014).
Figure 2.
Aging induces a diminished adaptive immune response following SARS-CoV-2 infection
(A) Total count of cells collected from the lung after Ficoll separation showing the overall recruitment of immune-enriched cells.
(B) Flow cytometry of total frequency of CD45+ hematopoietic cells in the lung from young and older hamsters at 7 dpi.
(C and D) Frequency of total CD3+ T cells, as in CD3+ MHCII cells (C) and CD4+ T cells, as in CD4+CD3+ cells, and CD8+ T cells, as in CD4−CD3+ cells (D) from young and older hamster lungs at 7 dpi.
(E) Frequency of CD4+ T cell subsets as Th1 (CD3+MHCII−CD4+CXCR3+) with in CD4+ T cells.
(F) Frequency of CD8+ T cell subsets as CXCR3+CD8+ T cells (CD3+ MHCII−CD4− CXCR3+). ∗p < 0.05 ∗∗p < 0.01 ∗∗∗p < 0.001 ∗∗∗∗p < 0.0001.
Increase of suppressor T cells in the airways of golden hamsters correlates with the induction of IL-17 and neutrophil recruitment
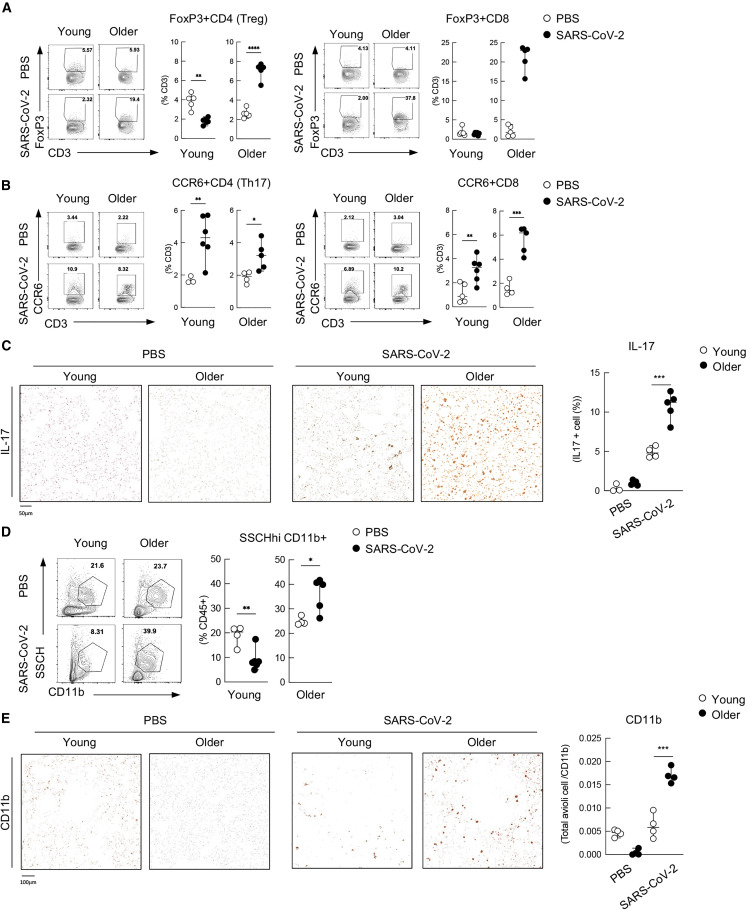
Given the decrease in Th1 and total CD4+ T cell frequencies in the respiratory tract of older hamsters (Figures 2C–2E), we next characterized suppressor T cells as they prevent T cell proliferation (Veiga-Parga et al., 2013). To this end, we defined the two populations of suppressor T cells as FoxP3+CD4+ T cells (here denoted as Tregs) and FoxP3+CD8+ T cells. Cohorts of young versus older hamsters were infected with 1,000 PFUs of SARS-CoV-2, and immune cell lineage populations in the lung were analyzed at 7 dpi. Because TGF-β is predominantly secreted by FoxP3+CD8+ T cells and Tregs (Joosten and Ottenhoff, 2008), we determined these cellular populations by flow cytometry and found that older hamsters showed a profound increase in frequency of these aforementioned cells (Figure 3A). These data could be further validated by examining the transcript levels of Tgfb, which revealed a 5-fold increase in transcript levels (Figure S1A). Because TGF-β is known to be involved in tissue repair (D'Alessio et al., 2009), upregulation in the older cohort seemed at odds with our Ki67 data (Figure 1F). These data could be the result of a loss in TGF-β signaling, as our RNA-seq data implicated downregulation of receptor subunit expression, showing a significant loss approaching one order of magnitude (Figure S4A). Together, these data suggest that older hamsters have elevated levels of FoxP3+CD8+ T cells and Tregs that generate increasing levels of TGF-β in an environment that lacks its receptor expression. These results also are in agreement with findings that TGF-β signaling induces a negative feedback loop responsible for receptor downregulation (Stroschein et al., 1999).
Figure 3.
Increase of suppressor T cells in the airways of golden hamsters correlates with the induction of IL-17 and neutrophil recruitment
(A) Flow cytometry of FoxP3+ Tregs (CD3+MHCII−CD4+FoxP3+), on the left, and FoxP3+ CD8+ T cells (CD3+ MHCII−CD4− FoxP3+), on the right.
(B) Flow cytometry of Th17 (CD3+MHCII−CD4+CCR6+) cells and CCR6+CD8+ T cells (CD3+ MHCII−CD4−CCR6+) in the lung from young and older hamsters at 7 dpi.
(C) Histology of IL-17+ cells in the lung from PBS-treated or infected hamsters at 7 dpi. A representative histology (left) along with the dataset of the frequency with three to five individuals are shown (right). For IHC-protein-expressed cell quantification, sections of each analyzed slide were quantified by Qupath software.
(D) Frequency of SSCHhi CD11b+ presumed neutrophils population in the CD45+ cells in the lung from young and older hamsters at 7 dpi.
(E) Histology of CD11b+ cells in the lung from PBS-treated or infected hamsters at 7 dpi. A representative histology (left) along with the dataset of the frequency with three to five individuals are shown (right). ∗p < 0.05 ∗∗p < 0.01 ∗∗∗p < 0.001 ∗∗∗∗p < 0.0001. For IHC-protein-expressed cell quantification, sections of each analyzed slide were quantified by Qupath software.
The observed enrichment of TGF-β, IL-1β, and IL-6 in the older cohort would generate an IL-17-producing T cell differentiation environment (Garg et al., 2014; Palacios-Pedrero et al., 2021). Given this, we investigated whether we see an increase of the levels of IL-17 in response to SARS-CoV-2 in older hamsters. To this end, we performed flow cytometry for IL-17-producing CD4+ and CD8+ cellular subsets by focusing on the frequencies of CCR6+CD4+ and CCR6+CD8+ T cells. These data found no significant age-related difference among CCR6+CD4+ T cells but CCR6+CD8+ T cells were elevated in the older population (Figure 3B). To verify IL-17 expression in the lung, we next performed immunohistochemistry on PBS-treated or SARS-CoV-2 infected hamsters at 7 dpi (Figure 3C). Examination of the lung parenchyma demonstrated a dramatic difference in IL-17 production between young and older animals not only in the lymphocyte but also in the epithelia cells, corroborating our flow data on CCR6+CD8+ T cells (Figures 3B and 3C). Because IL-17 signaling results in the activation of NF-κB and the subsequent induction of chemokines and proinflammatory cytokines, we propose that IL-17 production from T cells and epithelial cells contributes to the subsequent recruitment of neutrophils to the respiratory tract that has been characterized both in COVID-19 patients and in the hamster model (Hoagland et al., 2021; Rendeiro et al., 2021). To verify the age-related differences in neutrophil frequency, we assessed the levels of CD11b-expressing cells and found that, despite a lower baseline level in the older population, this same cohort showed significantly elevated levels of neutrophils in response to SARS-CoV-2 (Figure 3D). These data could be further corroborated by immunohistostaining for CD11b, which stains for neutrophils in addition to monocytes, macrophages, and DCs (Figure 3E). To further implicate neutrophils, we additionally stained these same tissues for myeloperoxidase (MPO), which yielded comparable results (Tostanoski et al., 2020) (Figure S3).
While the golden hamster model has been shown to recapitulate many aspects of COVID-19 biology, we next sought to corroborate our findings using clinical data (Figure S4B). To this end, human COVID-19 cadaver lung tissue was obtained and analyzed by RNA-seq. Stratifying normalized gene counts confirmed that IL-17 induction was statistically correlated with age. Numerous IL-17 target genes also trended in the same direction, including Il6, Cxcl2, and Cxcl1 (Figures S4B and S4C). Together, these data suggest that the observed phenotype in hamsters can be extended to human patient data.
Aged hamsters generate fewer GC B cells, resulting in reduced potency of neutralizing antibodies
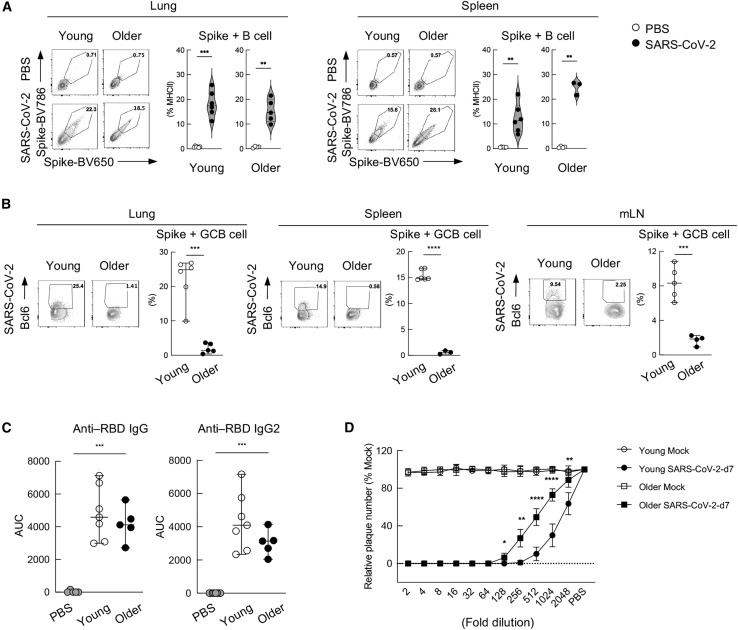
Given the increase in Treg populations and the diminished T cell response in aged animals (Figures 2D, 2E, and 3A), we next hypothesized that there may be additional B cell defects in older hamsters. Characterization of total B cell numbers as defined by MHCII+CD3 staining, demonstrated no significant differences in cell frequency when comparing young versus older hamsters (Figure S5A). Similarly, no discernable age-related differences were noted when examining class switch of B cells (Figure S5B). This trend was also consistent in SARS-CoV-2 spike (here denoted as S)-specific B cells from young and older hamster lung and spleen at 7dpi (Figure 4A). To assess B-cell-mediated affinity maturation between older cohorts, we determined the frequency of S-positive GC B cells as in Bcl6+ B cells (Haniuda et al., 2020; Kitano et al., 2011). These efforts identified a dramatic diminishment of GC B cells in the older cohort in the lung, spleen, and draining lymph node—tissues previously demonstrated to generate GC B cells (Figure 4B) (Poon et al., 2021).
Figure 4.
Older hamsters generate fewer GC B cells, resulting in reduced potency of neutralizing antibodies
(A) Frequency of SARS-CoV-2 S-specific B cells were shown by staining B cells with biotin-conjugated spike and streptavidin-BV650 and -BV786. The result in the lungs and spleen at 7 dpi are shown. A representative FACS plot (left) along with the dataset of the frequency with three to seven individuals are shown (right).
(B) Frequency of SARS-CoV-2 S-specific GC B cells are shown by MHC+CD3−spike+ Bcl6+ in the lungs (left), spleen (middle), and lung draining mediastinal lymph node (mLN) (right) at 7 dpi. A representative FACS plot (left) along with the dataset of the frequency with three to six individuals are shown (right).
(C) IgG and IgG2 antibody titer against SARS-CoV-2 receptor binding domain (RBD) from the young and older hamster serum after SARS-CoV-2 infection at 7 dpi. A dataset of five to seven individuals are shown (right).
(D) Plaque reduction assay of the serum from young or older hamsters after SARS-CoV-2 infection at 7 dpi. SARS-CoV-2 was incubated with 2-fold serial dilutions of sera, and antibody-virus mixtures containing 100 PFUs of SARS-CoV-2 were inoculated into Vero-E6 cells to perform the plaque assay. The 50% plaque reduction neutralization test (PRNT50) is defined as the highest serum dilution resulting in 50% reduction in plaque-forming units. The PRNT50 of each sample was calculated using a non-linear regression analysis for a four-parameter logistic sigmoidal curve. PRTN50: young, 1,549.00; older, 514.00. ∗p < 0.05 ∗∗p < 0.01 ∗∗∗p < 0.001 ∗∗∗∗p < 0.0001.
Because S-specific B cell numbers were unchanged despite a significant decrease in GC B cell populations (Figures 4A and 4B), we subsequently focused on antibody quantity and quality (Figures 4C and 4D). ELISA-based quantification of anti-RBD (receptor binding domain of S)-specific IgG and IgG2 antibodies in serum revealed no significant age-related difference in antibody production (Figure 4C). However, when the same serum samples were tested for their potential to reduce infectivity of SARS-CoV-2, the older population showed a reduction in function, as measured by a 50% plaque reduction neutralization test (PRNT50) (Figure 4D). Here, we find that the ability of older animals to neutralize virus is diminished by more than 60% compared with the younger cohort of hamsters (dilutions of 1,549 versus 514 PRNT50, respectively). These data suggest that the elevated levels of suppressor T cells in older animals may contribute to decreased GCs and thus, suboptimal affinity maturation of B cells.
Discussion
The studies presented here were performed to determine the molecular basis of age-related differences and their impact on SARS-CoV-2 pathology. We find that the comparison between young and older hamsters infected with SARS-CoV-2 revealed a diminished innate response to infection that was slower in both kinetics and magnitude. Surprisingly, the diminished response to infection did not equate to a greater level of SARS-CoV-2 replication, which is in agreement with previous aged hamster studies and findings that animals lacking an IFN-I response show similar viral RNA levels in the lung, suggesting that virus antagonism may influence this dynamic significantly (Boudewijns et al., 2020; Imai et al., 2020). However, it is worth noting that older animals had impaired NF-κB signaling at baseline, which has also been shown to be required for SARS-CoV-2 replication, which may contribute to this phenotype (Nilsson-Payant et al., 2021).
Independent from the innate immune response to SARS-CoV-2, here we also focus on the adaptive arm of our antiviral defenses. As observed in both our RNA-seq and flow cytometry data, we observed an impaired adaptive immune response in SARS-CoV-2 infected older hamsters. While this phenotype is most certainly multifactorial, we demonstrated that the frequency of suppressor T cells expands in the population with age—possibly explaining the diminished humoral response. Generally, Treg negatively regulates a variety of immune responses to self-antigens and plays a role in maintenance of immune tolerance and normal functioning (Grover et al., 2021). While an increase in Tregs among the elderly likely contributes to the prevention of autoimmune diseases, in the context of SARS-CoV-2 infection, it negatively impacts the ability to combat the infection. In support of these findings, an increased number of Tregs have been associated with increased severity to virus infection in both human and mouse models (Garg et al., 2014; Jagger et al., 2014; Raynor et al., 2012). The mechanism of age-related enhancement of Treg populations is still not fully understood, but epigenetics has been suggested to contribute to this process (Garg et al., 2014; Rocamora-Reverte et al., 2020). Taken together, our findings, in addition to independent corroborating data, suggest that the robustness of the adaptive response diminished with age, making it more difficult to recover from SARS-CoV-2 infection (Imai et al., 2020; Ohno et al., 2021; Osterrieder et al., 2020; Rosenke et al., 2020; Selvaraj et al., 2021).
In addition to the contribution of suppressor T cells to the aged immune response, we also find that in older hamsters, SARS-CoV-2 infection results in enhanced TGF-β-producing cells. Enhanced populations of these cells could possibly lead to induction of IL-17-producing cells and then to the recruitment of neutrophils to the lung—a hallmark of COVID-19 (Blanco-Melo et al., 2020). These data are in agreement with recent findings implicating elevated levels of TGF-β from senescent cells, which we also find to be increased in number in aged animals (Lorenzo et al., 2022). This biology would explain why we see a significant increase of neutrophils in the respiratory tract of older individuals, as TGF-β signaling induces IL-17, which in turn mediates neutrophil recruitment. This enhanced recruitment of neutrophils likely contributes to the severe pathology that correlates with age (Bradley et al., 2012; Kulkarni et al., 2019; Perrone et al., 2008). This concept is further supported by the recent finding showing a correlation between IL-17 expression and the severity of COVID-19 from human peripheral blood mononuclear cells (PBMCs) (Kuchroo et al., 2022). In all, these data could also be extrapolated to justify the therapeutic use of IL-17 antagonists as a putative treatment option, although this would require further testing and validation.
Lastly, longitudinal hamster studies allowed us to conclude that older hamsters generate antibodies with lower SARS-CoV-2 neutralization function as compared with their young counterparts, which is correlated with human reports (Collier et al., 2021). One of the reasons could be the increased induction of Tregs in older hamsters. Tregs are known to suppress both T and B cell function, resulting in the suppression of the generation of GC B cells and a diminishment in the potency of SARS-CoV-2 neutralization antibodies. These findings would suggest that viral clearance and the response to SARS-CoV-2 infection and protection against re-infection are both diminished in the older population and are likely contributing to the selective morbidity and mortality observed during this pandemic. Detailed analysis of how age affects adaptive immune response in terms of the protection from secondary infection needs to be further analyzed.
Limitations of the study
The hamster model has proven incredibly useful in characterizing the biology of COVID-19. Moreover, recently developed methods from our group made it possible to determine the details of the adaptive immune responses to SARS-CoV-2 in this model system (Horiuchi et al., 2021). These studies required some level of unconventional methods to monitor host immunology in the absence of certain commercial reagents. Flow cytometry analysis of T and B cell populations were only possible with limited number of commercially available anti-hamster antibodies; therefore, we were also not able to identify B-cell-specific or CD8+ T cell populations. For example, anti-CD19 or -CD20 antibodies for golden hamsters were not available to study B cells. In this study, we compromised by subjecting anti-major histocompatibility complex class II (MHCII) to separate presumed B cell populations. Anti-Ly6G antibody was also not available to separate neutrophils in golden hamsters. For this reason, we were only able to use either CD11b or MPO as a cell population that includes neutrophils. All experiments involving viral infections and harvest were carried out in a Centers for Disease Control and Prevention/US Department of Agriculture (CDC/USDA)-approved biosafety level 3 (BSL-3) facility. Older hamsters grow in size with age and need to be housed individually. Therefore, due to the limited capacity of the BSL-3 cage system, our n numbers could only range from 3 to 5/cohort.
STAR★Methods
Key resources table
| REAGENT or RESOURSE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FC block antibody (2.4G2) | BD Bioscience | Cat# 553142: RRID:AB_394657 |
| BV510-anti-CXCR3 (CXCR3-173) | BD Bioscience | Cat# 745033: RRID:AB_2742661 |
| BV711 -anti-Syrian golden hamster IgG2 (G192-3) | BD Biosciences | Cat# 745460: RRID:AB_2743003 |
| PE-anti-Bcl6 (K112-91) | BD Bioscience | Cat# 561522: RRID:AB_10717126 |
| Alexa 647- anti-MHCll (14-4-4s) | Biolegend | Cat# 10211: RRID:AB_493214 |
| Alexa 700-anti-FoxP3 (FJK-16s) | Thermo Fisher | Cat# 56-5773-82: RRID:AB_1210557 |
| APC/Fire750 -anti-CD4 (GK1.5) | Biolegend | Cat# 100460: RRID:AB_2572111 |
| BV421-anti-CXCR5 (L138D7) | Biolegened | Cat# 145512: RRID:AB_2562128 |
| Alexa 594-anti-Syrian golden hamster IgG | Invitrogen | Cat# A-21113: RRID:AB_2535762 |
| FITC-anti-CD3 (CD3-12) | BioRad | Cat# MCA1477A488 |
| PE-anti-CCR6 (140706) | R&D Systems | Cat# FAB590P-100 |
| CD45 Antibody (HASA25A) | Novus | Cat# NBP2-60909 |
| anti-Ki67 (B56) | BD Bioscience | Cat# 556003: RRID:AB_396287 |
| anti-IL-17 | abcam | Cat# ab79056:RRID:AB_1603584 |
| anti-CD11b (M1/70.15.11.5) | Miltenyi | Cat# 130-115-811: RRID:AB_2727205 |
| anti-myeloperoxidase | Dako | Cat# A0398 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fix Buffer I | BD Biosciences | Cat# 557870 |
| Perm/Wash Buffer | BD Biosciences | Cat# 557885 |
| Alexa Fluor 594 Antibody Labeling Kit | Invitrogen | Cat# A20185 |
| Fix Buffer I | BD Biosciences | Cat# 557870 |
| Perm/Wash Buffer | BD Biosciences | Cat# 557885 |
| Alexa Fluor 594 Antibody Labeling Kit | Invitrogen | Cat# A20185 |
| TRIzol Reagent | Thermo Fisher | Cat# 15596026 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat# 20020594 |
| Bacterial and virus strain | ||
| SARS-CoV-2 (USA-WA1/2020) | BEI Resources | Cat# NR-52281 |
| Experimental models: Cell lines | ||
| Vero-E6 cell | ATCC | Cat# CRL-1586 |
| Experimental models: Organisms/strains | ||
| LVG Golden Syrian hamster | Charles River Laboratories | Strain# 049 |
| Software | ||
| GraphPad Prism 9.0 | GraphPad Software | https://www.graphpad.com |
| FlowJo (v10.3.0) | FlowJo | http://www.graphpad.com |
| Microsoft Excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel |
| Qupath | Opensource | https://qupath.github.io |
| Other | ||
| gentleMAC Octo Dissociator with Heaters | Miltenyi | Cat# 130-096-427 |
| Attune Flow Cytometer | Thermo Fisher | N/A |
| Lysing Matrix A, 2 mL tube | MP Biomedicals | Cat# 6910050 |
| FastPrep-24 5G bead beating grinder and lysis system | MP Biomedicals | Cat# 6005500 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Benjamin R. tenOever (benjamin.tenoever@nyulangone.org).
Materials availability
All materials and reagents will be made available upon installment of a material transfer agreement (MTA).
Experimental model and subject details
Animal model
Yong (6–9 week old) or older (40 + week old) male Syrian golden hamsters were purchased from Charles River Laboratories and housed in a temperature-controlled environment with 12 h of light per day at the Center for Comparative Medicine and Surgery (CCMS) at the Icahn School of Medicine at Mount Sinai (New York, NY, USA). All experiments involving viral infections were carried out in a CDC/USDA-approved BSL-3 facility at CCMS and animals were transferred into the facility 4 days prior to onset of experiments. All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai (IACUC-2016-0438). Hamsters were anesthetized by intraperitoneal injection with 200 ul of a ketamine/xylazine solution (3:1) [100 mg/kg]. 100 ul of PBS or 103 pfu of SARS-CoV-2 was administered intranasally for each mock vs infected condition, respectively.
Hamsters were euthanized by intraperitoneal injection of Pentobarbital. Blood was collected from bleeding aorta during necropsy. Lung lobes from each hamster were collected and separated for FACS analysis, immunohistological analysis, plaque assay and RNA isolation. For plaque assay and RNA isolation, lung tissues were homogenized in Lysing Matrix A homogenization tubes (MP Biomedicals) in a FastPrep-24 5G bead beating grinder and lysis system (MP Biomedicals). Lung lobes for plaque assay were homogenized with an amount of PBS according to their weight. For RNA isolation, lung lobes were homogenized with 1 mL of TRIzol (Invitrogen).
Cell lines
Vero-E6 cells (kidney epithelial cells from African green monkey, ATCC, CRL-1586) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 5% FBS (Atlanta Biologicals) and 1% penicillin/streptomycin (Millipore Sigma) at 37°C and 5% CO2. Cells rested for the presence of mycoplasma biweekly using MycoAlert Mycoplasma Detection Kit (lonza).
Viruses
SARS-CoV-2, isolate USA-WA1/2020 (NR-52281) was deposited by the Center for Disease Control and Prevention (Atlanta, GA, USA). SARS-CoV-2 was propagated in Vero-E6 cells with DMEM supplemented with 5% FBS. Virus stocks were sequenced to ensure maintenance of the Furin cleavage site.
Transcriptome analysis of human samples
The plot was made with ggplot2 (v3.3.3) and ggpubr (v0.4.0) R packages. Pearson correlation coefficient was computed and regression line was drawn as described elsewhere (47). Count matrices were obtained from database of Genotypes and Phenotypes dbGAP (accession #38851 and ID phs002258.v1.p1).
Method details
Virus-based assays
Virus titers of SARS-CoV-2 were determined by means of plaque assay in Vero-E6 cells. 200 pfu of SARS-CoV-2 (100 ul) was incubated with two-fold serial dilutions of sera (100 ul) for 1h at room temperature. 100 ul of antibody-virus mixtures containing 100 pfu of SARS-CoV-2 were inoculated into Vero-E6 cells to perform the plaque assay. At 48h post-infection, plaques were visualized and counted.
RNA-seq analyses
1 μg of total RNA from hamster lung was enriched for polyadenylated RNA species and prepared for short-read next-generation sequencing using the TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. Sequencing libraries were sequenced on an Illumina NextSeq 500 platform. Fastq files were generated using bcl2fastq (Illumina) and aligned to the Syrian golden hamster genome (MesAur 1.0, ensembl) using the RNA-Seq Alignment application (Basespace, Illumina). Differential gene expression was determined using the DESeq2 pipeline (Love MI, Huber W, Anders S (2014). “Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.” Genome Biology, 15, 550. https://doi.org/10.1186/s13059-014-0550-8). All genes with a p-adjusted value (p-adj) < 0.1 were classified as differentially expressed genes. Number of reads mapping to the viral genome (GenBank: MN985325.1) was performed using bowtie2 (65). Gene set enrichment analysis was performed with GSEA java application GSEA_4.1.0 (made available by the Broad Institute and UC San Diego) (https://www.nature.com/articles/ng1180). GSEA was conducted as a pre-ranked analysis using a ranking statistic of -log10(p value)/sign(log2FoldChange). Gene sets used for analysis were part of Hallmark and curated human phenotype and gene ontology gene sets maintained as part of MSigDB (v7.4). All visualizations of RNA-sequencing differential expression data were created in R using ggplot2 and gplots packages. Gene set enrichment plots were adapted from VisualizeRNAseq (https://github.com/GryderArt/VisualizeRNAseq).
Immune profiling in golden hamsters by flow cytometry
Lung cells were collected upon perfusion with 30 mL PBS to avoid contamination of lymphocytes from the blood. After collection, lungs were dissociated by Miltenyi Gentle MACS with the lung dissociation Kit protocol. Dissociated lungs were incubated in 37°C for 30 min with 2 mL Collagenase (1 mg/mL)/DNase (0.5 mg/mL) and passed through 70 mm nylon mesh (FALCON #352350). For total immune cell-enriched cell count, homogenized lungs were further subjected to Ficol separation (1200Gx20min). For FACS analysis, cells were incubated with FC block antibody (2.4G2: BD Bioscience) following the staining with APC/Fire750 -anti-CD4 (GK1.5; Biolegend), Alexa 647- anti-MHCll (14-4-4s; ThermoFisher), BV421-anti-CXCR5 (L138D7; Biolegened), BV510-anti-CXCR3 (CXCR3-173: BD Biosciences), PE-anti-CCR6 (140,706; R&D Systems), BV711 -anti-Syrian golden hamster IgG2 (G192-3; BD Biosciences), Alexa 594-anti-Syrian golden hamster IgG (Invitrogen) extracellularly, anti-CD45 Antibody (HASA25A; Novus). anti-CD45 Antibody was labeled by Alexa Fluor 594 Antibody Labeling Kit (Invitrogen). For intranuclear staining, cells were fixed for 30 min at 4°C with Fix Buffer I (BD Biosciences) and were then incubated for overnight at 4°C, PE-anti-Bcl6 (K112-91; BD Biosciences), FITC-anti-CD3 (CD3-12; BioRad), Alexa 700-anti-FoxP3 (FJK-16s; ThermoFisher) in Perm/Wash Buffer (BD Biosciences). All samples were incubated with LIVE/DEAD Fixable yellow (Invitrogen) to exclude dead cells from the analysis. Cells were acquired on an Attune (ThermoFisher). The expression of each molecule was assessed with FlowJo software (TreeStar).
Antigen-specific B cell detection assay
To identify SARS-CoV-2 S-specific B cells, recombinant SARS-CoV-2 S protein were biotin-labeled (EZ-Link Sulfo-NHS-LC-Biotinylation kit, ThermoFisher). Ficoll separated cells collected from either lungs or spleens were stained with biotin labeled SARS-CoV-2 S protein with the combination of antibodies. Biotin was further detected by streptavidin-BV786 and -BV650.
Immunohistochemistry
Lung tissues were harvested following perfusion and fixed in 10% neutral buffered formalin for 24–48 hours. Fixed tissues were then embedded in paraffin and sectioned into 8 microns thick onto slides. Each slide was deparaffinized/rehydrated and then subjected to Immune staining by anti-Ki67 (BD Bio: B56), anti-IL-17 (abcam: ab79056), anti-CD11b (Miltenyi: M1/70.15.11.5), and anti-myeloperoxidase (MPO) (Dako, A0398). Second antibody with HRP was used following DISCOVERY OmniMap ChromoMap DAB RUO (Roche) for utilized for development. For H&E staining hematoxylin (Gill’s formula, Vector Laboratories, Cat# H3401) and eosin Y (Sigma Aldrich, Cat# E4009) was sued according to manufacturer’s instructions. For IHC protein expression quantification, four sections of each analyzed slide were quantified by Qupath software.
Anti-RBD hamster IgG ELISA
The ELISA protocol was adapted from a previously established protocol (Amanat et al., 2020). 96-well plates (Immulon 4 HBX, Thermo Fisher) were coated with 50 mL of 2 mg/mL recombinant RBD in PBS at 4°C overnight. After overnight incubation, the coating solutions were removed, and the plates were blocked with 100 mL of 3% non-fat milk (americanBio) prepared in 0.1% Tween 20 containing PBS (PBS-T) for 1 h at room temperature. To reduce the risk of containing live virus, serum samples were heated 1 h for 56°C before use and was handled in a BSL-3 facility. Serial dilution of serum samples and dilution of secondary antibody was done in 1% non-fat milk prepared in PBS-T. After blocking, solutions were removed and 100 mL of diluted serum was added following 2 h incubation at room temperature. After the solutions were removed, the wells were washed with 250 mL PBS-T three times. 100 mL of 1:7500 diluted anti-Syrian hamster IgG HRP (ThermoFisher) secondary antibody was added and incubated for 1 h at room temperature. After the solutions were removed, the wells were washed with 250 mL PBS-T three times. Once the wells were completely dry, 100 mL of SIG- MAFAST o-phenylenediamine dihydrochloride (Millipore Sigma) solution was added and the reaction was stopped with 30 mL of 3M HCl. The optical density at 490 nm was measured and the concentrations of the antibody were analyzed by area under curve (AUC) using Prism 8 (GraphPad).
Quantification and statistical analysis
Significance was determined by two-tailed unpaired t test using Prism software (GraphPad). A value of p < 0.05 was considered significant.
Acknowledgments
We wish to thank the Zegar Family Foundation for supporting this research. In addition, we wish to thank Dr. J. Lim for use of her flow cytometry instrument and Dr. F. Krammer for providing the recombinant SARS-CoV-2 spike protein used to define SARS-CoV-2-specific B cells.
Author contributions
B.R.T. supervised the project. K.O. and S.H. designed and performed the experiments. J.F. performed bioinformatics analysis of the hamster samples. R.E.S. performed bioinformatics analysis of the clinical samples. K.O., S.H., and B.R.T. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 9, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111002.
Supplemental information
Data and code availability
The accession number of the RNA-sequencing datasets reported in this paper can be found using NCBI GEO accession number GSE161200. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Allen I.C., Scull M.A., Moore C.B., Holl E.K., McElvania-TeKippe E., Taxman D.J., Guthrie E.H., Pickles R.J., Ting J.P.Y. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.E., Sutherland T.E., Rückerl D. IL-17 and neutrophils: unexpected players in the type 2 immune response. Curr. Opin. Immunol. 2015;34:99–106. doi: 10.1016/j.coi.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler C.F., Wang X., Mühlberger E., Volchkov V., Paragas J., Klenk H.D., García-Sastre A., Palese P. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U S A. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzi G., Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S., et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley L.M., Douglass M.F., Chatterjee D., Akira S., Baaten B.J.G. Matrix metalloprotease 9 mediates neutrophil migration into the airways in response to influenza virus-induced toll-like receptor signaling. PLoS Pathog. 2012;8:e1002641. doi: 10.1371/journal.ppat.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu S., Goraya M.U., Maarouf M., Huang S., Chen J.L. Host immune response to influenza A virus infection. Front. Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C.C., Korinek M., Cheng W.J., Hwang T.L. Targeting neutrophils to treat acute respiratory distress syndrome in coronavirus disease. Front. Pharmacol. 2020;11:572009. doi: 10.3389/fphar.2020.572009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., Ferreira I.A.T.M., Kotagiri P., Datir R.P., Lim E.Y., Touizer E., Meng B., Abdullahi A., CITIID-NIHR BioResource COVID-19 Collaboration. Kingston N., Graves B., et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. 2021;596:417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couper K.N., Blount D.G., Riley E.M. IL-10: the master regulator of immunity to infection. J. Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- D'Alessio F.R., Tsushima K., Aggarwal N.R., West E.E., Willett M.H., Britos M.F., Pipeling M.R., Brower R.G., Tuder R.M., McDyer J.F., King L.S. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J. Clin. Invest. 2009;119:2898–2913. doi: 10.1172/jci36498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der S.D., Zhou A., Williams B.R.G., Silverman R.H. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl. Acad. Sci. U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S.K., Delaney C., Toubai T., Ghosh A., Reddy P., Banerjee R., Yung R. Aging is associated with increased regulatory T-cell function. Aging Cell. 2014;13:441–448. doi: 10.1111/acel.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover P., Goel P.N., Greene M.I. Regulatory T cells: regulation of identity and function. Front. Immunol. 2021;12:750542. doi: 10.3389/fimmu.2021.750542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cao W., Zhu Y. Immunoregulatory functions of the IL-12 family of cytokines in antiviral systems. Viruses. 2019;11:772. doi: 10.3390/v11090772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniuda K., Fukao S., Kitamura D. Metabolic reprogramming induces germinal center B cell differentiation through Bcl6 locus remodeling. Cell Rep. 2020;33:108333. doi: 10.1016/j.celrep.2020.108333. [DOI] [PubMed] [Google Scholar]
- Hernandez-Vargas E.A., Wilk E., Canini L., Toapanta F.R., Binder S.C., Uvarovskii A., Ross T.M., Guzmán C.A., Perelson A.S., Meyer-Hermann M. Effects of aging on influenza virus infection dynamics. J. Virol. 2014;88:4123–4131. doi: 10.1128/jvi.03644-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D.A., Møller R., Uhl S.A., Oishi K., Frere J., Golynker I., Horiuchi S., Panis M., Blanco-Melo D., Sachs D., et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity. 2021;54:557–570.e5. doi: 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi S., Oishi K., Carrau L., Frere J., Møller R., Panis M., tenOever B.R. Immune memory from SARS-CoV-2 infection in hamsters provides variant-independent protection but still allows virus transmission. Sci. Immunol. 2021;6:eabm3131. doi: 10.1126/sciimmunol.abm3131. [DOI] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. A virological view of innate immune recognition. Annu. Rev. Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger A., Shimojima Y., Goronzy J.J., Weyand C.M. Regulatory T cells and the immune aging process: a mini-review. Gerontology. 2014;60:130–137. doi: 10.1159/000355303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten S.A., Ottenhoff T.H. Human CD4 and CD8 regulatory T cells in infectious diseases and vaccination. Hum. Immunol. 2008;69:760–770. doi: 10.1016/j.humimm.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Kaneko N., Kurata M., Yamamoto T., Morikawa S., Masumoto J. The role of interleukin-1 in general pathology. Inflamm. Regen. 2019;39:12. doi: 10.1186/s41232-019-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Greten F.R. NF-κB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- Kato H., Fujita T. Autoimmunity caused by constitutive activation of cytoplasmic viral RNA sensors. Cytokine Growth Factor Rev. 2014;25:739–743. doi: 10.1016/j.cytogfr.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Khalil B.A., Elemam N.M., Maghazachi A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021;19:976–988. doi: 10.1016/j.csbj.2021.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Koyama S., Ishii K.J., Coban C., Akira S. Innate immune response to viral infection. Cytokines. 2008;43:336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Kuchroo M., Huang J., Wong P., Grenier J.C., Shung D., Tong A., Lucas C., Klein J., Burkhardt D.B., Gigante S., et al. Multiscale PHATE identifies multimodal signatures of COVID-19. Nat. Biotechnol. 2022;40:681–691. doi: 10.1038/s41587-021-01186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni U., Zemans R.L., Smith C.A., Wood S.C., Deng J.C., Goldstein D.R. Excessive neutrophil levels in the lung underlie the age-associated increase in influenza mortality. Mucosal Immunol. 2019;12:545–554. doi: 10.1038/s41385-018-0115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo E.C., Torrance B.L., Keilich S.R., Al-Naggar I., Harrison A., Xu M., Bartley J.M., Haynes L. Senescence-induced changes in CD4 T cell differentiation can be alleviated by treatment with senolytics. Aging Cell. 2022;21:e13525. doi: 10.1111/acel.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Zhang H., Dauphars D.J., He Y.W. A potential role of interleukin 10 in COVID-19 pathogenesis. Trends Immunol. 2021;42:3–5. doi: 10.1016/j.it.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major J., Crotta S., Llorian M., McCabe T.M., Gad H.H., Priestnall S.L., Hartmann R., Wack A. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellett L., Khader S.A. S100A8/A9 in COVID-19 pathogenesis: impact on clinical outcomes. Cytokine Growth Factor Rev. 2021;63:90–97. doi: 10.1016/j.cytogfr.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D., Teo Q., Eggermont D., Lee H.H.Y., Thery F., Ho J., van Leur S.W., Ng W.W.S., Siu L.Y.L., Beling A., et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 2021;22:1416–1427. doi: 10.1038/s41590-021-01035-8. [DOI] [PubMed] [Google Scholar]
- Narasaraju T., Tang B.M., Herrmann M., Muller S., Chow V.T.K., Radic M. Neutrophilia and NETopathy as key pathologic drivers of progressive lung Impairment in patients with COVID-19. Front. Pharmacol. 2020;11:870. doi: 10.3389/fphar.2020.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson-Payant B.E., Uhl S., Grimont A., Doane A.S., Cohen P., Patel R.S., Higgins C.A., Acklin J.A., Bram Y., Chandar V., et al. The NF-κB transcriptional footprint is essential for SARS-CoV-2 replication. J. Virol. 2021;95:Jvi0125721. doi: 10.1128/JVI.01257-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., Fontanet A., Cauchemez S., Salje H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- Ohno M., Sasaki M., Orba Y., Sekiya T., Masum M.A., Ichii O., Sawamura T., Kakino A., Suzuki Y., Kida H., et al. Abnormal blood coagulation and kidney damage in aged hamsters infected with severe acute respiratory syndrome coronavirus 2. Viruses. 2021;13:2137. doi: 10.3390/v13112137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Pedrero M.Á., Osterhaus A.D.M.E., Becker T., Elbahesh H., Rimmelzwaan G.F., Saletti G. Aging and options to halt declining immunity to virus infections. Front. Immunol. 2021;12:681449. doi: 10.3389/fimmu.2021.681449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A., Iwasaki A. Type I and type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone L.A., Plowden J.K., García-Sastre A., Katz J.M., Tumpey T.M. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E., Atkins E., Nakayama T., Yoshie O., Groves I., Alcami A., Sinclair J. NF-κB-Mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86. J. Virol. 2008;82:4250–4256. doi: 10.1128/jvi.02156-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon M.M.L., Rybkina K., Kato Y., Kubota M., Matsumoto R., Bloom N.I., Zhang Z., Hastie K.M., Grifoni A., Weiskopf D., et al. SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci. Immunol. 2021;6:eabl9105. doi: 10.1126/sciimmunol.abl9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynor J., Lages C.S., Shehata H., Hildeman D.A., Chougnet C.A. Homeostasis and function of regulatory T cells in aging. Curr. Opin. Immunol. 2012;24:482–487. doi: 10.1016/j.coi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendeiro A.F., Ravichandran H., Bram Y., Chandar V., Kim J., Meydan C., Park J., Foox J., Hether T., Warren S., et al. The spatial landscape of lung pathology during COVID-19 progression. Nature. 2021;593:564–569. doi: 10.1038/s41586-021-03475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocamora-Reverte L., Melzer F.L., Würzner R., Weinberger B. The complex role of regulatory T cells in immunity and aging. Front. Immunol. 2020;11:616949. doi: 10.3389/fimmu.2020.616949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Meade-White K., Letko M., Clancy C., Hansen F., Liu Y., Okumura A., Tang-Huau T.L., Li R., Saturday G., et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microb. Infect. 2020;9:2673–2684. doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S., Ashour J., Maruyama T., Altenburg A.F., Cragnolini J.J., Bilate A., Avalos A.M., Kundrat L., García-Sastre A., Ploegh H.L. Type I interferon imposes a TSG101/ISG15 checkpoint at the Golgi for glycoprotein trafficking during influenza virus infection. Cell Host Microbe. 2013;14:510–521. doi: 10.1016/j.chom.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott N.R., Swanson R.V., Al-Hammadi N., Domingo-Gonzalez R., Rangel-Moreno J., Kriel B.A., Bucsan A.N., Das S., Ahmed M., Mehra S., et al. S100A8/A9 regulates CD11b expression and neutrophil recruitment during chronic tuberculosis. J. Clin. Invest. 2020;130:3098–3112. doi: 10.1172/jci130546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj P., Lien C.Z., Liu S., Stauft C.B., Nunez I.A., Hernandez M., Nimako E., Ortega M.A., Starost M.F., Dennis J.U., Wang T.T. SARS-CoV-2 infection induces protective immunity and limits transmission in Syrian hamsters. Life Sci. Alliance. 2021;4:e202000886. doi: 10.26508/lsa.202000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.C., Panda A., Joshi S.R., Qian F., Allore H.G., Montgomery R.R. Dysregulation of human Toll-like receptor function in aging. Ageing Res. Rev. 2011;10:346–353. doi: 10.1016/j.arr.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol C.L., Luster A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015;7:a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Li S. The role of ubiquitination in NF-κB signaling during virus infection. Viruses. 2021;13:145. doi: 10.3390/v13020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout-Delgado H.W., Du W., Shirali A.C., Booth C.J., Goldstein D.R. Aging promotes neutrophil-induced mortality by augmenting IL-17 production during viral infection. Cell Host Microbe. 2009;6:446–456. doi: 10.1016/j.chom.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein S.L., Wang W., Zhou S., Zhou Q., Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Parga T., Sehrawat S., Rouse B.T. Role of regulatory T cells during virus infection. Immunol. Rev. 2013;255:182–196. doi: 10.1111/imr.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Salinas L., Verdugo-Rodriguez A., Rodriguez L.L., Borca M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019;10:1057. doi: 10.3389/fmicb.2019.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H., et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/s1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Riedel K., Lynch P., Chien C.Y., Montelione G.T., Krug R.M. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. Rna. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Weiner A.I., Neupauer K.M., de Mello Costa M.F., Palashikar G., Adams-Tzivelekidis S., Mangalmurti N.S., Vaughan A.E. Regeneration of the pulmonary vascular endothelium after viral pneumonia requires COUP-TF2. Sci. Adv. 2020;6:eabc4493. doi: 10.1126/sciadv.abc4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number of the RNA-sequencing datasets reported in this paper can be found using NCBI GEO accession number GSE161200. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.