Significance
Human α2-macroglobulin (hα2M) is an ∼720-kDa homotetrameric particle with pan-peptidase inhibitory functions that transits between an open native conformation and a closed induced state, in which endopeptidases are trapped upon cleavage of an accessible bait region. We determined the molecular mechanism of this function through eight cryo–electron microscopy (cryo-EM) structures, which revealed that the hα2M subunits are organized in two flexible modules that undergo independent expanded-to-compact transitions. In the induced state, a reactive thioester bond triggers covalent linking of the proteinase, and a receptor-binding domain is exposed on the tetramer surface for binding to its specific cellular receptor for internalization and clearance from circulation. These results elucidate the long-awaited molecular mechanism of a historical suicidal inhibitory trap.
Keywords: α2-macroglobulin, proteinase, blood proteostasis, multifunctional complex, conformational states
Abstract
Human α2-macroglobulin (hα2M) is a multidomain protein with a plethora of essential functions, including transport of signaling molecules and endopeptidase inhibition in innate immunity. Here, we dissected the molecular mechanism of the inhibitory function of the ∼720-kDa hα2M tetramer through eight cryo–electron microscopy (cryo-EM) structures of complexes from human plasma. In the native complex, the hα2M subunits are organized in two flexible modules in expanded conformation, which enclose a highly porous cavity in which the proteolytic activity of circulating plasma proteins is tested. Cleavage of bait regions exposed inside the cavity triggers rearrangement to a compact conformation, which closes openings and entraps the prey proteinase. After the expanded-to-compact transition, which occurs independently in the four subunits, the reactive thioester bond triggers covalent linking of the proteinase, and the receptor-binding domain is exposed on the tetramer surface for receptor-mediated clearance from circulation. These results depict the molecular mechanism of a unique suicidal inhibitory trap.
The α2-macroglobulins are large multidomain proteins found in animals and selected colonizing bacteria (1–8). The best characterized is human α2-macroglobulin (hα2M), a 1,451-residue protein built of 11 domains (hα2M domain nomenclature shown in SI Appendix, Fig. 1A), which is produced by several cell types including macrophages, astrocytes, and hepatocytes. Four protomers associate to an ∼720-kDa polyglycosylated dimer of disulfide-linked homodimers, (hα2M)4 (SI Appendix, Fig. 1B), which is the largest nonimmunoglobulin protein in human plasma and constitutes 2 to 4% of its total protein content (1, 9). Its multiple molecular functions include endopeptidase inhibition, as well as sequestration and transport of growth factors, cytokines, and hormones (10, 11). It is also an acute-phase reactant in rodents and a chaperone that binds misfolded or inactivated proteins and has many more moonlighting functions, such as transglutamination and zinc/copper binding (4, 8, 12).
These disparate functions explain the universal physiological significance of hα2M: It is part of the innate immune response against pathogens (13, 14) and a major hemostatic regulator of the cardiovascular system through its anticoagulant, procoagulant, and antifibrinolytic activities (4). It is an early marker of cardiac hypertrophy, as well as a potential diagnostic marker for myocardial infarct and for HIV patients with cardiac pathologies (5). In amyloidoses, it is prophylactic, as it binds the major component of β-amyloid deposits, the Aβ peptide, and mitigates its neurotoxicity and fibrillogenic capacity (8). In addition, it has anti-inflammatory, signaling, and apoptotic properties; is engaged in growth and tissue remodeling; and protects joint cartilage (15, 16). Its deregulation contributes to most major human diseases including Alzheimer’s disease, AIDS, inflammatory diseases, diabetes, arthritis, tumor growth and progression, and cardiovascular conditions (8, 15, 17). No total hα2M deficiency has been described in humans, which supports the idea that its absence is embryonically lethal (4, 18–20).
The best-characterized molecular function of hα2M is its indiscriminate inhibitory action on endopeptidases irrespective of catalytic class, whether endogenous or exogenous. This hallmarks it as a unique pan-endopeptidase inhibitor (1, 4, 5, 7, 8, 21). In the cardiovascular system, it inhibits thrombin, factor Xa, activated protein C, plasma kallikrein and kallikrein-related peptidases, and plasmin (4, 17). As part of the humoral defense barrier, it inhibits bacterial and viral proteolytic virulence factors such as pseudolysin, HIV-1 proteinase, clostripain, and vibriolysin, as well as snake-venom proteinases (22, 23). Finally, by inhibiting neutrophil elastase, matrix metalloproteinases, and ADAM/adamalysin metallopeptidases, it participates in inflammatory processes and tissue turnover (15, 16).
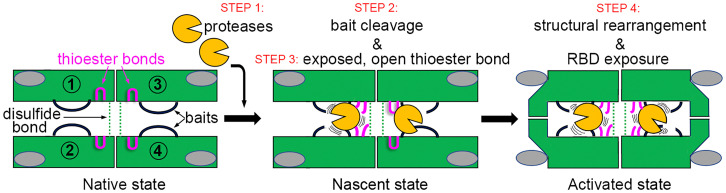
Whereas the vast majority of peptidase inhibitors act through reversible “lock-and-key” mechanisms that sterically block the active-site cleft of target enzymes (24, 25), hα2M operates according to a unique suicidal Venus flytrap (7, 26) or trap-hypothesis mechanism (1), by which prey peptidases diffuse into an unreacted or native (hα2M)4 tetramer (Fig. 1, step 1). Once inside, peptidases access and cleave a flexible, multitarget bait region (Fig. 1, step 2) within an exposed bait-region domain (BRD; SI Appendix, Fig. 1). Cleavage leads to a nascent state of the inhibitor, in which a buried reactive β-cysteinyl-γ-glutamyl thioester bond becomes exposed on a thioester domain (TED). The bond is attacked by surface lysine amines of the prey peptidase, which thus becomes covalently bound to the inhibitor through an ε-lysyl-γ-glutamyl crosslink (Fig. 1, step 3). Bait-region cleavage also triggers a large, irreversible structural rearrangement of the tetramer, leading to a reacted, induced, or activated state, which engulfs the peptidase without disturbing its active site, similarly to insect capture by the Venus flytrap plant (Fig. 1, step 4). A large reorganization of (hα2M)4 can also be observed by treatment of the native species with nucleophilic chemicals such as methylamine (MA), which yields an activated species with open thioester bonds but intact bait regions, unable to inhibit peptidases (27–30). Native and activated states can be distinguished because they have different sedimentation coefficients and mobilities in native gel electrophoresis (13).
Fig. 1.
Overview of hα2M action based on biochemical analysis. In native (hα2M)4, the four intact bait regions (black) are exposed in the enclosed cavity (prey chamber), while the thioester bonds (pink) and RBDs (gray ovals) are hidden. Monomer pairs 1 and 2, as well as 3 and 4, are disulfide-linked protomers (dashed green lines); pairs 1 and 3, as well as 2 and 4, are vicinal protomers; and pairs 1 and 4, as well as 2 and 3, are diagonally opposite protomers. Each protomer thus has a vicinal, a disulfide-linked, and an opposite protomer. The mechanism of action is illustrated in four steps that result in three states as described in the text.
Once within the closed trap, the peptidase no longer cleaves large protein substrates but is still accessible to small inhibitors and substrates through openings in the tetrameric particle. Moreover, the four receptor-binding sequences of the receptor-binding domains (RBDs), which are cryptic in the native and nascent species (31), become exposed on the tetrameric particle surface (32) and are recognized by cell-surface receptors such as the low-density lipoprotein receptor-related protein (LRP1) for receptor-mediated endocytosis (Fig. 1, step 4) (33). Inside the cell, the peptidase:inhibitor complex is cleared in the lysosomes within minutes of complex formation (34).
This sequence of events has been established through painstaking biochemical analyses for decades, but its molecular determinants remain unknown. Attempts have been undertaken to study the structure of tetrameric hα2M and mammalian orthologs to provide the molecular determinants of this inhibitory mechanism. Nonetheless, owing to the large size and intrinsic flexibility of hα2M, the lack of suitable recombinant expression systems for large-scale production of homogeneous functional protein, and the structural heterogeneity of samples purified from natural sources, only two small domains (the RBD and the macroglobulin-like domain MG2) have so far been described by X-ray crystallography (35–39). For full-length assemblies, 20- to 40-Å resolution maps derived from electron microscopy (EM) of negative-stained or vitrified samples and a crystallographic map of a hypothetically MA-activated (hα2M)4 to 10-Å resolution have shown morphological variations of the native and activated states of the hα2M tetramer, with no details at the domain level (29, 40–43). Moreover, the 4.3-Å resolution crystal structure of activated (hα2M)4 showed conserved structural features with the proteolytically activated C3b complement component (derived from the native C3 factor) (26). The most recent structural analyses of native hα2M are limited to homology models calculated from the native C3 complement component and docking in low-resolution EM maps (26, 44).
Thus, this study addresses three questions relevant to hα2M biochemistry: (i) the structure of the native (hα2M)4 complex when poised to trap plasma endopeptidases and that of the peptidase-activated inhibitor; (ii) the conformational changes and the intermediates on the path between the native and activated states; and (iii) how large proteases are entrapped in a cavity with insufficient volume a priori. We used authentic human protein to determine eight cryo-EM structures that represent functional states of (hα2M)4, unbound and in complex with physiologically relevant endopeptidases. The resulting structures show striking conformational rearrangement and provide detailed insight into the molecular mechanism of a unique sequential inhibitory mechanism.
Results and Discussion
Cryo-EM Structure Analysis of the (hα2M)4 Complex.
Authentic (hα2M)4 from human plasma was vitrified and imaged by cryo-EM (SI Appendix, Fig. 2A). A total of ∼1,625,000 particles collected on 300-keV FEI Titan Krios microscopes were automatically picked. Owing to the intrinsic flexibility and heterogeneity of the structures, they were subjected to several rounds of exhaustive two-dimensional (2D) and three-dimensional (3D) classification (SI Appendix, Fig. 2 B and C). These resulted in five distinct cryo-EM structures at different states of reaction, to resolutions spanning 4.5 to 7.3 Å as estimated by the values at which the Fourier shell correlation (FSC) coefficient equals 0.143 (SI Appendix, Fig. 3 A–E and Table 1).
To obtain a homogeneous population of fully activated (hα2M)4 as a control, a purified sample of mostly native protein was incubated with trypsin, and its structure was determined to 3.6-Å resolution (SI Appendix, Figs. 3F and 4). This structure enabled unambiguous assignment of the polypeptide chain of the whole particle, which gives rise to a C2 tetramer featuring a compact cage with several openings of variable size and a large inner cavity, the prey chamber.
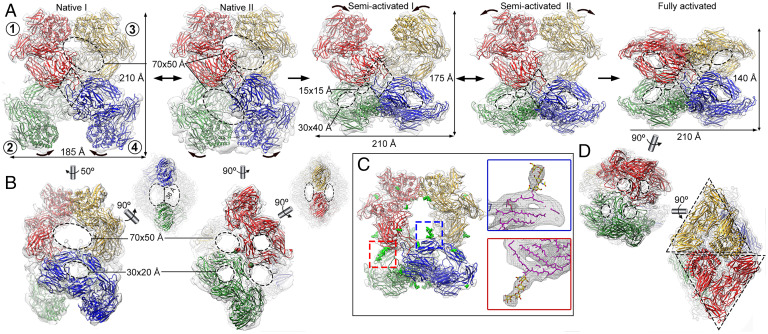
Based on protomer conformations, either expanded or compact, on their relative arrangement within the tetramers and on the volume and occupancy of the prey chambers in the distinct structures (i.e., the presence of entrapped proteinases in its interior chamber), we identified five major states of (hα2M)4, which we termed native I and II, semi-activated I and II, and fully activated, which corresponded to distinct reaction intermediates (Fig. 2A). With maximal dimensions of 210 × 185 × 150 Å, the native tetramer is substantially larger than the activated form, which spans 140 × 210 × 140 Å (Fig. 2A) and provides an explanation for the higher electrophoretic mobility of the latter (45). The native states have four large and four small openings of 70 × 50 Å and 30 × 20 Å, respectively, and enclose a prey chamber of ∼600 nm3, whereas the activated state has 12 small openings of 30 × 40 Å and encloses an ∼300-nm3 prey chamber (Fig. 2 A, B, and D).
Fig. 2.
Cryo-EM structures of (hα2M)4 functional states. (A) Five functional states of (hα2M)4 from human plasma are isolated from cryo-EM analysis, termed native I (Left) and II (Center Left), semi-activated I (Center) and II (Center Right), and activated (Right). Protomer nomenclature [1 (red), 2 (green), 3 (yellow), and 4 (blue)] as described in Fig. 1. The red protomer has a disulfide-linked (green), a vicinal (yellow), and an opposite neighbor (blue). Native I and II states have protomers with expanded conformation and are in an equilibrium, at which vicinal dimers are in a distal (native I) or proximal (native II) position (curved arrows). After proteolytic activation, the native state becomes semi-activated states I and II, and one vicinal dimer is built of protomers in compact conformation. Semi-activated states I and II, which correspond to the nascent state described in the literature (46), evolve to the activated state, shown in “H-view,” in which all protomers are compact. Openings are indicated as dashed ovals. Tethering loops of opposite protomers (1, 4) are framed in a dashed rhombus. (B) Additional views of the native I (Left) and II (Right) complexes of A. The latter highlights the disulfide-linked residues between protomers 1 and 2 (red and green spheres). Dashed ellipses denote openings. (C) The semi-activated II complex highlights the N-linked glycosylation sites as green sticks (Left). Magnified view of the red and blue boxes that correspond to the same glycan bound to MG4 Asn396 in the compact protomer 4 (blue) and in the expanded protomer 1 (red). In the Insets, protein backbones are shown as magenta sticks, glycan chains as stick models with yellow carbon atoms. (D) The activated state is shown in end view (Top) and X-view (Bottom), in which the triangular prism profile for each protomer is framed. Dashed ellipses denote openings.
Each (hα2M)4 protomer has a vicinal, a disulfide-linked, and an opposite neighbor (Fig. 2A, Left). In the native and semi-activated states, one vicinal dimer was solved to much better resolution than the other (SI Appendix, Fig. 3). For the native I and II states, resolution of the upper pair of vicinal protomers was sufficient to determine unambiguously their subunit organization (Fig. 2A, red and yellow). By contrast, the intrinsic flexibility of the lower vicinal protomers (∼11- to 22-Å resolution) required development of a strategy that combined rigid and flexible domain refinement (Fig. 2A, green and blue). The semi-activated I state behaved similarly to native states, with the upper pair more flexible than the lower pair (8- to 25-Å versus 3.5- to 12-Å resolution). Finally, the semi-activated II and activated states showed density with more features, which allowed accurate modeling of the four polypeptide chains (SI Appendix, Fig. 5; see Materials and Methods for details). Each hα2M subunit has eight surface-located, protruding N-linked glycans at positions 55, 70, 247, 396, 410, 869, 991, and 1424 (Fig. 2C, indicated for semi-activated state II, green sphere model; see UniProt entry P01023 for sequence numbering). Structural alignment of compact or expanded protomers in all states showed rmsd values <1.4 Å, indicating high structural similarity in the respective conformations.
The native (I and II), semi-activated (I and II), and activated states were captured at an ∼1:2:1 ratio, although in a specific preparation (preparation P2) more than half the total particles were initially classified as native (SI Appendix, Fig. 2). These data underpin the existence of a substantial population (25 to 42%) of intrinsically activated (hα2M)4 in preparations assumed to be mainly native, following long-established standard biochemical purification procedures (27).
The Functional Tetrameric States Are Built of Expanded and Compact Protomer Conformations.
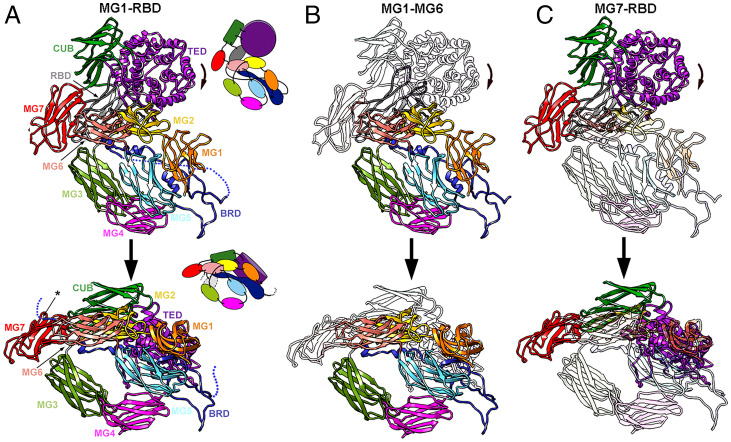
Each hα2M subunit structure consists of 11 or 10 domains for the expanded and compact conformations, respectively, as the C-terminal RBD is flexible and thus not assignable in the latter (Fig. 3A and SI Appendix, Fig. 1). The first seven domains (MG1-MG7) are concatenated ∼110-residue β-sandwiches with a three- and a four-strand antiparallel β-sheet (Fig. 3A). Domains MG1-MG6 form an N-terminal module, which is similarly arranged in all structures as a 1.5-turn right-handed superhelix that encircles an ellipsoidal opening of 30 × 20 Å (Fig. 2B, dashed ellipses). The 127-residue BRD is an extended, flexible domain inserted between the fourth and fifth β-strands of MG6 (SI Appendix, Fig. 1A); the domains downstream of MG6 form a C-terminal module that adopts two different conformations: expanded or compact. MG7 acts as a hinge domain that connects MG1-MG6 with the 116-residue CUB domain (acronym of complement protein subcomponents C1r/C1s, urchin embryonic growth factor, and bone morphogenetic protein 1), which consists of two four-stranded antiparallel β-sheets. The 315-residue helical TED domain is inserted between the third and fourth β-strands of CUB and includes the scissile thioester bond. TED adopts a thick disk-like structure that consists of six concentric β-hairpins arranged as a six-fold α-propeller around a central shaft (Fig. 3A). The thioester bond is formed between the cysteine side-chain sulfur and the glutamine side-chain carbonyl of the conserved motif Cys972-Gly-Glu-Gln975. Finally, the C-terminal domain of this module is the RBD, a variant of the MG fold with an extra α-β-α unit.
Fig. 3.
Functional states of (hα2M)4 are built of expanded and compact protomers. (A) Spatial domain organization of the expanded (Top) and compact (Bottom) protomer conformations in front view (as in Fig. 2A). The BRD (dark blue) contains either the flexible, intact (Top), or the cleaved (Bottom) bait region (dashed line). The visible end of the BRD is pinpointed by an asterisk. Insets, diagrams of the approximate domain organization of the conformers. (B and C) Regions equivalent to A, highlighting the N-terminal module (domains MG1-MG6) (B) and the C-terminal module (domains MG7-RBD) (C).
In all five functional states identified in the native preparation, the hα2M monomers adopt either a compact or an expanded conformation for each pair of vicinal dimers. In native states I and II, all subunits are expanded (Fig. 2A). In the native I state, the TED domains of vicinal protomers (within the top dimer) contact each other through their respective fifth helices (Fig. 2 A, Left; subunits 1 and 3). By contrast, the protomers are dynamic in the bottom dimer (Fig. 2 A, Left; subunits 2 and 4) and swing around the interface between subunits 1 and 2 and subunits 3 and 4. These subunits encompass the interprotomeric disulfide bond (Fig. 2B, indicated in the native II state, red and green spheres) as well as BRD-mediated contacts between opposite monomers of subunits 1 and 4 and subunits 3 and 2 (Fig. 2A, dashed rhombuses). As a result, the fifth helices of the bottom TED domains can either be separated by 36 Å (Fig. 2 A and B, Left; green and blue protomers) or in contact, as found in the top domains (Fig. 2 A, Center-Left; green and blue protomers). Both expanded protomers, in a distal or proximal position, share an almost identical conformation (rmsd < 0.7 Å). This yields the native II state, which is quasi-symmetric for the top and bottom dimers as the bottom dimer is resolved at lower resolution, indicating it is more flexible or dynamic than the top dimer. In semi-activated states I and II, which correspond to the nascent state (46) (Fig. 1), the bottom vicinal subunits adopt a compact conformation (Fig. 2 A, Center and Center-Right; green and blue protomers), while the top vicinal subunits are dynamic and very similar to the native states. In the fully activated state, all subunits show a compact conformation (Fig. 2 A, Right, and Movie S1).
Whereas the architecture of the N-terminal module is equivalent in the expanded and compact conformations and merely undergoes a 22° rotation around MG4 (Fig. 3B), the C-terminal modules are coordinately shifted, with additional 45° and 55° rotations for CUB and TED, respectively, around the hinge-domain MG7 (Fig. 3C and Movie S2). Following this conformational change, all domains except MG3 and MG4 are arranged as a triangular prism in the compact conformation, the hallmark structural motif of the activated state (Fig. 2D, dashed triangles). Vicinal TEDs interact through their fifth and third α-helix in the expanded and compact conformation, respectively. TED also contacts MG4 of the respective disulfide-linked subunit, as well as MG1s and BRDs of the respective vicinal subunit.
The conformational rearrangement between the expanded and compact conformations implies notable changes in the contact points within the top and bottom dimers of vicinal protomers (SI Appendix, Table 2). Whereas the internal cavity in the native tetramer is framed by MG1-MG5, BRD, TED, and a small part of RBD, in the activated tetramer only MG1-MG5 and TED face the prey chamber as the exposed surfaces of MG1, MG2, and MG5 are substantially reduced. All domains have a very similar fold in both conformations, except for TED. The four MG4 domains in the equatorial plane of the tetramer are nearly invariant in all functional states despite the massive structural changes that underlie the transition. Together with the BRD tethering loops (see next section), these domains form a constant belt responsible for the structural integrity of the tetramer in either state (SI Appendix, Fig. 6).
The Major Players: BRD, TED, and RBD.
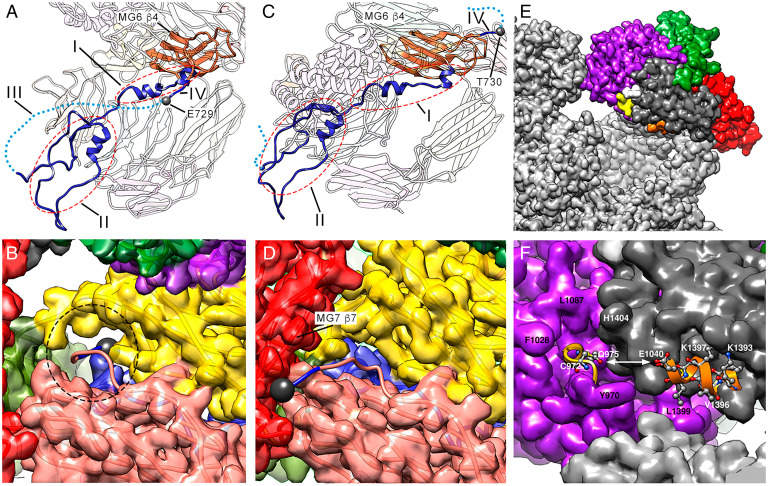
The BRD spans 75 Å and contains little regular secondary structure. It consists of four regions in the expanded conformation (Fig. 4A, I to IV). Region I (Gln602-Leu627) is an N-terminal extended segment that is linked to the third β-strand of MG6 and contains two short α-helices. Region II (Thr628-Gln694) is a compact region that includes the tethering loop (Ile643-Cys689) and interacts with the symmetric region of the opposite protomer. Region III (Pro690-Thr728) is the 39-residue bait segment that is disordered and invisible in our maps. Finally, region IV (Glu729-Arg732) is the C-terminal segment connected with the fourth β-strand of MG6 on the outer surface of the protomer through one of the openings in the expanded conformation. This opening is framed by MG6, MG2, MG3, and the loop connecting MG2 with MG3, and it allows passage of an intact polypeptide chain (Fig. 4B, dashed circle; SI Appendix, Fig. 5). In the compact conformation (Fig. 4C), the beginning of the C-terminal segment at Thr730 is on the outer surface and interacts with the seventh strand of MG7. A small rearrangement in this region causes MG7 to fold over MG6, which almost occludes the opening (Fig. 4D and SI Appendix, Fig. 6). The BRD-MG6 connecting segment thus regulates the expanded-to-compact transition by acting as a trigger that must be cleaved for displacement of the new N terminus to the exterior surface. This is consistent with the assumption that MA-activated (hα2M)4, whose bait region is intact, adopts a structure distinct from the peptidase-activated state (32). These results further indicate that the reported crystal structure of induced (hα2M)4 (SI Appendix, Table 3), originally thought to be a noncleaved MA-induced variant (26), cannot contain an intact bait region but is a cleaved, proteinase-induced species. Its overall conformation is equivalent to the structure of the fully peptidase-activated state (see below and Fig. 2 A, Right).
Fig. 4.
The major players in the conformational shift: BRD, TED, and RBD. (A) Unprocessed BRD (blue) seen from inside the prey chamber in the expanded conformation of the native I state (upper vicinal protomers 1 or 3). Regions I, II, III (dashed line), and IV are indicated. MG6 β4 (orange) is shown, bound to region IV (Glu729-Arg732). Glu729 is highlighted (gray sphere). (B) Closeup of the unprocessed segment Glu729-Arg732 passing through the opening framed by MG6 (pink), MG2 (yellow), MG3 (green), and the MG2-MG3 connecting loop (Glu729, gray sphere). The dashed ellipse pinpoints the passage of the polypeptide chain through a narrow aperture in the expanded conformation. (C) The proteolytically processed BRD seen from inside the prey chamber in the compact conformation of the activated state in any of the protomers (1–4), in which region III (dashed line) is discontinuous (similar view as A). The last visible residue is Thr730 (gray sphere). (D) Closeup of the processed segment Thr730-Arg732 (blue), which interacts with the seventh β-strand of MG7 on the outer surface (Thr730, gray sphere). The opening seen in B is occluded. (E) In the expanded conformation of the native I state, the TED thioester bond (yellow) and α2 helix of the RBD (orange) are 16 Å apart and are buried in the structure. (F) Closeup of the region shown in E. The thioester bond is in a local cavity surrounded by the large hydrophobic side chains of Tyr970, Phe1028, and Leu1087 (from TED) and His1404 (from RBD). Helix α2 from the RBD is partially hidden by segment Ser1428-Thr1432 from the domain’s seventh β-strand.
At the C terminus, protomers in expanded conformation, both in activated and semi-activated states, have a well-defined RBD surrounded by MG7, CUB, and TED (Fig. 4E, dark gray; and SI Appendix, Fig. 5). The second α-helix of RBD, which includes the receptor-binding region (47), points toward the prey chamber and is occluded (Fig. 4 E and F, orange). Following activation by cleavage of the bait region, the substantial rearrangement of MG7-CUB-TED-RBD disrupts the interaction among these domains. In the activated (hα2M)4 structures, this causes the RBD to project away from the outer surface and become flexible and thus disordered, except for a small, blurred density for its first three or four residues (SI Appendix, Fig. 7). The receptor-binding region thus becomes accessible for interaction with LRP1. In previous structural studies, the RBD was visible in the crystal structure of activated (hα2M)4 for only one of the four subunits, probably due to tetramer–tetramer interactions within the crystal, and obviously in a physiologically irrelevant position (26).
The TED thioester bond is 16 Å from the second RBD α-helix in the expanded conformation of the native I state (Fig. 4 E and F, yellow). Small nucleophiles such as MA can access and open the thioester bond without BRD processing. This would be sensed by the RBD, resulting in exposure of its receptor binding sequence to trigger removal of this inactivated hα2M complex. Alternatively, these two critical regions might be adjacent in the highly porous structure of the native complex to remain simultaneously inaccessible to the external environment. Access to TED is limited in the expanded conformation, as it is found in a cleft surrounded by bulky hydrophobic side chains (Phe1024, Leu1087, Tyr970, and His-1404) (Fig. 4F). Moreover, the glycan bound to Asn1424 is nearby (Fig. 4E, white), which might prevent thioester-bond opening by surface amines of circulatory proteins and other substances. In contrast, in the compact conformation, thioester bonds are accessible and face the prey chamber and are thus prepared for reaction with surface lysines of the prey (see below).
We did not identify a native state in which both vicinal dimers displayed simultaneously noninteracting TED domains (Fig. 2 A, Left; green and blue protomers), which suggests that this intermediate is unstable or very short-lived and thus cannot be isolated by cryo-EM. To communicate the separated, interacting TED positions, two small contact areas between vicinal dimers are critical: the opposite BRD tethering loops and the MG3 and MG4 domains. They are symmetrically crosslinked through intermolecular disulfide bonds (Cys278 of MG3 with Cys431 of MG4; SI Appendix, Figs. 1 and 5). The structural integrity of the native tetramer resides only on the tethering loop-mediated, noncovalent interaction. This TED rigid-body rearrangement might capture nonspecific circulatory proteins, which are conveyed to the prey chamber. Most of them will not interact with the hα2M subunits and will be released directly through the 70 × 50 Å openings, which in turn might also be the direct entrance of many soluble proteins. By contrast, those with endopeptidolytic activity will cleave the bait region and trigger conformational rearrangement from the expanded to the compact conformation, which halves the volume of the prey chamber and restricts the openings to maximally 30 × 40 Å, thus preventing prey from escaping. This mechanism facilitates scanning of many plasma proteins, and only endopeptidases that must be quickly withdrawn from the circulation would be “swallowed.”
The Symmetric Activated State.
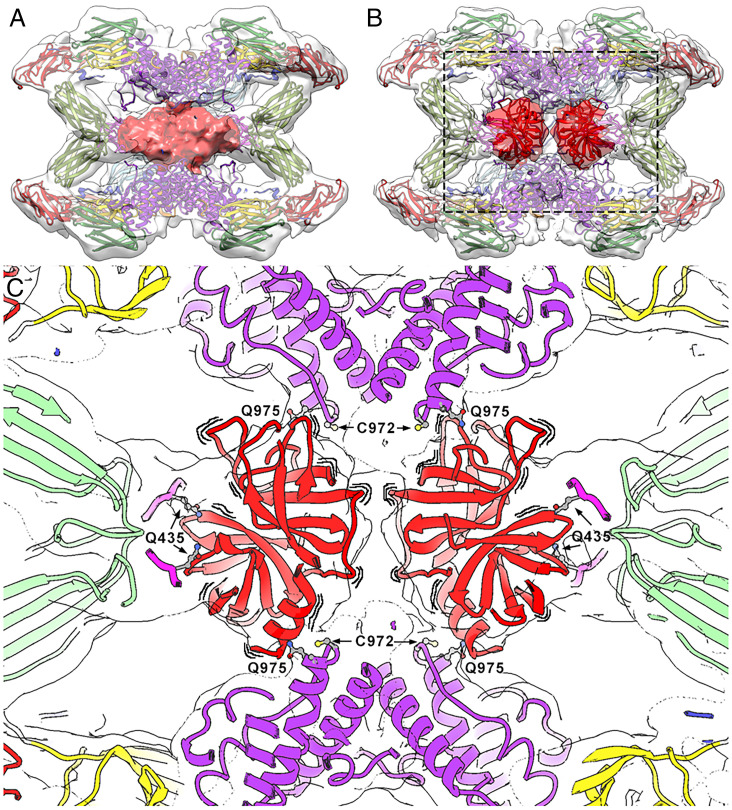
Intrinsically activated (hα2M)4 was resolved as a single complex in which the heterogeneous proteinases trapped were resolved as a featureless density that occupied the prey chamber (Fig. 5A, red surface). Since the purified samples contained 25 to 55% native (hα2M)4, we analyzed a sample activated with bovine trypsin in vitro (SI Appendix, Fig. 4). The tetramer in the resulting compact conformation was refined to 3.6-Å resolution (SI Appendix, Fig. 3F), and the density corresponding to the trapped proteinases was resolved as two independent volumes (Fig. 5B, red surface). This result indicates that each disulfide-linked dimer participates in the binding of one trypsin molecule, in accordance with an inhibition stoichiometry of two proteinase molecules per inhibitor tetramer (13). Although the entrapped peptidases are probably not static inside the cavity, they could be tentatively docked (223 residues; Protein Data Bank [PDB] 1MTS) into these two densities (Fig. 5 B and C, red ribbons). The density corresponding to the two peptidase moieties, in both intrinsically and trypsin-activated (hα2M)4, suggests intimate contact with the two symmetric MG4 loops, in particular with Gln435, and with Gln975 of the vicinal subunit thioester bonds for covalent binding (Fig. 5C).
Fig. 5.
The symmetrically activated state. (A) Intrinsically activated (hα2M)4 from plasma. The front half of the map (transparent surface) was removed to visualize the heterogeneous proteinase density (red). (B) Trypsin-activated (hα2M)4 in a similar view as in A, with two trypsin molecules tentatively docked into the density (dark red), which is clearly resolved in two separate volumes (clear red). However, the exact orientation of the molecules cannot be determined due to the lowish resolution of the map. (C) Closeup of the boxed region in B. To highlight that the caged trypsin molecules are dynamically oriented, wavy lines are shown around the corresponding red ribbons. Location of major functional residues are indicated in the activated complex. Trypsin is fixed to the TED thioester bonds formed between Cys972 and Gln975 from vicinal protomers. Residues Gln435 of each MG4 moiety are indicated. Color code: MG2, yellow; MG3, green; MG4, pink; TED, purple; trypsin, red.
The Subunit-Mediated Activated State.
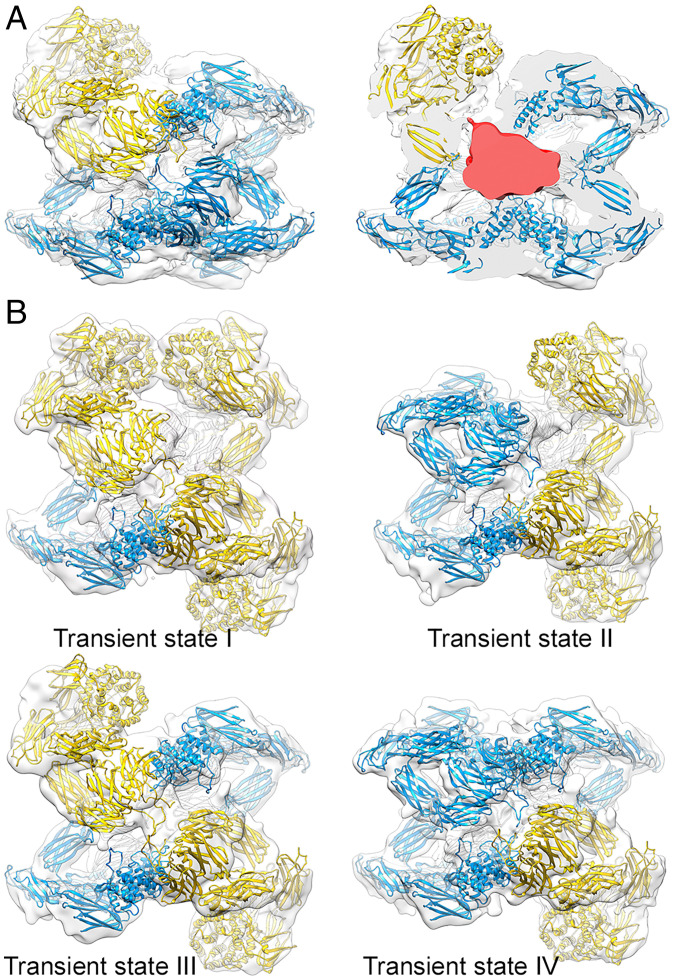
(hα2M)4 was also activated in vitro with plasmin, a 791-residue protease that cannot be accommodated in the internal cavity of the activated tetramer but is nevertheless inhibited efficiently by (hα2M)4 following cleavage after Arg719 (48). Whereas trypsin-incubated (hα2M)4 adopted an activated state in which the four subunits were in the same compact conformation, plasmin-incubated (hα2M)4 was resolved in two activated states, with four (plasmin-activated I state) and three (plasmin-activated II state) compact subunits (SI Appendix, Figs. 3 G and H and 8), which account for 79% and 21% of the activated complexes, respectively (Fig. 6A). The density in the prey chamber attributable to a bound prey was much larger in the complex with three compact subunits (Fig. 6 A, Right), which suggests the absence of the large peptidase in the particles with four compact protomers. We thus hypothesize that acquisition of the compact conformation for the fourth subunit is prevented by steric hindrance through parts of the peptidase protruding from the inhibitor tetramer, even though the bait region is cleaved. Plasmin-activated II state therefore provides a structural explanation for the inhibitory potential of (hα2M)4 for endopeptidases larger than the prey chamber.
Fig. 6.
The subunit-mediated activated state. (A) State I of plasmin-activated (hα2M)4 with three compact (blue) and an expanded (yellow) subunit (Left). After the front half of the map was removed, an asymmetric, large density corresponding to plasmin becomes evident (red) (Right). (B) Analysis by symmetry expansion of native (hα2M)4 reveals arrangements with one (Top Left), two (Top Right and Bottom Left), and three (Bottom Right) compact subunits (blue).
We sought to determine whether both subunits within a vicinal pair must simultaneously undergo the transition from the expanded to the compact conformation. We thus reanalyzed native (hα2M)4 complexes using a computational approach that included symmetry expansion of the previously calculated map, with imposed C2 symmetry and subsequent 3D classification to detect unique features of each subunit (SI Appendix, Fig. 2D and Table 4). In addition to the two native states built of expanded subunits and to the fully activated state with four compact subunits, we isolated tetramer arrangements with one, two, and three activated subunits, which made up 22% of the tetramers included in the 3D classification and were probably partially activated intermediates (Fig. 6B). We hypothesize that once a first bait region is cleaved by the prey proteinase, the transition from the expanded to the compact conformation exposes the thioester bond to covalently fix the peptidase. This would cause the bait region of the vicinal subunit to be cleaved with higher probability, as the prey cannot escape from the trap, which would make the second protomer become compact. In some cases, however, the peptidase would not be immediately bound by the thioester bond and would thus gain access to the bait regions of the opposite and/or disulfide-linked subunit. These cleavages would produce species with three compact protomers. These intermediate structures indicate that conformational transition for each subunit is only dependent on cleavage of its own bait region, independently of the other subunits.
Molecular Mechanism of (hα2M)4 Function.
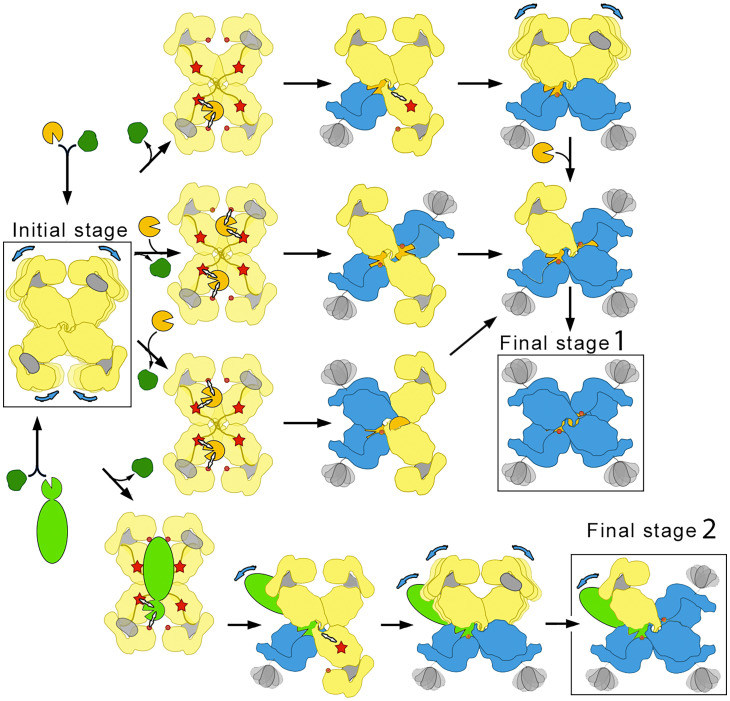
Based on the structures obtained in this study, we propose a molecular mechanism of plasma endopeptidase inhibition by (hα2M)4 (Fig. 7). The native, unreacted tetramer, which is built of subunits in expanded conformation, is extremely porous and flexible and comprises four large 50 × 70–Å openings that would fuse to yield a large, irregular cavity. The openings are framed by five of the eight N-linked glycans, which contribute to thermal stability and high solubility in blood plasma but not to the inhibitory capacity of hα2M (49). We hypothesize that this structure could act as a sieving complex in a crowded plasma environment, which is primed with four internal bait regions exposed to the particle lumen for testing of prey endopeptidases. Processing of the bait region leads to a conformational rearrangement of the particular protomer from an expanded to a compact conformation; this exposes the reactive thioester bond from TED for covalent entrapping of the peptidase and the RBD for recognition by its cognate receptor for subsequent endocytosis. Native subunits become activated as they are cleaved, although for large trapped proteinases, steric constraints or clashes with the prey might prevent the structural transition in some of the processed hα2M subunits. This dynamic complex is stabilized throughout this peptidase inhibition process by an inalterable structural belt formed by the MG4 domains and the BRD tethering loops.
Fig. 7.
Mechanism of irreversible protease inhibition by (hα2M)4. In the initial stage, native (hα2M)4 (four yellow expanded subunits) is a molecular sieve of plasma proteins (dark green, light green, and gold). Only peptidases (gold and light green) are retained in the internal cavity after processing of the bait region (red star), which can include two small peptidases (gold) or one large peptidase (light green). The thioester bond (red dot) becomes accessible to covalently fix the prey peptidase, which triggers the expanded-to-compact transition of the protomer (yellow-to-blue) and exposure of the flexible RBD (gray) on the surface, ready for cognate receptor binding (compact blue subunits). As the prey remains active within the trap, it can still cleave a second, third, and fourth protomer to yield the distinct intermediates and, ultimately, the final fully compact stage 1 for one or two small peptidases. In the case of large peptidases (bottom row), peptidase regions protruding from the prey chamber prevent complete transition, giving rise to an asymmetric final stage 2 that corresponds to the penultimate step of the pathway for small peptidases. Blue arrows denote domain flexibility.
A comparison with the mechanism derived for Escherichia coli α2-macroglobulin (ECAM) shows substantial differences despite a generally similar domain architecture in both inhibitors (50). While ECAM is monomeric and anchored to the cytosolic membrane of the bacterium facing the periplasm, hα2M is tetrameric and secreted to the plasma in humans. In the former, covalent linkage and steric hindrance of peptidases thus inhibit activity, but only against very large substrates. This modus operandi has been dubbed a “snap-trap mechanism.” In contrast, hα2M inhibition is elicited through physical entrapment in a large cage following a “Venus flytrap mechanism,” in which preys are still active against small substrates and inhibitors that can enter the cage through several apertures.
In conclusion, our structural studies report the molecular determinants of the mechanism of action of (hα2M)4, a universal pan-peptidase inhibitor that has been the subject of considerable biochemical and biophysical study since its discovery in 1946 (51). We document a stepwise Venus flytrap mechanism unique among peptidase inhibitors in its irreversibility and versatility, which enables it to sequester endogenous and exogenous peptidases from plasma and remove them from the circulation. Given the role of these complexes in numerous essential processes, knowledge of their structure provides a starting point for further studies of the many functions of (hα2M)4.
Materials and Methods
Cryo-EM Data Collection.
Wild-type authentic native (hα2M)4 was isolated from thawed frozen plasma from healthy human donors, which was de-identified prior to use in this study. The protein was purified, assessed for peptidase-inhibition competence as described (15, 26, 49), and verified to be equivalent to protein purified from fresh plasma in functional and physiological assays. Aliquots of pure protein (5 μL) were diluted to 0.1 mg/mL, applied to R2/2 300 mesh acetone vapor-treated copper grids, and vitrified using a Leica EM CPC cryofixation unit. Data were collected on FEI Titan Krios electron microscopes operated at 300 kV, and images were recorded with Gatan K2-summit cameras in counting mode using the EPU Automated Data Acquisition Software for Single Particle Analysis (Thermo Fisher Scientific). The total number of recorded movies, nominal magnification, calibrated pixel size at the specimen level, total exposure, exposure per frame, and defocus range for each specimen are described in SI Appendix, Table 1.
Image Processing.
Movies were drift-corrected and dose-weighted with Motioncor2 (52), and contrast transfer function values were estimated with CTFFIND4.1 (53) using non–dose-weighted micrographs. All subsequent image processing was with RELION 2.1 (54, 55) within Scipion (56), unless otherwise stated. The data processing workflows for (hα2M)4 complexes purified from plasma, as well as for trypsin- and plasmin-treated complexes, are described in SI Appendix, Figs. 3, 5, and 9, respectively. Particle statistics are indicated in SI Appendix, Table 1. Class averages from preliminary datasets were used as templates for subsequent automated particle picking with Gautomatch (written by Kai Zhang, https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/). Particles were then extracted, normalized, and subjected to several rounds of reference-free 2D classification to discard particles from 2D classes that did not show secondary structural elements. Selected particles were 3D classified, imposing C2 symmetry for plasma-purified and trypsin-treated (hα2M)4 complexes and C1 symmetry for plasmin-treated complexes. Classes representing equivalent conformational states were pooled and included in a 3D auto-refinement. Particles assigned to a native state were submitted to an additional round of 3D classification without alignment to identify and refine particles corresponding to native I and II states. To identify and classify intermediate conformations between the (hα2M)4 major states in plasma, the C2 symmetry of these states was expanded (57) and particles were subjected to 3D classification without alignment (particle processing is indicated in SI Appendix, Table 4). Classes representing equivalent conformational states were pooled and included in a 3D auto-refinement. Local resolution was estimated using MonoRes (58) and unsharpened maps treated by local resolution-based sharpening in LocalDeblur (59).
Model Building and Refinement.
The hα2M crystal structure (26) (PDB 4ACQ) was first docked manually as a rigid body into the trypsin-activated locally sharpened density map and then subjected to real-space fitting with the Fit_in_Map routine of Chimera (60). A first step of real-space refinement was performed with Phenix (61) applying global minimization, local grid search, and atomic displacement parameter refinement protocols. The model was then rebuilt manually in Coot (62) to optimize the fit to the density for one set of disulfide-linked subunits (protomers 1 and 2). This asymmetric unit was then C2-symmetrized and further refined in Phenix with similar options as above and with secondary structure, noncrystallographic symmetry, side chain rotamer, and Ramachandran restraints, as well as with hydrogens in riding positions. This model was used as a starting model to build the compact subunits and was fitted into the naturally activated and plasmin-activated I sample maps, as well as in densities corresponding to protomers 2, 3, and 4 of plasmin-activated II, as a starting point for model building of these states. In Chimera, activated protomer 1 coordinates were flexible fitted in the native I protomer 1 density, considering each domain as a rigid body. These docked domains were used as the starting point for model building of the prototype of an expanded subunit, whose coordinates were fitted in expanded protomers of native I (protomer 2), native II (protomers 1 and 2), semi-activated I (protomer 1), semi-activated II (protomer 1), and plasmin-activated II (protomer 1). Fitted coordinates were checked manually in Coot and C2 symmetrized (to generate protomers 3 and 4). The first step of real-space refinement was performed in Phenix, with morphing and simulated annealing options, followed by the steps described above. Refinement statistics are listed in SI Appendix, Table 1.
Model Validation and Analysis.
The FSC curves between model and map after local sharpening (Model versus Map) are shown in SI Appendix, Fig. 10. For cross-validation against overfitting, the atoms in the final atomic models were displaced by 0.5 Å in random directions using Phenix. The shifted coordinates were then refined against one of the half-maps (work set) in Phenix using the same procedure as for refinement of the final model. The other half-map (test set) was not used in refinement for cross-validation. FSC curves of the refined shifted model against the work set (FSCwork) and against the test set (FSCtest) are shown in SI Appendix, Fig. 10. The FSCwork and FSCtest curves do not diverge markedly, consistent with the absence of overfitting in the final models. The quality of the atomic model was assessed by analysis of the basic protein geometry, Ramachandran plots, and clash analysis and validated with Coot and MolProbity (63), as implemented in Phenix, and with the Worldwide PDB OneDep System (https://deposit-pdbe.wwpdb.org/deposition/). Graphics were produced using University of California San Francisco Chimera.
Re-refinement of the Crystallographic Structure of Peptidase-Activated hα2M.
To obtain a more accurate model of the reported crystal structure of peptidase-activated hα2M with complete side chains and correct glycan structure, the X-ray diffraction data [PDB 4ACQ (26)] were reprocessed to 4.2-Å resolution with current versions of the XDS (64) and XSCALE programs (65) (SI Appendix, Table 3). A test set for Rfree monitoring was chosen in thin shells with SFTOOLS within the CCP4 suite of programs (66). The coordinates of the hα2M part of the cryo-EM model of the complex with trypsin were superimposed onto the original crystallographic coordinates and refined in reciprocal space against the newly processed data with Phenix and Buster/TNT (67); this included hydrogens (set to zero occupancy) at riding positions for the protein residues, noncrystallographic symmetry restraints, and translation/libration/screw-rotation refinement. This refinement alternated with manual model building with Coot (SI Appendix, Table 3). The eight glycosylation sites of each protomer were rebuilt to match the recently determined glycan structure of hα2M (68).
Supplementary Material
Acknowledgments
We are grateful to R. Bonet of the IBMB Protein Purification Service and to C. Mark for editorial assistance. We thank members of the Proteomics Facility of the Centro de Investigaciones Biológicas (CIB–Consejo Superior de Investigaciones Cientificas [CSIC], Madrid) for protein identification; Rocío Arranz and Javier Chichón of the Cryo-EM Centro Nacional de Biotecnología/CIB-CSIC facility (Madrid) in the context of the CRIOMECORR project (ESFRI-2019-01-CSIC-16); the Diamond Light Source for access to its cryo-EM facility (proposals EM15997 and BI22006); the Astbury BioStructure Laboratory at the University of Leeds for help with cryo-EM data acquisition; and the European Synchrotron Radiation Facility for microscope time (proposal MX-2154). We thank personnel of the High Performance Computing Unit of the Instituto de Salud Carlos III Unidad de Tecnologías de la Información y Comunicación. This work was supported by grants from the Spanish Ministries of Economy and Competitivity (Grant No. BFU2017-88736-R) and of Science and Innovation (Grant No. PID2020-113287RB-I00) and the Comunidad Autónoma de Madrid (Grant No. P2018/NMT-4389) to J.R.C. and by grants from Catalan and Spanish public and private agencies (Grant Nos. BFU2019-107725-RB-I00 and 2017SGR00003; Fundació “La Marató de TV3” Grant No. 201815) to F.X.G.-R. T.G. acknowledges a Juan de la Cierva research contract (JCI-2012-13573) from the Ministry of Economy (MINECO) and S.R.M. a Ph.D.-fellowship (BES2016-076877) from the Ministry of Science and Innovation. The Structural Biology Unit of the Molecular Biology Institute of Barcelona (IBMB) was a María de Maeztu Unit of Excellence (2015 to 2019) and the Centro Nacional de Biotecnología is a Severo Ochoa Center of Excellence (MINECO award SEV 2017-0712), as awarded by the Spanish Ministry of Economy, Industry and Competitiveness. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200102119/-/DCSupplemental.
Data Availability
The atomic coordinates and cryo-EM density maps were deposited in the PDB and EM Data Bank with codes 7O7L and EMD-12747 (native I); 7O7M and EMD-12748 (native II); 7O7N and EMD-12750 (semi-activated I); 7O7O and EMD-12751 (semi-activated II); 7O7P and EMD-12752 (activated); 7O7Q and EMD-12753 (trypsin-activated); 7O7R and EMD-12754 (plasmin-activated I); and 7O7S and EMD-12755 (plasmin-activated II). The cryo-EM density maps of intermediate structures with one, two (two maps), and three activated monomers, corresponding to transient I to IV states, were deposited in the EM Data Bank with codes EMD-12941, EMD-12942, EMD-12943, and EMD-12944. The final X-ray crystallography model was likewise deposited at the PDB (PDB 6TAV) and supersedes PDB entry 4ACQ.
References
- 1.Barrett A. J., Starkey P. M., The interaction of α2-macroglobulin with proteinases. Characteristics and specificity of the reaction, and a hypothesis concerning its molecular mechanism. Biochem. J. 133, 709–724 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budd A., Blandin S., Levashina E. A., Gibson T. J., Bacterial α2-macroglobulins: Colonization factors acquired by horizontal gene transfer from the metazoan genome? Genome Biol. 5, R38 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doan N., Gettins P. G. W., α-Macroglobulins are present in some gram-negative bacteria: Characterization of the α2-macroglobulin from Escherichia coli. J. Biol. Chem. 283, 28747–28756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ignjatovic V., Mertyn E., Monagle P., The coagulation system in children: Developmental and pathophysiological considerations. Semin. Thromb. Hemost. 37, 723–729 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Rehman A. A., Ahsan H., Khan F. H., α-2-Macroglobulin: A physiological guardian. J. Cell. Physiol. 228, 1665–1675 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Wong S. G., Dessen A., Structure of a bacterial α2-macroglobulin reveals mimicry of eukaryotic innate immunity. Nat. Commun. 5, 4917 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Goulas T., et al. , Structural and functional insight into pan-endopeptidase inhibition by α2-macroglobulins. Biol. Chem. 398, 975–994 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Seddighi S., Varma V., Thambisetty M., α2-macroglobulin in Alzheimer’s disease: New roles for an old chaperone. Biomarkers Med. 12, 311–314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starkey P. M., Barrett A. J., Inhibition by α-macroglobulin and other serum proteins. Biochem. J. 131, 823–831 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonias S. L., et al. , α 2-Macroglobulin and the α 2-macroglobulin receptor/LRP. A growth regulatory axis. Ann. N. Y. Acad. Sci. 737, 273–290 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Chu C. T., Pizzo S. V., Interactions between cytokines and α 2-macroglobulin. Immunol. Today 12, 249 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Liu N., et al. , Transcuprein is a macroglobulin regulated by copper and iron availability. J. Nutr. Biochem. 18, 597–608 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sottrup-Jensen L., α-Macroglobulins: Structure, shape, and mechanism of proteinase complex formation. J. Biol. Chem. 264, 11539–11542 (1989). [PubMed] [Google Scholar]
- 14.Armstrong P. B., Role of α2-macroglobulin in the immune response of invertebrates. Invertebrate Surviv. J. 7, 165–180 (2010). [Google Scholar]
- 15.Liu C. J., The role of ADAMTS-7 and ADAMTS-12 in the pathogenesis of arthritis. Nat. Clin. Pract. Rheumatol. 5, 38–45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Troeberg L., Nagase H., Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 1824, 133–145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaller J., Gerber S. S., The plasmin-antiplasmin system: Structural and functional aspects. Cell. Mol. Life Sci. 68, 785–801 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borth W., Alpha 2-macroglobulin, a multifunctional binding protein with targeting characteristics. FASEB J. 6, 3345–3353 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Roberts R. C., Protease inhibitors of human plasma. Alpha-2-macroglobulin. J. Med. 16, 129–224 (1985). [PubMed] [Google Scholar]
- 20.Zucker S., Lysik R. M., Zarrabi M. H., Fiore J. J., Strickland D. K., Proteinase-alpha 2 macroglobulin complexes are not increased in plasma of patients with cancer. Int. J. Cancer 48, 399–403 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Ferrer I., Marrero A., Gomis-Rüth F. X., Goulas T., α2-Macroglobulins: Structure and function. Subcell. Biochem. 83, 149–183 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Kisselev A. F., von der Helm K., Human immunodeficiency virus type 1 proteinase is rapidly and efficiently inactivated in human plasma by α 2-macroglobulin. Biol. Chem. Hoppe Seyler 375, 711–714 (1994). [PubMed] [Google Scholar]
- 23.Sánchez E. F., Flores-Ortiz R. J., Alvarenga V. G., Eble J. A., Direct fibrinolytic snake venom metalloproteinases affecting hemostasis: Structural, biochemical features and therapeutic potential. Toxins (Basel) 9, 392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laskowski M. Jr., Kato I., Protein inhibitors of proteinases. Annu. Rev. Biochem. 49, 593–626 (1980). [DOI] [PubMed] [Google Scholar]
- 25.Rawlings N. D., Peptidase inhibitors in the MEROPS database. Biochimie 92, 1463–1483 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Marrero A., et al. , The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew. Chem. Int. Ed. Engl. 51, 3340–3344 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Sottrup-Jensen L., Petersen T. E., Magnusson S., A thiol-ester in α 2-macroglobulin cleaved during proteinase complex formation. FEBS Lett. 121, 275–279 (1980). [DOI] [PubMed] [Google Scholar]
- 28.Travis J., Salvesen G. S., Human plasma proteinase inhibitors. Annu. Rev. Biochem. 52, 655–709 (1983). [DOI] [PubMed] [Google Scholar]
- 29.Qazi U., Gettins P. G., Strickland D. K., Stoops J. K., Structural details of proteinase entrapment by human alpha2-macroglobulin emerge from three-dimensional reconstructions of Fab labeled native, half-transformed, and transformed molecules. J. Biol. Chem. 274, 8137–8142 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Armstrong P. B., Quigley J. P., α2-Macroglobulin: An evolutionarily conserved arm of the innate immune system. Dev. Comp. Immunol. 23, 375–390 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Debanne M. T., Bell R., Dolovich J., Characteristics of the macrophage uptake of proteinase-α-macroglobulin complexes. Biochim. Biophys. Acta 428, 466–475 (1976). [DOI] [PubMed] [Google Scholar]
- 32.Delain E., et al. , The molecular organization of human alpha 2-macroglobulin. An immunoelectron microscopic study with monoclonal antibodies. J. Biol. Chem. 263, 2981–2989 (1988). [PubMed] [Google Scholar]
- 33.Andersen O. M., et al. , Specific binding of α-macroglobulin to complement-type repeat CR4 of the low-density lipoprotein receptor-related protein. Biochemistry 39, 10627–10633 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Imber M. J., Pizzo S. V., Clearance and binding of two electrophoretic “fast” forms of human α2-macroglobulin. J. Biol. Chem. 256, 8134–8139 (1981). [PubMed] [Google Scholar]
- 35.Jenner L., Husted L., Thirup S., Sottrup-Jensen L., Nyborg J., Crystal structure of the receptor-binding domain of α2-macroglobulin. Structure 6, 595–604 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Huang W., Dolmer K., Liao X., Gettins P. G. W., NMR solution structure of the receptor binding domain of human α(2)-macroglobulin. J. Biol. Chem. 275, 1089–1094 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Xiao T., DeCamp D. L., Sprang S. R., Structure of a rat α1-macroglobulin receptor-binding domain dimer. Protein Sci. 9, 1889–1897 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janssen B. J. C., et al. , Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 437, 505–511 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Doan N., Gettins P. G. W., Human α2-macroglobulin is composed of multiple domains, as predicted by homology with complement component C3. Biochem. J. 407, 23–30 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen G. R., Koch T. J., Dolmer K., Sottrup-Jensen L., Nyborg J., Low resolution X-ray structure of human methylamine-treated α2-macroglobulin. J. Biol. Chem. 270, 25133–25141 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Boisset N., Taveau J. C., Pochon F., Lamy J., Similar architectures of native and transformed human α2-macroglobulin suggest the transformation mechanism. J. Biol. Chem. 271, 25762–25769 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Kolodziej S. J., Schroeter J. P., Strickland D. K., Stoops J. K., The novel three-dimensional structure of native human α2-macroglobulin and comparisons with the structure of the methylamine derivative. J. Struct. Biol. 116, 366–376 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Kolodziej S. J., Wagenknecht T., Strickland D. K., Stoops J. K., The three-dimensional structure of the human alpha 2-macroglobulin dimer reveals its structural organization in the tetrameric native and chymotrypsin alpha 2-macroglobulin complexes. J. Biol. Chem. 277, 28031–28037 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Harwood S. L., et al. , Structural investigations of human A2M identify a hollow native conformation that underlies its distinctive protease-trapping mechanism. Mol. Cell. Proteomics 20, 100090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett A. J., Brown M. A., Sayers C. A., The electrophoretically ‘slow’ and ‘fast’ forms of the α 2-macroglobulin molecule. Biochem. J. 181, 401–418 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sottrup-Jensen L., Petersen T. E., Magnusson S., Trypsin-induced activation of the thiol esters in alpha 2-macroglobulin generates a short-lived intermediate (‘nascent’ alpha 2-M) that can react rapidly to incorporate not only methylamine or putrescine but also proteins lacking proteinase activity. FEBS Lett. 128, 123–126 (1981). [DOI] [PubMed] [Google Scholar]
- 47.Jensen G. A., et al. , Binding site structure of one LRP-RAP complex: Implications for a common ligand-receptor binding motif. J. Mol. Biol. 362, 700–716 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Sottrup-Jensen L., et al. , Primary structure of the ‘bait’ region for proteinases in alpha 2-macroglobulin. Nature of the complex. FEBS Lett. 127, 167–173 (1981). [DOI] [PubMed] [Google Scholar]
- 49.Goulas T., Garcia-Ferrer I., García-Piqué S., Sottrup-Jensen L., Gomis-Rüth F. X., Crystallization and preliminary X-ray diffraction analysis of eukaryotic α2-macroglobulin family members modified by methylamine, proteases and glycosidases. Mol. Oral Microbiol. 29, 354–364 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Ferrer I., et al. , Structural and functional insights into Escherichia coli α2-macroglobulin endopeptidase snap-trap inhibition. Proc. Natl. Acad. Sci. U.S.A. 112, 8290–8295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohn E. J., et al. , Preparation and properties of serum and plasma proteins; a system for the separation into fractions of the protein and lipoprotein components of biological tissues and fluids. J. Am. Chem. Soc. 68, 459–475 (1946). [DOI] [PubMed] [Google Scholar]
- 52.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez-Leiro R., Scheres S. H. W., A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de la Rosa-Trevín J. M., et al. , Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Scheres S. H., Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Vilas J. L., et al. , MonoRes: Automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Ramírez-Aportela E., et al. , Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen E. F., et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smart O. S., et al. , Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68, 368–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clerc F., et al. , Human plasma protein N-glycosylation. Glycoconj. J. 33, 309–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and cryo-EM density maps were deposited in the PDB and EM Data Bank with codes 7O7L and EMD-12747 (native I); 7O7M and EMD-12748 (native II); 7O7N and EMD-12750 (semi-activated I); 7O7O and EMD-12751 (semi-activated II); 7O7P and EMD-12752 (activated); 7O7Q and EMD-12753 (trypsin-activated); 7O7R and EMD-12754 (plasmin-activated I); and 7O7S and EMD-12755 (plasmin-activated II). The cryo-EM density maps of intermediate structures with one, two (two maps), and three activated monomers, corresponding to transient I to IV states, were deposited in the EM Data Bank with codes EMD-12941, EMD-12942, EMD-12943, and EMD-12944. The final X-ray crystallography model was likewise deposited at the PDB (PDB 6TAV) and supersedes PDB entry 4ACQ.