Abstract
The BRI1-EMS suppressor 1 (BES1)/brassinazole-resistant 1(BZR1) transcription factors play crucial roles in plant growth, development, and stress response. However, little is known about the function of maize’s BES1/BZR1s. In this study, the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes were cloned from maize’s inbred line, B73, and they were functionally evaluated by analyzing their expression pattern, subcellular localization, transcriptional activation activity, as well as their heterologous expression in Arabidopsis, respectively. The results of the qRT-PCR showed that the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes were predominantly expressed in the root, and their expression was significantly down-regulated by drought stress. The ZmBES1/BZR1-3 and ZmBES1/BZR1-9 proteins localized in the nucleus but showed no transcriptional activation activity as a monomer. Subsequently, it was found that the heterologous expression of the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes in Arabidopsis decreased drought tolerance, respectively. The transgenic lines showed a more serious wilting phenotype, shorter root length, lower fresh weight, and higher relative electrolyte leakage (REL) and malondialdehyde (MDA) content compared to the control under drought stress. The RNA-sequencing data showed that the 70.67% and 93.27% differentially expressed genes (DEGs) were significantly down-regulated in ZmBES1/BZR1-3 and ZmBES1/BZR1-9 transgenic Arabidopsis, respectively. The DEGs of ZmBES1/BZR1-3 gene’s expressing lines were mainly associated with oxidative stress response and amino acid metabolic process and enriched in phenylpropanoid biosynthesis and protein processing in the endoplasmic reticulum. But the DEGs of the ZmBES1/BZR1-9 gene’s expressing lines were predominantly annotated with water deprivation, extracellular stimuli, and jasmonic acid and enriched in phenylpropanoid biosynthesis and plant hormone signal transduction. Moreover, ZmBES1/BZR1-9 increased stomatal aperture in transgenic Arabidopsis under drought stress. This study indicates that ZmBES1/BZR1-3 and ZmBES1/BZR1-9 negatively regulate drought tolerance via different pathways in transgenic Arabidopsis, and it provides insights into the underlying the function of BES1/BZR1s in crops.
Keywords: maize, BES1/BZR1s, transcription factor, abiotic stress, drought
1. Introduction
Drought stress is one of the major environmental stimuli with deleterious effects on plant growth and development [1]. To survive under these changes, plants have developed multifaceted strategies at the morphology, physiology, and photosynthesis levels during the evolution process [2,3,4,5]. Among these cues, the induction of various transcription factors (TFs) regulating the expression of downstream genes is one of the most important approaches to plant stress responses [6,7]. The BRI1-EMS1 suppressor (BES1)/brassinazole-resistant 1 (BZR1) TFs are a kind of plant-specific TF family [8,9,10]. BES1/BZR1s are activated by brassinosteroid (BR); then, they bind to the E-box (CANNTG) or BRRE (CGTGT/CG) element of the downstream gene’s promoter to regulate their transcription and transduce the BR signal [9,11,12,13].
Previous studies have found that BES1/BZR1s are involved in various biological and developmental processes and in the stress response of plants. For instance, BES1 promotes the transcription of BEE1 (BR ENHANCED EXPRESSION 1) to control the flowering of the blue light signaling pathway [14]. BZR1 also enhances photosynthesis through upregulating the transcription of Calvin cycle genes, including FBA1, RCA1, FBP5, and PGK1 [15]. BZR1 and BES1 are antagonized by C3H15 to regulate the expression of the cell elongation-associated target gene, SMALL AUXIN-UP RNA 15 (SAUR15), to control cell elongation [16]. Likewise, BZR1 directly binds to the promoter of the HPPD (4-HYDROXYPHENYLPYRUVATE DIOXYGENASE) gene to suppress its expression, resulting in decreasing carotenoid production [17]. In addition, BES1/BZR1s interact with other proteins to regulate the transcription of downstream genes. The interaction of BZR1 and ARR (ARABIDOPSIS RESPONSE REGU LATORs) positively promotes ovule initiation and seed production [18]. BZR1 interacts with SPL9 to coordinately regulate the downstream genes’ expression to regulate the vegetative phase change and cell elongation [19]. In Arabidopsis, BES1 interacts with drought-induced RD26 and WRKY54 TFs to negatively regulate its drought response [20,21]; but TaBZR2 and ZmBES1/BZR1-5, the BES1/BZR1 family member from wheat and maize, positively regulate drought response by activating downstream genes, respectively [22,23]. Besides, the BES1/BZR1 family members promote heat and freezing tolerance via directly regulating the expression of HSF (heat shock factor), CBF (C-repeat binding factor), and COR (cold-regulated) genes, respectively [24,25]. However, the function of BES1/BZR1 in crops remains obscure.
In previous studies, it has been demonstrated that there are eleven ZmBES1/BZR1 TFs in the maize genome [26,27]. However, only ZmBES1/BZR1-2 (Zm00001d039439) and ZmBES1/BZR1-5 (Zm00001d053975) are functionally validated in regulating seed size and stress response [23,28,29]. The function of other ZmBES1/BZR1 genes remains unknown. In this study, the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes were cloned, and their function was evaluated by analyzing their expression pattern under drought stress, subcellular localization, transcriptional activation activity, as well as the heterologous expression in transgenic Arabidopsis.
2. Results
2.1. Expression Patterns of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 in Maize
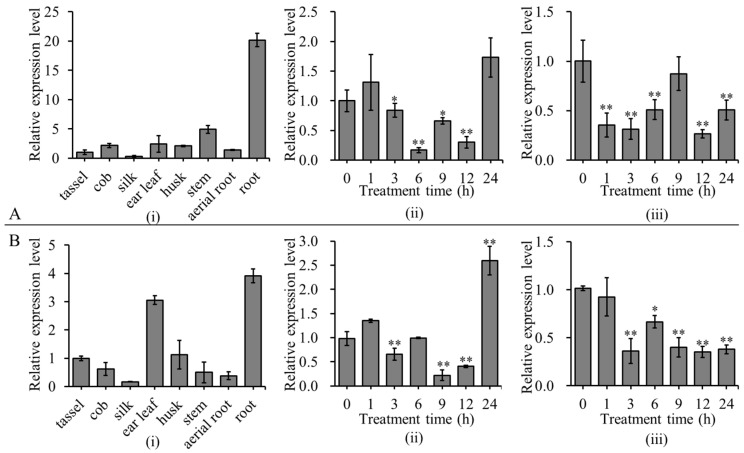
To analyze the tissue-specific expression and stress-responsive expression of ZmBES1/BZR1-3 and ZmBES1/BZR1-9, a quantitative real-time PCR (qRT-PCR) was performed. As shown in Figure 1, the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes were predominantly expressed in maize roots; likewise, they were highly expressed in leaves. In addition, the expression of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes under drought stress showed a similar pattern. Their expression in the shoot and root of seedlings was both significantly inhibited by drought stress. The expression of ZmBES1/BZR1-3 reached the lowest level in the shoot and root at 6 h and 12 h of treatment, respectively. The expression of ZmBES1/BZR1-9 reached the lowest level in the shoot and root at 9 h and 12 h of treatment, respectively. However, the expression of ZmBES1/BZR1-9 was significantly up-regulated at 24 h of treatment. The result suggests that ZmBES1/BZR1-3 and ZmBES1/BZR1-9 might function in regulating drought tolerance.
Figure 1.
Expression patterns of ZmBES1/BZR1-3 (A) and ZmBES1/BZR1-9 (B). (i) Expression pattern in different tissues of maize. (ii); (iii) expression pattern in the shoot and root under drought stress, respectively. The three-leaf old seedlings of maize’s inbred line, B73, were subjected to 16% PEG-6000 to mimic drought stress. The relative expression level was calculated and normalized using the 2−ΔΔCT method of the CFX Manger™ software version 2.0 (Bio-Rad, Berkeley, CA, USA). * and ** represent p < 0.05 and p < 0.01 by the Student’s t-test, respectively.
2.2. ZmBES1/BZR1-3 and ZmBES1/BZR1-9 Localized in the Nucleus
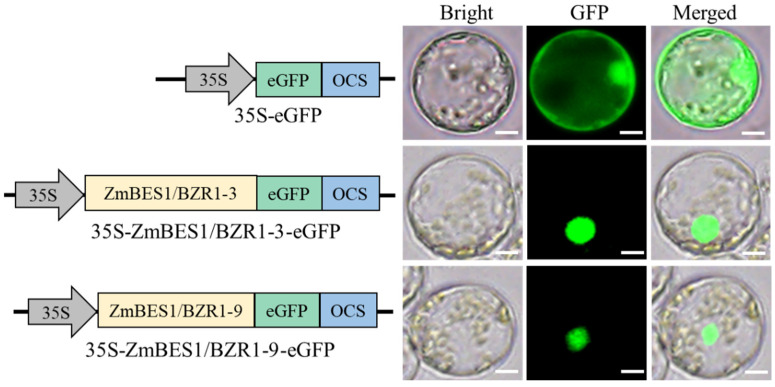
BES1/BZR1 TF accumulates in the nucleus to regulate the downstream gene’s expression. Therefore, the open reading frame (ORF) of ZmBES1/BZR1-3 and ZmBES1/BZR1-9, without a stop codon, was amplified and inserted into pC2300-35S-eGFP fused to eGFP and used to transient the expression in maize’s mesophyll protoplasts. The result exhibited that the green fluorescence (GFP) was observed in the whole cell, including the cytoplasm and nucleus of protoplasts transformed by 35S-eGFP’s empty vector. However, the GFP was only observed in the nucleus of protoplasts transformed by 35S-ZmBES1/BZR1-3-eGFP and 35S-ZmBES1/BZR1-9-eGFP, respectively (Figure 2).
Figure 2.
Subcellular localization. The ORF of ZmBES1/BZR1-3 and ZmBES1/BZR1-9, without a stop codon, was fused to eGFP and transiently expressed in maize’s mesophyll protoplasts for 12 h. The scale bar was 20 μm.
2.3. ZmBES1/BZR1-3 and ZmBES1/BZR1-9 Showed no Transcriptional Activation Activity in Yeast
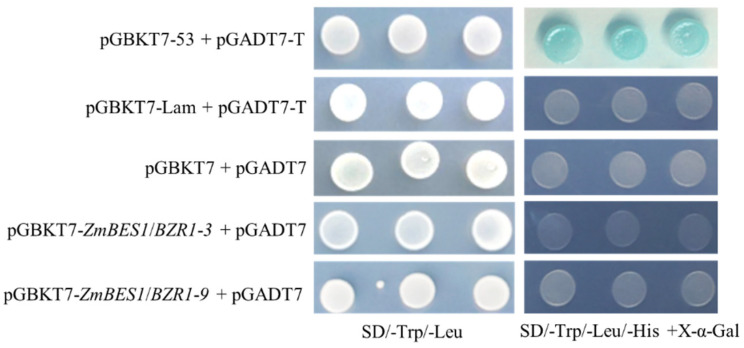
To verify the transcriptional activation activity of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 proteins, the ORF of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 was cloned, inserted into pGBKT7, and transformed into yeast AH109 with pGADT7, respectively. The results exhibited that the yeast strains, co-transformed by pGBKT7-ZmBES1/BZR1-3 with pGADT7, pGBKT7-ZmBES1/BZR1-9 with pGADT7, as well as the positive control (pGBKT7-53 with pGADT7-T) and negative control (pGBKT7-Lam with pGADT7-T, and pGBKT7 with pGADT7), grew normally and produced clones on SD/-Leu/-Trp plates, respectively. However, only the yeast strains that co-transformed pGBKT7-53 with pGADT7-T (positive control) could grow and turn blue on SD/-Leu/-Trp/-His with X-α-Gal plates. The yeast strains co-transformed by pGBKT7-ZmBES1/BZR1-3 with pGADT7, pGBKT7-ZmBES1/BZR1-9 with pGADT7, as well as the negative control could not grow on SD/-Leu/-Trp/-His with X-α-Gal plates (Figure 3). This finding suggests that ZmBES1/BZR1-3 and ZmBES1/BZR1-9 have no transcriptional activation activity as a monomer in yeast.
Figure 3.
The transcriptional activation assay. The pGBKT7-ZmBES1/BZR1-3 with pGADT7, and pGBKT7-ZmBES1/BZR1-9 with pGADT7, were co-transformed into yeast, respectively. The co-transformation of pGBKT7-53 with pGADT7-T was used as the positive control. The co-transformation pGBKT7-Lam with pGADT7-T and pGBKT7 with pGADT7 were used as the negative control.
2.4. ZmBES1/BZR1-3 and ZmBES1/BZR1-9 Negatively Regulated Drought Tolerance in Arabidopsis
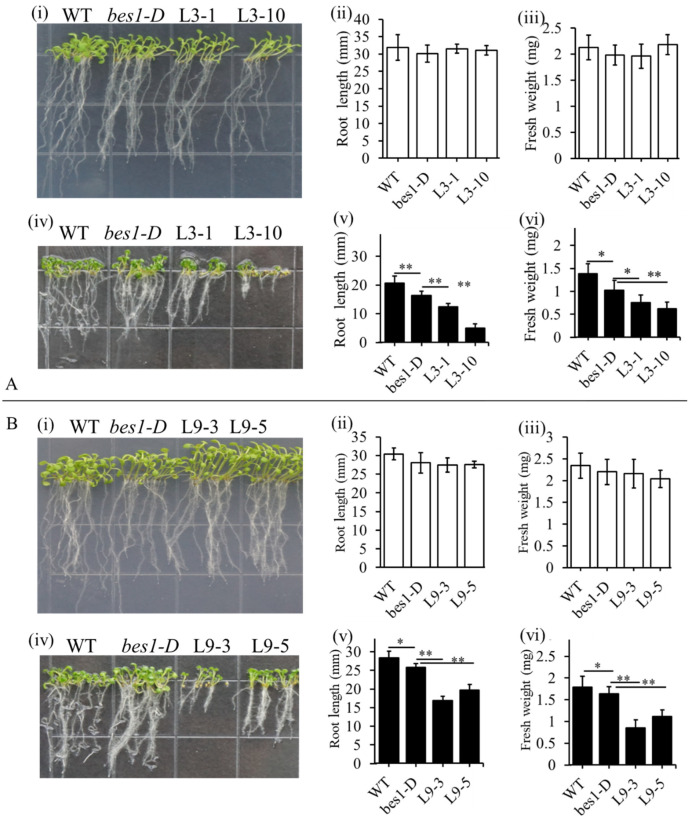
To investigate the function of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 in the drought stress response, they were transformed into bes1-D, the Arabidopsis AtBES1 mutant, for phenotyping under the stress treatment. As shown in Figure 4, all transgenic lines expressing ZmBES1/BZR1-3 (L3-1 and L3-10) or ZmBES1/BZR1-9 (L9-3 and L9-5) showed no difference compared to bes1-D and the wild type (WT) on the 1/2 MS plates. However, under drought stress mimicked by a mannitol treatment, the transgenic lines L3-1, L3-10, L9-3, and L9-5 were much more sensitive to drought stress compared to the bes1-D seedlings that were more sensitive than WT, although all of the plants were inhibited by stress. The L3-1 and L3-10 lines showed significant differences compared to bes1-D under a 200 mM mannitol treatment, while L9-3 and L9-5 exhibited differences compared to bes1-D under a 150 mM mannitol treatment. The root length and fresh weight of the L3-1, L3-10, L9-3, and L9-5 lines were both significantly lower than bes1-D on the 1/2 MS plates with mannitol. Meanwhile, the root length and fresh weight of bes1-D were significantly lower than the WT.
Figure 4.
The phenotype of transgenic plants under drought stress mimicking by mannitol. (A) Phenotype of ZmBES1/BZR1-3 expressing lines. (B) The phenotype of the ZmBES1/BZR1-9 expressing lines. (i), (ii), (iii) The phenotype, root length, and fresh weight of every line on 1/2 MS plates, respectively. (iv), (v), (vi) The phenotype, root length, and fresh weight of every line on 1/2 MS plates with mannitol, respectively. The seeds of every line were surfaced-sterilized, planted on 1/2 MS plates without (control) or with mannitol, and vertically cultured for three weeks. All data are means (±SD) of three biological replicates. L3-1 and L3-10 are homozygous lines expressing ZmBES1/BZR1-3. L9-3 and L9-5 are homozygous lines expressing ZmBES1/BZR1-9. bes1-D, untransformed mutant. WT, wild type. * and ** represents p < 0.05 and p < 0.01, respectively.
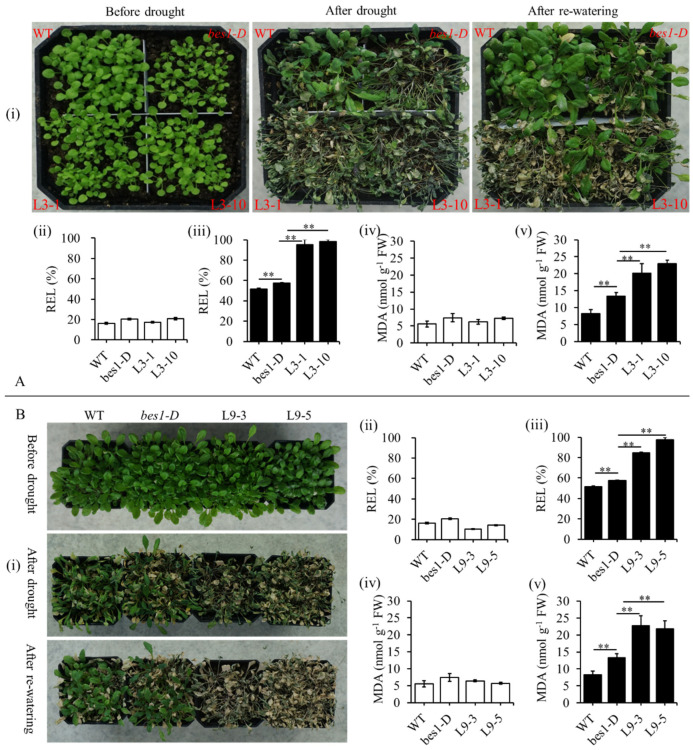
Subsequently, the seeds of the L3-1, L3-10, L9-3, L9-5, bes1-D, and WT were sown into the soil for drought treatment. As shown in Figure 5, before the treatment, all of the plants grew well and showed a vigorous phenotype. The relative electrolyte leakage (REL) and malondialdehyde (MDA) content of all of the plants kept a low level and showed no significant difference. Whereas, after three weeks of drought stress without watering, the L3-1, L3-10, L9-3, and L9-5 lines showed serious a wilting phenotype, but bes1-D and WT showed slighter and greener phenotype compared to the transgenic lines. The REL and MDA content of the L3-1, L3-10, L9-3, and L9-5 lines was significantly higher than bes1-D and the WT, although all of them were increased after the drought treatment. Meanwhile, the bes1-D was more sensitive to drought stress than the WT.
Figure 5.
The phenotype of transgenic plants under the drought treatment. (A) The phenotype of the ZmBES1/BZR1-3 expressing lines. (B) The phenotype of the ZmBES1/BZR1-9 expressing lines. (i) The phenotype, (ii), (iv) the REL and MDA content before treatment, respectively. (iii), (v) The REL and MAD content after treatment, respectively. The seeds of every line were planted on the soil, cultured in the chamber, and irrigated at 4-day intervals. The 20-day-old seedlings were used for the drought treatment by holding water and subsequently monitored for the stress symptoms for three weeks. All of the values are means (±SD) of three biological replicates. L3-1, L3-10, L9-3, and L9-5 are transgenic lines; bes1-D, mutant. WT, wild type. ** represents p < 0.05 and p < 0.01, respectively.
The above results indicate that the expression of the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes both decrease the drought tolerance in transgenic Arabidopsis.
2.5. ZmBES1/BZR1-3 and ZmBES1/BZR1-9 Inhibited Transcription of Stress-Related Genes in Transgenic Arabidopsis
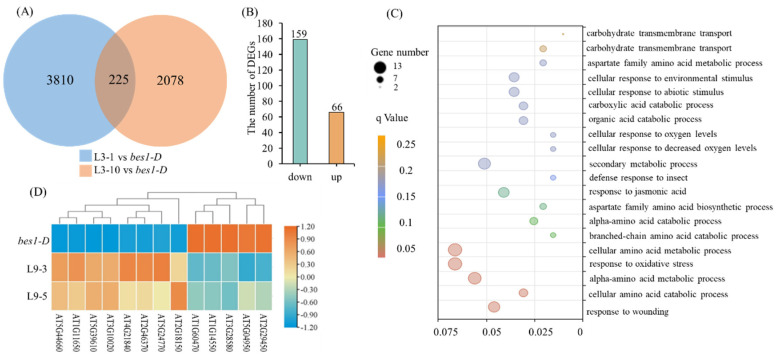
To explore the potential mechanism of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 negatively regulating the drought tolerance, the differentially expressed genes (DEGs) in the transgenic lines, compared to bes1-D, were unraveled by the RNA-sequencing (RNA-seq). In transgenic Arabidopsis with the ZmBES1/BZR1-3 gene, there were 225 common DEGs shared by the L3-1 and L3-10 lines (Figure 6A and Table S1 in the Supplementary Materials). Among these common DEGs, 70.67% DEGs (159) were down-regulated, and 29.33% DEGs (66) were up-regulated, respectively (Figure 6B). Gene Ontology (GO) analysis exhibited that the common DEGs were mainly enriched in the oxidative stress response and amino acid metabolic process (Figure 6C and Table S2). In addition, the KEGG enrichment analysis indicated that these common DEGs participated in phenylpropanoid biosynthesis and protein processing in the endoplasmic reticulum (Figure S1).
Figure 6.
DEGs of transgenic Arabidopsis with the ZmBES1/BZR1-3 gene compared to bes1-D. (A) DEGs of L3-1 and L3-10 compared to bes1-D, respectively. (B) The common DEGs are shared by L3-1 and L3-10. (C) GO analysis of common DEGs. (D) The heat map of the 13 DEGs responds to oxidative stress.
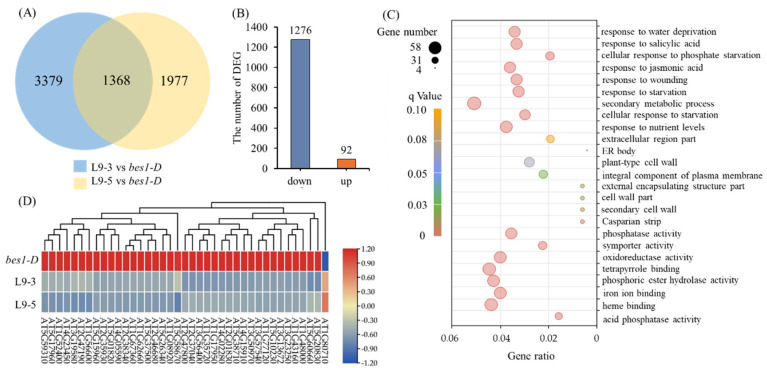
In transgenic Arabidopsis with the ZmBES1/BZR1-9 gene, there were 1368 common DEGs shared by the L9-3 and L9-5 lines (Figure 7A and Table S3). Among these DEGs, 93.27% of the DEGs (1276) were down-regulated, and only 6.73% of the DEGs (92) were up-regulated compared to bes1-D, respectively (Figure 7B). Furthermore, the result of the GO analysis showed that 39, 43, and 41 DEGs were annotated with response to water deprivation, extracellular stimuli, and jasmonic acid, respectively (Figure 7C and Table S4). Among the 39 DEGs that respond to water deprivation, 38 DEGs were down-regulated, and only one DEG was up-regulated (Figure 7D). Specifically, 12 down-regulated DEGs were well confirmed to positively regulate drought stress, including four genes (AT2G46680, AT2G47800, AT4G23450, and AT5G37500) positively regulate stomatal aperture, and one up-regulated DEG (AT1G80710) negatively regulates drought stress (Table S3). KEGG enrichment analysis showed that the DEGs were mainly enriched in phenylpropanoid biosynthesis and the plant hormone signal transduction pathways (Figure S2).
Figure 7.
DEGs of transgenic Arabidopsis with the ZmBES1/BZR1-9 gene compared to bes1-D. (A) DEGs of L9-3 and L9-5 compared to bes1-D, respectively. (B) The common DEGs are shared by L9-3 and L9-5. (C) GO analysis of common DEGs. (D) The heat map of the 39 DEGs responds to water deprivation.
2.6. ZmBES1/BZR1-9 Increased Stomatal Aperture
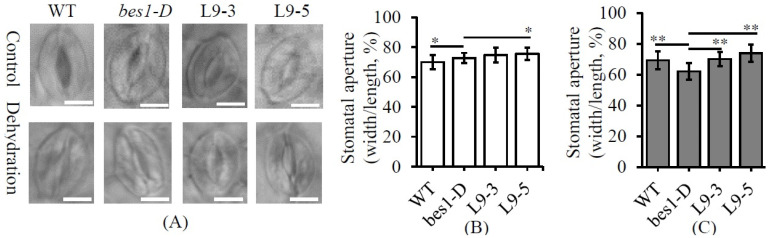
Due to the four stomatal development genes, including AT4G23450 (AtAIRP1), AT4G23450 (AtMRP4), AT2G46680 (AtHB7), and AT5G37500 (AtGORK), were significantly down-regulated in the ZmBES1/BZR1-9 transgenic lines, the stomatal phenotype of L9-3 and L9-5 was monitored. As shown in Figure 8, under an optimal condition, the stomata exhibited an opened status, and the stomatal aperture of L9-5 was significantly larger than bes1-D. However, after 1 h of dehydration, most of the stomata of L9-3 and L9-5 were kept open. The stomatal aperture of L9-3 and L9-5 was greatly bigger than that of bes1-D, respectively. Likewise, the stomatal aperture of bes1-D was greatly smaller than the WT. These results suggest that enhanced stomatal aperture was one cause of drought sensitivity in the L9-3 and L9-5 plants.
Figure 8.
The stomatal phenotype. (A) The stomatal morphology. (B) The stomatal aperture before dehydration. (C) The stomatal aperture after 1 h of dehydration. The leaf of every line was detached for 1 h of dehydration treatment and used to measure the stomatal aperture. All values are means (±SD) of three biological replicates. L9-3 and L9-5 are transgenic lines. bes1-D, mutant. WT, wild type. * and ** represents p < 0.05 and p < 0.01, respectively. The scale bar was 10 μm.
3. Discussion
The BES1/BZR1 family has been identified as the TF in several plant species and accumulates in the nucleus to activate or inhibit the downstream genes’ expression [26,30,31,32,33]. In the present study, we found that the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 proteins localized in the nucleus and had no transcriptional activation activity (Figure 2 and Figure 3), which implies that ZmBES1/BZR1-3 and ZmBES1/BZR1-9 may function as TFs to regulate the downstream genes via requiring co-factors. Previous studies show that BES1/BZR1 interacts with other TFs to co-regulate their target genes’ expression [19,20,21,22]. Likewise, in our previous study, ZmBES1/BZR1-5 was also found to have no transcriptional activation as a monomer but functioned via homodimerization [28].
The gene expression patterns can provide information about gene function [34]. We found that the expression of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 in maize was significantly inhibited by drought stress, although ZmBES1/BZR1-9 was up-regulated at 24 h of treatment (Figure 1). It suggests that the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 function in drought tolerance. Because the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 shared a highly conserved bHLH domain, the characterised domain of BES1/BZR1, with Arabidopsis BES1/BZR1s (Supplementary Figure S3) [9,23,26,28]; hence, they were heterologously expressed in the bes1-D mutant. After the evaluation of the drought tolerance of transgenic Arabidopsis, it’s found that the heterologous expression of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 in the bes1-D mutant both increase drought sensitivity of the transgenic lines (Figure 5). The bes1-D mutant also exhibited more sensitivity to drought stress than the WT in the study (Figure 4 and Figure 5). It was previously reported that bes1-D negatively regulates drought tolerance [20]. However, TaBZR2 and ZmBES1/BZR1-5 were reported to regulate the drought tolerance positively [22,23]. These findings mean that the BES1/BZR1 family members evolve a functional diversity and play dual roles in drought response. In addition, ZmBES1/BZR1-3 and ZmBES1/BZR1-9 were predominantly expressed in maize roots (Figure 1). The transgenic lines, likewise, exhibited shorter roots after mimicking drought stress (Figure 4). This concluded that ZmBES1/BZR1-3 and ZmBES1/BZR1-9 suppressed the root development, which resulted in an increase in drought sensitivity. Adjusting the root system architecture is one important clue for plants to conquer stress due to the root being the first defensive line to adapt to their environment [35,36]. Moreover, bes1-D had a shorter meristem size in the root [37]. BES1 regulates the vascular cell development in the root apex and root growth mediated by the type one protein phosphatases [37,38]. Besides, BES1 acts to promote seedling growth in the auxin pathway [39].
Although ZmBES1/BZR1-3 and ZmBES1/BZR1-9 showed similar expression patterns (Figure 1), ZmBES1/BZR1-3 regulated the expression of the oxidative-stress responsive genes (Figure 6), while ZmBES1/BZR1-9 inhibited the water response genes’ expression (Figure 7), suggesting that they act on different signaling. These DEGs were enriched in phenylpropanoid biosynthesis, a protein processing in the ER, and hormone signal transduction (Figures S1 and S2), which showed positive roles in the drought stress response [5,40,41]. Among the 13 oxidative-stress responsive DEGs of the ZmBES1/BZR1-3 expressing lines, the up-regulated gene, AT5G39610 (ORE1 or NAC092), negatively regulates oxidative stress [42,43], and the down-regulated gene AT1G60470 (Galactinol synthase, GolS) positively improves the drought tolerance [44,45]. Moreover, the transgenic lines with the ZmBES1/BZR1-9 gene also showed the stomatal insensitivity to drought compared to bes1-D, while the stomatal aperture of the bes1-D plants was smaller than the WT under stress (Figure 8). It could be explained that AtBES1 regulated drought tolerance via acting on the RD26 and WRKY TFs [21,22]. However, ZmBES1/BZR1-9 inhibited the expression of AT4G23450 (AtAIRP1) and AT2G47800 (AtMRP4), and the AT2G46680 (AtHB7) and AT5G37500 (AtGORK) genes (Figure 7 and Table S2), which has been proved to positively regulate drought tolerance via a coordinating stomatal movement [46,47]. Previous studies confirmed that many genes in the BR signaling pathway played a positive role in controlling the stomatal open [48]. Recently, BZR1 was found to promote the sugar-promoted expression of β-AMYLASE1 (BAM1), which was responsible for starch degradation in the guard cells and increased the effects of BR on the stomatal opening [49,50].
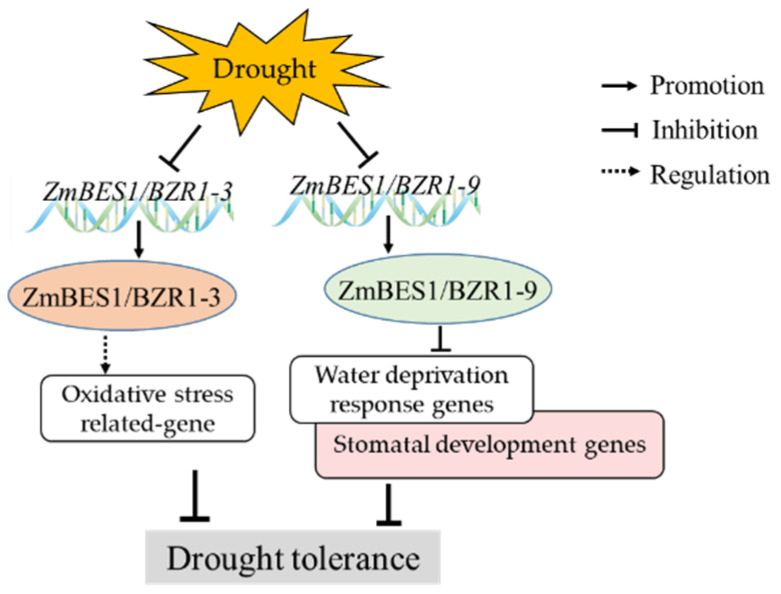
In conclusion, the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes were cloned from maize and confirmed to be inhibited by drought stress in the present study. The ZmBES1/BZR1-3 and ZmBES1/BZR1-9 proteins both localized in the nucleus and showed no transcriptional activation activity as the monomer. However, the heterologous expression of the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes negatively regulate drought tolerance in Arabidopsis via different pathways (Figure 9).
Figure 9.
Diagram of the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 regulated the network in the drought response.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The seeds of the maize’s inbred line, B73, were germinated in filter paper, then transplanted into a Hoagland’s solution for a hydroponic culture under 16 h of light at 28 °C/8 h and in the dark at 25 °C. As described by Sun et al. [23], at the three-leaf old stage, the seedlings of the same size were subjected to 16% PEG-6000 to mimic drought stress. At 0, 1, 3, 6, 9, 12, and 24 h of treatment, the shoot and root were sampled, frozen in liquid nitrogen, and stocked at −80 °C for an RNA isolation. Meanwhile, the tassel, cob, silk, ear leaf, husk, stem, aerial root, and root of B73 were sampled for an RNA extraction, respectively. The seeds of the Arabidopsis BES1 gene mutant (bes1-D, CS65988) were purchased from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/, accessed on 8 August 2019) and grown in a greenhouse at 22 °C and 60–70% relative humidity under a 10 h light/14 h dark photoperiod.
4.2. Expression Analysis of the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 Gene
The total RNA was extracted from the above samples by an RNAiso plus kit (TaKaRa, Dalian, China), removed genomic DNA by digesting with RNAse-free DNAse (TaKaRa, Dalian, China), quantified using NanoDropTM OneC (ThermoScientific, Waltham, MA, USA), and reversely transcribed into cDNA according to the PrimeScriptTM reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s instruction, respectively. The specific primer pairs of D3-F (5′-CGCTATCGTGTGGAACCTGA-3′) with D3-R (5′-CACGAGACCGTAACTCCGTC-3′), and D9-F (5′-GCTTACAACGGCCTCAGCTA-3′) with D9-R (5ʹ-GTAGCGGG-ACAGGTTGTTGA-3′), were designed using the Primer-BLAST at the NCBI website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome, accessed on 28 March 2020), synthesized at Sangon Biotech (Chendu, China), and used to amplify the 98 and 192 bp of the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 genes, respectively. A pair of primers, TUB-F (5′-CTACCTCACGGCATCTGCTATGT-3′) and TUB-R (5′-GTCACACACACTCGACTTCACG-3′), were designed, synthesized, and used to amplify the 139 bp of the ZmTUB gene, which was used as an internal reference. According to the methods of Sun et al. [23], the qRT-PCR was performed in the Bio-Rad CFX96TM Real-Time PCR system with a TransScript® II Two-Step RT-PCR SuperMix (Transgen, Beijing, China) according to the manufacturer’s instructions.
4.3. Subcellular Localization
The ORF sequence without a stop codon of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 was cloned from a cDNA sample using the primers PC-3F (5′-CGGGATCCATGGAGGGAGGGGTAGGAGCGGGAGGAAG-3′; the underlined sequences are the BamH I sites) with PC-3R (5′-ACGCGTCGACCGAAAACCTTTCTTGCTGCTCCCTCATAG-3′; the underlined sequences are the Sal I sites), and PC-9F (5′-TCCCCCGGGATGAACGGGGGAGAAGGAGAAGGAG-3′ the underlined sequences are the Sma I sites) with PC-9R (5′-GGACTAGTAGCGCGGTCAGCGCCGG-3′; the underlined sequences are the Spe I sites), respectively. The PCR products and pC2300-35S-eGFP plasmids were digested using BamH I/Sal I and Sma I/Spe I, respectively. Then, the ORF of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 was inserted into the BamH I/Sal I sites, and the Sma I/Spe I sites of pC2300-35S-eGFP to generate 35S-ZmBES1/BZR1-3-eGFP and 35S-ZmBES1/BZR1-9-eGFP, respectively. According to the methods described by Fu et al. [51], the 20 g leaves from the etiolated seedlings were cut into strips and digested into a 50 mL tube containing a 10 mL enzyme solution with 1.5% cellulase R10 (Yakult, Japan), 0.3% macerozyme R10 (Yakult, Japan), 0.6 M D-mannitol (Sigma, MO, USA), 10 mM MES (pH 5.7) (Sigma), 1 mM CaCl2 (Sigma), and 0.1% BSA (Sigma) for 6 h at 28 °C. The protoplasts were collected by centrifugating at 100× g and resuspended using a W5 solution (pH 5.7) with 154 mM NaCl (Sigma), 125 mM CaCl2 (Sigma), 5 mM KCl (Sigma), and 2 mM MES (Sigma). Then, the maize’s mesophyll protoplasts were used to transiently express the ZmBES1/BZR1-3 and ZmBES1/BZR1-9 proteins. The 35S-ZmBES1/BZR1-3-eGFP, 35S-ZmBES1/BZR1-9-eGFP, and 35S-eGFP plasmid (control) were transformed into the 100 μL protoplasts, respectively. Subsequently, the protoplasts were incubated at 25 °C in the dark for 12 h, sampled, and used for GFP fluorescence imaging using the confocal microscope LSM800 (Carl Zeiss, Oberkochen, Germany).
4.4. Transcriptional Activation Activity Assay in Yeast
The transcriptional activation activity assay was performed as described by Lu et al. [52]; the ORF sequence of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 was cloned from a cDNA sample using primers BD-3F (5′-AGAATTCATGGAGGGAGGGGTAGGAG-3′; the underlined sequences are the EcoR I sites) with BD-3R (5′-CGGGATCCTCACGAAAACCTTTCTTGCTGC-3′; the underlined sequences are the BamH I sites), and BD-9F (5′-TTACAATCATATGATGAACGGGGGAGAAGGA-3′; the underlined sequences are the Nde I sites) with BD-9R (5′-TGAATTCTCAAGCGCGGTCAGC-3ʹ; the underlined sequences are the EcoR I sites), respectively. The PCR products and pGBKT7 plasmids were digested using EcoR I/BamH I and Nde I/EcoR I, respectively. Then, the ORF of ZmBES1/BZR1-3 and ZmBES1/BZR1-9 was inserted into the EcoR I/BamH I and Nde I/EcoR I sites of pGBKT7 to generate pGBKT7-ZmBES1/BZR1-3 and pGBKT7-ZmBES1/BZR1-9, respectively. The pGBKT7-ZmBES1/BZR1-3 and pGBKT7-ZmBES1/BZR1-9 with the combination of pGADT7 were co-transformed into the yeast strain, AH109, respectively, and were cultured on SD/-Leu/-Trp and SD/-Leu/-Trp/-His/+X-α-Gal. The co-transformation of pGBKT7-53 and pGADT7-T was used as the positive control. The co-transformation of pGBKT7-Lam and pGADT7-T, as well as pGBKT7 and pGADT7, was used as the negative control. The yeast strains were cultured 3 days at 28 °C and used for phenotyping.
4.5. Plant Transformation and Phenotyping
The above constructs of the 35S-ZmBES1/BZR1-3 and 35S-ZmBES1/BZR1-9 plasmids were transformed into the Agrobacterium tumefaciens strain GV3101. Subsequently, the Arabidopsis plants (bes1-D mutant, CS65988) were transformed by the GV3101 strains with ZmBES1/BZR1-3 and ZmBES1/BZR1-9 through the floral-dipping method, respectively. The seeds of the T0 plants were collected, surface-sterilized using 75% ethanol for 1 min and in 5% NaClO for 5 min, washed three times with sterile distilled water, then planted on 1/2 MS plates with 50 mg/L kanamycin (Sigma, Saint Louis, MO, USA) for screening of positive transformants. The homozygous lines without the segregation of kanamycin-resistance in T3 were screened using the same method. The homozygous lines L3-1 and L3-10 of ZmBES1/BZR1-3 expressing Arabidopsis, L9-3, and L9-5 of ZmBES1/BZR1-3 expressing Arabidopsis were selected for the next studies.
The seeds of the homozygous lines L3-1, L3-10, L9-3, and L9-5, and the WT and bes1-D were surfaced-sterilized, sown on 1/2 MS plates with 0, 150, and 200 mM mannitol and vertically cultured for three weeks. Then the phenotype was photographed. The root length and fresh weight of every line were measured, respectively. Meanwhile, the seeds of every line were grown in soil in a growth chamber and irrigated at 4-day intervals. The 20-day-old seedlings were divided into four groups. Three of them were used for drought treatment by holding water and subsequently monitored for stress symptoms. Two weeks later, two groups of seedlings treated by drought with one group of seedlings under optimal conditions were used to determine the REL and MDA content according to the methods of Sun et al. [23] and Yu et al. [53], respectively. Another group of seedlings was treated for three weeks, then re-watered with a recovery time of three days and used to photograph wilting phenotype.
4.6. RNA-Seq Analysis
For the RNA-seq analysis, the total RNA was extracted from the 14-day-old seedlings of bes1-D, L3-1, L3-10, L9-3, and L9-10 using an RNAprep Pure Plant Kit (Tiangen, Beijing, China), respectively. The RNA’s integrity was evaluated by Bioanalyzer 2100 (Agilent, CA, USA) (RIN ≥ 7, 28S/18S ≥ 1.5). According to the methods of Sun et al. [23], the RNA-seq library was constructed using a VAHTSTM mRNA-seq V2 Library Prep Kit and sequenced using a novaseq6000 system at the Beijing Novogene Technology Company (Beijing, China). The sequencing data were evaluated, filtered, mapped to the Arabidopsis genome (TAIR 10), and used for assembling the transcripts and calculating the gene expression using FastQC (version 0.11.2), Trimmomatic (version 0.36) hisat2, and StringTie, respectively [54,55]. The DEGs were analyzed using the DESeq2 R package (1.20.0) with a p-value < 0.05 and |FoldChange| > 1. GO, and the KEGG enrichment analysis of the DEGs was implemented by the clusterProfiler.
4.7. Measurement of Stomatal Aperture
A batch of the rosette leaves of the three-week-old seedlings from L9-3, L9-5, bes1-D, and the WT were detached and immediately used for the measurement of the stomatal aperture. Another batch of leaves from the same plants was detached and dehydrated for 1 h at room temperature. The stomatal apertures in the epidermal peels were observed under the Confocal microscope LSM800 (Carl Zeiss, Oberkochen, Germany). For each line, five leaves from ten plants were selected for the assay. Forty stomata per leaf were measured for the stomatal width and length using ImageJ software (https://imagej.nih.gov/ij/, accessed on 19 January 2021), as described by Huque et al. [56].
4.8. Statistical Analysis
All of the experiments were carried out in three replicates. The data are the mean ± standard deviation (SD) of three biological replicates. The significance was analyzed in the Student’s t tests. The * and ** represents p < 0.05 and p < 0.01, respectively.
Acknowledgments
The authors thank the technical support from the Key Laboratory of Biology and Genetic Improvement of Maize in the Southwest Region.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23116025/s1.
Author Contributions
Conceptualization, W.F. and H.Y.; methodology, W.F., Y.L. (Yuan Liu) and Y.C.; software, W.F.; formal analysis, F.S., X.Z. and Q.Y.; investigation, Y.Z. and H.Z.; writing—original draft preparation, W.F.; writing—review and editing, H.Y. and X.Z.; supervision, W.L., Y.L. (Yanli Lu) and X.Z.; project administration, F.F. and H.Y.; funding acquisition, H.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Sichuan Science and Technology Program (2022YFH0067), the National Nature Science Foundation of China (32001552), the Key Research and Development Project of Chengdu (2021-YF05-02024-SN).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sofia A., de Almeida A.M., da Silva A.B., da Silva J.M., Paula A., Santos D., Fevereiro P., de Sousa Araujo S. Abiotic Stress—Plant Responses and Applications in Agriculture. IntechOpen; London, UK: 2013. Abiotic Stress Responses in plants: Unraveling the complexity of genes and networks to survive. [DOI] [Google Scholar]
- 2.Zong N., Li X.J., Wang L., Wang Y., Wen H.T., Li L., Zhang X., Fan Y.L., Zhao J. Maize ABP2 enhances tolerance to drought and salt stress in transgenic Arabidopsis. J. Integr. Agric. 2018;17:2379–2393. doi: 10.1016/S2095-3119(18)61947-1. [DOI] [Google Scholar]
- 3.Wang C., Lu G., Hao Y., Guo H., Guo Y., Zhao J., Cheng H. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta. 2017;246:453–469. doi: 10.1007/s00425-017-2704-x. [DOI] [PubMed] [Google Scholar]
- 4.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J.K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirilae V., Kauppinen K. Stress signal transduction: Components, pathways and network integration. Der Hautarzt Z. für Dermatol. Venerol. Und Verwandte Geb. 2006;10:3–29. [Google Scholar]
- 7.Singh K.B., Foley R.C., Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z.Y., Wang Q., Chong K., Wang F., Wang L., Bai M., Jia C. The brassinosteroid signal transduction pathway. Cell Res. 2006;16:427–434. doi: 10.1038/sj.cr.7310054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 10.Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong H., Chu C. Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 2018;23:1016–1028. doi: 10.1016/j.tplants.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Li Q.F., Lu J., Yu J.W., Zhang C.Q., He J.X., Liu Q.Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta-Gene Regul. Mech. 2018;1861:561–571. doi: 10.1016/j.bbagrm.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z.-Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., et al. Nuclear-Localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/S1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang F., Gao Y., Liu Y., Zhang X., Gu X., Ma D., Zhao Z., Yuan Z., Xue H., Liu H. BES1-regulated BEE1 controls photoperiodic flowering downstream of blue light signaling pathway in Arabidopsis. New Phytol. 2019;223:1407–1419. doi: 10.1111/nph.15866. [DOI] [PubMed] [Google Scholar]
- 15.Yin X., Tang M., Xia X., Yu J. BRASSINAZOLE RESISTANT 1 mediates brassinosteroid-induced calvin cycle to promote photosynthesis in tomato. Front. Plant Sci. 2022;12:811948. doi: 10.3389/fpls.2021.811948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai G., Qi G., Wang D., Zhuang Y., Xu H., Bai Z., Bai M.-Y., Hu R., Wang Z., Zhou G., et al. The CCCH zinc finger protein C3H15 negatively regulates cell elongation by inhibiting brassinosteroid signaling. Plant Physiol. 2022;189:285–300. doi: 10.1093/plphys/kiac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park T.K., Kang I.A., Park C.H., Roh J., Lee S.H., Kim M., Jin E., Kim S.K., Kim T.W. Inhibition of 4-HYDROXYPHENYLPYRUVATE DIOXYGENASE expression by brassinosteroid reduces carotenoid accumulation in Arabidopsis. J. Exp. Bot. 2022;73:1415–1428. doi: 10.1093/jxb/erab475. [DOI] [PubMed] [Google Scholar]
- 18.Zu S.H., Jiang Y.T., Chang J.H., Zhang Y.J., Xue H.W., Lin W.H. Interaction of brassinosteroid and cytokinin promotes ovule initiation and increases seed number per silique in Arabidopsis. J. Integr. Plant Biol. 2022;64:702–716. doi: 10.1111/jipb.13197. [DOI] [PubMed] [Google Scholar]
- 19.Wang L., Yu P., Lyu J., Hu Y., Han C., Bai M.Y., Fan M. BZR1 physically interacts with SPL9 to regulate the vegetative phase change and cell elongation in arabidopsis. Int. J. Mol. Sci. 2021;22:10415. doi: 10.3390/ijms221910415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye H., Liu S., Tang B., Chen J., Xie Z., Nolan T.M., Jiang H., Guo H., Lin H.Y., Li L., et al. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 2017;8:14573. doi: 10.1038/ncomms14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Nolan T.M., Ye H., Zhang M., Tong H., Xin P., Chu J., Chu C., Li Z., Yina Y. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 2017;29:1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui X.Y., Gao Y., Guo J., Yu T.F., Zheng W.J., Liu Y.W., Chen J., Xu Z.S., Ma Y.Z. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019;180:605–620. doi: 10.1104/pp.19.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun F., Yu H., Qu J., Cao Y., Ding L., Feng W., Bin Khalid M.H., Li W., Fu F. Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Int. J. Mol. Sci. 2020;21:996. doi: 10.3390/ijms21030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Ye K., Shi Y., Cheng J., Zhang X., Yang S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant. 2017;10:545–559. doi: 10.1016/j.molp.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Albertos P., Dündar G., Schenk P., Carrera S., Cavelius P., Sieberer T., Poppenberger B. Transcription factor BES1 interacts with HSFA1 to promote heat stress resistance of plants. EMBO J. 2022;41:e108664. doi: 10.15252/embj.2021108664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H., Feng W., Sun F., Zhang Y.Y., Qu J.T., Liu B., Lu F., Yang L., Fu F., Li W. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018;86:235–249. doi: 10.1007/s10725-018-0424-2. [DOI] [Google Scholar]
- 27.Manoli A., Trevisan S., Quaggiotti S., Varotto S. Identification and characterization of the BZR transcription factor family and its expression in response to abiotic stresses in Zea mays L. Plant Growth Regul. 2018;84:423–436. doi: 10.1007/s10725-017-0350-8. [DOI] [Google Scholar]
- 28.Sun F., Ding L., Feng W., Cao Y., Lu F., Yang Q., Li W., Lu Y., Shabek N., Fu F., et al. Maize transcription factor ZmBES1/BZR1-5 positively regulates kernel size. J. Exp. Bot. 2021;72:1714–1726. doi: 10.1093/jxb/eraa544. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Guo W., Du D., Pu L., Zhang C. Overexpression of a maize BR transcription factor ZmBZR1 in Arabidopsis enlarges organ and seed size of the transgenic plants. Plant Sci. 2020;292:110378. doi: 10.1016/j.plantsci.2019.110378. [DOI] [PubMed] [Google Scholar]
- 30.Su D., Xiang W., Wen L., Lu W., Shi Y., Liu Y., Li Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021;21:161. doi: 10.1186/s12870-021-02933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song X., Ma X., Li C., Hu J., Yang Q., Wang T., Wang L., Wang J., Guo D., Ge W., et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genom. 2018;19:346. doi: 10.1186/s12864-018-4744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y., Zhao N., Wang M., Zhou W., Guo J., Han C., Zhou C., Wang W., Wu S., Tang W., et al. Integrated regulation of periclinal cell division by transcriptional module of BZR1-SHR in Arabidopsis roots. New Phytol. 2022;233:795–808. doi: 10.1111/nph.17824. [DOI] [PubMed] [Google Scholar]
- 33.Zhao N., Zhao M., Tian Y., Wang Y., Han C., Fan M., Guo H., Bai M.Y. Interaction between BZR1 and EIN3 mediates signalling crosstalk between brassinosteroids and ethylene. New Phytol. 2021;232:2308–2323. doi: 10.1111/nph.17694. [DOI] [PubMed] [Google Scholar]
- 34.Bin Khalid M.H., Raza M.A., Yu H.Q., Khan I., Sun F.A., Feng L.Y., Qu J.T., Fu F.L., Li W.C. Expression, subcellular localization, and interactions of CPK family genes in Maize. Int. J. Mol. Sci. 2019;20:6173. doi: 10.3390/ijms20246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amtmann A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant. 2009;2:3–12. doi: 10.1093/mp/ssn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlova R., Boer D., Hayes S., Testerink C. Root plasticity under abiotic stress. Plant Physiol. 2021;187:1057–1070. doi: 10.1093/plphys/kiab392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradai M., Amorim-Silva V., Belgaroui N., Esteban Del Valle A., Chabouté M.E., Schmit A.C., Lozano-Duran R., Botella M.A., Hanin M., Ebel C. Wheat type one protein phosphatase participates in the brassinosteroid control of root growth via activation of bes1. Int. J. Mol. Sci. 2021;22:10424. doi: 10.3390/ijms221910424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salazar-Henao J.E., Lehner R., Betegón-Putze I., Vilarrasa-Blasi J., Caño-Delgado A.I. BES1 regulates the localization of the brassinosteroid receptor BRL3 within the provascular tissue of the Arabidopsis primary root. J. Exp. Bot. 2016;67:4951–4961. doi: 10.1093/jxb/erw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galstyan A., Nemhauser J.L. Auxin promotion of seedling growth via ARF5 is dependent on the brassinosteroid-regulated transcription factors BES1 and BEH4. Plant Direct. 2019;3:e00166. doi: 10.1002/pld3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geng D., Shen X., Xie Y., Yang Y., Bian R., Gao Y., Li P., Sun L., Feng H., Ma F., et al. Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Hortic. Res. 2020;7:102. doi: 10.1038/s41438-020-0324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes-Impellizzeri S., Moreno A.A. The endoplasmic reticulum role in the plant response to abiotic stress. Front. Plant Sci. 2021;12:2582. doi: 10.3389/fpls.2021.755447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hye R.W., Jin H.K., Hong G.N., Pyung O.L. The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant Cell Physiol. 2004;45:923–932. doi: 10.1093/pcp/pch110. [DOI] [PubMed] [Google Scholar]
- 43.Balazadeh S., Siddiqui H., Allu A.D., Matallana-Ramirez L.P., Caldana C., Mehrnia M., Zanor M.I., Köhler B., Mueller-Roeber B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010;62:250–264. doi: 10.1111/j.1365-313X.2010.04151.x. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan F., Kopka J., Sung D.Y., Zhao W., Popp M., Porat R., Guy C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007;50:967–981. doi: 10.1111/j.1365-313X.2007.03100.x. [DOI] [PubMed] [Google Scholar]
- 45.Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 46.Ré D.A., Capella M., Bonaventure G., Chan R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine-tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014;14:150. doi: 10.1186/1471-2229-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosy E., Vavasseur A., Mouline K., Dreyer I., Gaymard F., Porée F., Boucherez J., Lebaudy A., Bouchez D., Véry A.A., et al. The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. USA. 2003;100:5549–5554. doi: 10.1073/pnas.0733970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue S.I., Iwashita N., Takahashi Y., Gotoh E., Okuma E., Hayashi M., Tabata R., Takemiya A., Murata Y., Doi M., et al. Brassinosteroid involvement in arabidopsis thaliana stomatal opening. Plant Cell Physiol. 2017;58:1048–1058. doi: 10.1093/pcp/pcx049. [DOI] [PubMed] [Google Scholar]
- 49.Li J.G., Fan M., Hua W., Tian Y., Chen L.G., Sun Y., Bai M.Y. Brassinosteroid and hydrogen peroxide interdependently induce stomatal opening by promoting guard cell starch degradation. Plant Cell. 2020;32:984–999. doi: 10.1105/tpc.19.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han C., Hua W., Li J., Qiao Y., Yao L., Hao W., Li R., Fan M., De Jaeger G., Yang W., et al. TOR promotes guard cell starch degradation by regulating the activity of β-AMYLASE1 in Arabidopsis. Plant Cell. 2022;34:1038–1053. doi: 10.1093/plcell/koab307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu J., Liu Q., Wang C., Liang J., Liu L., Wang Q. ZmWRKY79 positively regulates maize phytoalexin biosynthetic gene expression and is involved in stress response. J. Exp. Bot. 2018;69:497–510. doi: 10.1093/jxb/erx436. [DOI] [PubMed] [Google Scholar]
- 52.Lu X., Yang L., Yu M., Lai J., Wang C., McNeil D., Zhou M., Yang C. A novel Zea mays ssp. mexicana L. MYC-type ICE-like transcription factor gene ZmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2017;113:78–88. doi: 10.1016/j.plaphy.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Yu H.Q., Zhou X.Y., Wang Y.G., Zhou S.F., Fu F.L., Li W.C. A betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus enhances tolerance of Arabidopsis to high salt and drought stresses. Plant Growth Regul. 2017;83:265–276. doi: 10.1007/s10725-016-0245-0. [DOI] [Google Scholar]
- 54.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huque A.K.M.M., So W., Noh M., You M.K., Shin J.S. Overexpression of Atbbd1, Arabidopsis bifunctional nuclease, confers drought tolerance by enhancing the expression of regulatory genes in aba-mediated drought stress signaling. Int. J. Mol. Sci. 2021;22:2936. doi: 10.3390/ijms22062936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.