Abstract
Wastewater-based epidemiology (WBE) has proven to be a useful surveillance tool during the ongoing SARS-CoV-2 pandemic, and has driven research into evaluating the most reliable and cost-effective techniques for obtaining a representative sample of wastewater. When liquid samples cannot be taken efficiently, passive sampling approaches have been used, however, insufficient data exists on their usefulness for multi-virus capture and recovery. In this study, we compared the virus-binding capacity of two passive samplers (cotton-based tampons and ion exchange filter papers) in two different water types (deionised water and wastewater). Here we focused on the capture of wastewater-associated viruses including Influenza A and B (Flu-A & B), SARS-CoV-2, human adenovirus (AdV), norovirus GII (NoVGII), measles virus (MeV), pepper mild mottle virus (PMMoV), the faecal marker crAssphage and the process control virus Pseudomonas virus phi6. After deployment, we evaluated four different methods to recover viruses from the passive samplers namely, (i) phosphate buffered saline (PBS) elution followed by polyethylene glycol (PEG) precipitation, (ii) beef extract (BE) elution followed by PEG precipitation, (iii) no-elution into PEG precipitation, and (iv) direct extraction. We found that the tampon-based passive samplers had higher viral recoveries in comparison to the filter paper. Overall, the preferred viral recovery method from the tampon passive samplers was the no-elution/PEG precipitation method. Furthermore, we evidenced that non-enveloped viruses had higher percent recoveries from the passive samplers than enveloped viruses. This is the first study of its kind to assess passive sampler and viral recovery methods amongst a plethora of viruses commonly found in wastewater or used as a viral surrogate in wastewater studies.
Keywords: COVID-19 surveillance, Sewage sampling, Viral capture method, Public health risk, Environmental monitoring
Graphical abstract
1. Introduction
The COVID-19 pandemic, declared in March 2020, is responsible for millions of deaths worldwide (World Health Organization, 2021) and has caused major disruption to world trade and social wellbeing (Wei et al., 2021). The causative agent of COVID-19 is SARS-CoV-2, an enveloped, spherical, positive sense, single-stranded RNA (+ssRNA) virus that causes a wide-ranging infection, on a clinical spectrum from asymptomatic infection to severe flu-like symptoms. Positive SARS-CoV-2 cases are most often confirmed in individuals via lateral flow testing or by reverse transcription quantitative polymerase chain reaction (RT-qPCR), conducted in a diagnostic laboratory. Testing is usually performed when the infected person becomes symptomatic, therefore asymptomatic cases may go undiagnosed (Kronbichler et al., 2020). For population-level disease surveillance, wastewater-based epidemiology (WBE) may be used as a non-invasive alternative to clinical disease monitoring (Wade et al., 2022). Monitoring human pathogens through sewage has been used for the past 75 years, mainly focusing on enteric bacteria and viruses transmitted via the faecal-oral route, for example typhoid, poliovirus, enterovirus, and adenovirus (Gell et al., 1945; Moore, 1951; Sinclair et al., 2008). SARS-CoV-2-infected individuals have been evidenced to shed virus in their faeces at 101 to 108 genome copies (gc) g−1 (Cheung et al., 2020; Jeong et al., 2020; Zheng et al., 2020), several days before symptoms commence (Zhu et al., 2021). The shedding of SARS-CoV-2 viral RNA through faeces is independent of infection severity, and all infected individuals, including asymptomatic and pre-symptomatic cases shed through their faeces (Zheng et al., 2020; Gerrity et al., 2021), whilst viral shedding in urine is rare (Huang et al., 2020; Lo et al., 2020). Therefore, WBE has now been successfully introduced to monitor SARS-CoV-2 on a spatial and temporal level in many communities across the globe (Gonzalez et al., 2020; Kumar et al., 2020; La Rosa et al., 2020; Sherchan et al., 2020; Agrawal et al., 2021; Ahmed et al., 2021; Gerrity et al., 2021; Hillary et al., 2021). This approach enables the monitoring of large cities (Agrawal et al., 2021; Karthikeyan et al., 2021; Kumar et al., 2021; Saguti et al., 2021) or can be used for small-scale monitoring (Corchis-Scott et al., 2021). The emergence of WBE in viral monitoring has led to the development of different methods for sampling wastewater. Most of the research and surveillance has been conducted using grab or composite sewage samples (Shah et al., 2022), taken either manually or with an autosampler device. However, grab samples may not be representative due to diurnal variations in virus titres in wastewater, and the deployment of autosamplers is often not feasible for logistical or economic reasons. Therefore, alternative techniques for wastewater sampling are needed.
Passive sampling is a method stemming from Moore (1951) where an absorbent material is immersed in the wastewater stream to capture human pathogens (e.g. bacteria, viruses; Sikorski and Levine, 2020; Schang et al., 2021). However, their use is not limited to biological agents, as passive samplers are also used to monitor chemical agents and pollutants in the environment (Greenwood et al., 2007). Passive sampling works on the premise that the sampler material surface charge attracts and holds viruses over a prolonged period of time (Blanco et al., 2019). The use of passive samplers could lower the cost of WBE, by reducing sampling machinery and requires no electrical power or running costs. Over the course of the SARS-CoV-2 pandemic, different passive samplers have been explored, such as cotton gauze (Hayes et al., 2021a; Rafiee et al., 2021; Schang et al., 2021; Habtewold et al., 2022; Li et al., 2022b; Liu et al., 2022; Wang et al., 2022), cotton buds (Schang et al., 2021; Habtewold et al., 2022; Li et al., 2022b), cheese cloth (Hayes et al., 2021a), electromagnetic membranes (Hayes et al., 2021a, Hayes et al., 2021; Schang et al., 2021; Habtewold et al., 2022; Li et al., 2022a, Li et al., 2022), cellulose sponge (Hayes et al., 2021a) and tampons (Corchis-Scott et al., 2021; Bivins et al., 2022a; Li et al., 2022b). Their effectiveness at viral capture varies depending upon a number of factors, such as surface volume of the sampler, length of exposure time and material composition (as reviewed by Bivins et al., 2022a). As viral loads in wastewater can be low, the sampler should be designed to effectively concentrate the virus increasing detection sensitivity (Vincent-Hubert et al., 2017, Vincent-Hubert et al., 2021). This increased sensitivity may also allow early identification of low-level disease outbreaks, including SARS-CoV-2 as hypothetically discussed by Jiang et al. (2022). Furthermore, passive sampling has been shown to outperform grab samples, being more consistent with composite sampling (Bivins et al., 2022a), and thus more sensitive to changes in viral loads over time; however, this has only been evidenced for SARS-CoV-2 using Moore swab sampling (Rafiee et al., 2021). Recent studies have highlighted the use of in-situ passive samplers to detect enterovirus, human adenovirus (hAdV), and pepper mild mottle virus (PMMoV) and SARS-CoV-2 from wastewater (Li et al., 2022b). These studies investigated different sampler types, with an aim to develop cheaper, less time consuming sampling methods (Li et al., 2022b). However, the effectiveness of sample processing and viral recovery methods from passive samplers for a range of viruses has not been explored. A critical assessment of viral recovery from wastewater using different methodologies and sampler types is therefore required to improve WBE beyond SARS-CoV-2 monitoring and to enable the development of robust standardized protocols.
2. Methods
2.1. Study aims
This study aims to evaluate two passive sampler materials, Tampax Super Compak Tampon (Procter & Gamble Inc., Cincinnati, OH) and Whatman SG81 Si-cellulose ion exchange paper (Global Life Sciences Solutions USA, Marlborough, MA), alongside four different viral concentration and extraction methods (phosphate buffered saline (PBS) elution into polyethylene glycol (PEG) precipitation, beef extract (BE) elution into PEG precipitation, no elution in PEG precipitation, and direct nucleic acid extraction), with two water types (wastewater and deionised water (dH2O)), to identify the best material and method for viral recovery of SARS-CoV-2, Influenza A and B viruses (Flu-A & B), measles virus (MeV), norovirus GII (NoVGII), adenovirus (AdV), human faecal markers (crAssphage and pepper mild mosaic virus (PMMoV)), and a process control virus (Pseudomonas virus phi6 (Phi6)). To the authors knowledge this is the first time that a comprehensive study, looking at multiple viral targets, samplers and recovery methods has been undertaken.
2.2. Sample collection
On the 15th November 2021, 20 l of untreated sewage influent (hereby referred to as wastewater) was collected at the central wastewater treatment plant (WwTP) at 09.00 h located in Bangor, North Wales, UK (53°12′34.04″N, 4°10′58.56″W). This sampling time was chosen to reflect peak flow and aimed to capture the highest faecal load (Hillary et al., 2021). The WwTP serves a population of 40,000 people and is mainly composed of domestic wastewater with few industrial inputs. Samples of crude influent wastewater were taken from behind the primary screen (flow 285 l s−1) in polypropylene bottles and immediately transported to the laboratory at 4 °C for experimentation (within 5 km of the WwTP). At the time of collection, the pH of the wastewater was 7.28, the electrical conductivity was 605 μS cm−1, the turbidity was 226 NTU and the ammonium and phosphate concentrations were 1.3 mg N l−1 and 2.46 mg P l−1, respectively.
2.3. Viral stocks and spiking
For spiking, we used inactivated SARS-CoV-2 (kindly provided by Prof Andrew Weightman, Cardiff University), influenza A/California/07/2009 (H1N1) and B/Lee/40 strains (kindly provided by Dr. Eleanor Gaunt, University of Edinburgh), NoVGII in diluted and filtered faecal matter from a patient with confirmed norovirus infection (kindly provided by Dr. Lydia Drumwright, University of Cambridge) and MeV in the form of a vaccine (VWR International, USA). We also used the Pseudomonas spp. phi6 bacteriophage, which we cultured in-house as described in Kevill et al. (2022).
We created four groups of samples: spiked wastewater, unspiked wastewater, spiked dH2O, and unspiked dH2O. To achieve this, we prepared 2 × 3000 ml dH2O and 2 × 3000 ml wastewater aliquots. Subsequently, one set of each water type was spiked with SARS-CoV-2, Flu-A, Flu-B, MeV, NoVGII and Phi6 to reach the final concentration of approx. 104–105 genome copies (gc)/ml per virus. We used this specific concentration for spiking to enable the calculation of recoveries as low as 0.1 % for the precise comparison of methods. Wastewater was spiked as the water matrices is representative of a typical wastewater sample, and dH2O was spiked to allow for inhibition to be compared between the two sample types. The unspiked wastewater samples were used to determine the baseline of viruses that occur in wastewater (SARS-CoV-2, NoVGII, MeV, Flu A and B). The wastewater sample used for spiking was negative for MeV, Flu A and B, yet positive for SARS-CoV-2 and NoVGII, albeit at levels much lower than spiking concentrations (103 gc/sampler). The unspiked dH2O were used as negative controls. Aliquots of 100 ml of wastewater and dH2O were placed in sterile polypropylene copolymer (PCCO) centrifuge jars for experimentation. Each aliquot was made in triplicate, in four groups for each of the four viral recovery methods and passive sampler type (Tampax Super Compak Tampon and SG81 ion exchange Whatman paper).
2.4. Viral recovery methods
All samples were processed in a Containment level/Biosecurity level 2 (CL/BSL2) laboratory, in class II biological safety cabinets. For each of the methods and sample type (spiked or unspiked wastewater or dH2O), a Tampax Super Compak Tampon or 3 cm diameter circular, SG81 ion exchange Whatman paper (both now referred to as passive sampler) was placed into the corresponding 100 ml aliquot and left at room temperature (20 °C) for 1 h. The passive samplers were then recovered and transferred to plastic Ziploc® bags (SC Johnson & Son Inc., Racine, WI), and processed immediately as per the methods detailed below. This resulted in direct comparisons between spiked/unspiked sample and tampon/Whatman paper samplers.
Subsamples of 200 μl spiked and unspiked wastewater and dH2O were taken in triplicates at the start of the experiment. The nucleic acids were also extracted from these to allow baseline quantification of the viruses present.
2.4.1. Phosphate buffered saline (PBS) elution into polyethylene glycol (PEG) precipitation (PBS-PEG method)
Each of the passive samplers per sample type were saturated with 20 ml sterile PBS, pH 7.4. The PBS was then hand massaged into the passive sampler, the corner of the Ziploc® bag was cut and the liquid squeezed into a sterile 50 ml tube. The volume of each eluent was adjusted to 30 ml with PBS; this step ensured that the final concentration of PEG is consistent between samples. Samples were centrifuged at 3000g at 4 °C for 30 mins. The supernatant was then poured into a new sterile 50 ml centrifuge tube, without disturbing the pellet. A 10 ml aliquot of 40 % PEG8000 with 8 % NaCl (PEG-NaCl) solution was then added to each eluent to reach a final concentration of 10 % PEG and 2 % NaCl. The tubes were inverted several times to mix, followed by incubation at 4 °C for 16 h. After incubation, the samples were precipitated by centrifuging at 10,000g for 30 min at 4 °C. The supernatant was then discarded without disturbing the pellet. The pellet was then resuspended in 800 μl of NucliSens lysis buffer (bioMérieux SA, Marcy l'Etoile, France). The viral nucleic acids were extracted using the NucliSens extraction reagents (bioMérieux SA), as described previously (Farkas et al., 2021). The final volume of the eluent was 0.1 ml.
2.4.2. Beef extract (BE) elution into PEG precipitation (BE-PEG method)
The BE-PEG method is identical to the PBS-PEG method described above except the 20 ml of sterile PBS used to saturate the passive sampler was replaced with 20 ml of 3 % beef extract containing 0.5 M glycine (Lambertini et al., 2008). The BE solution was freshly prepared on the day of use and pH of adjusted to 7.0 using NaOH, prior to saturation of the passive sampler. The nucleic acid extraction method is also the same as that described for the PBS-PEG method.
2.4.3. No elution in PEG precipitation (no elution-PEG method)
The no elution method follows the PBS-PEG method described above except that PBS was not added to the passive sampler. Instead, the passive sampler was massaged and the solution it contained directly squeezed into a sterile 50 ml tube. The rest of the protocol remains the same. The nucleic acid extraction method is also the same as that described for the PBS-PEG method.
2.4.4. Direct extraction method
The direct extraction method differed between tampon and Whatman paper sampler. A 1 cm2 area was cut from the tampon whilst for the Whatman paper all of the 3 cm diameter circle was used. Subsequently, each material was placed into 50 ml sterile polypropylene tubes alongside 2 ml of NucliSens lysis buffer (bioMérieux SA). The samples were then vortexed for 10 s and incubated at room temperature for 10 min. The tampon fragment or Whatman paper was then removed from the solution and viral nucleic acid were extracted as described previously (Farkas et al., 2021).
2.5. Quantification of viral RNA/DNA
Due to the number of PCR targets, differing reaction chemistry and cycling conditions, details of each qPCR method and kit are displayed in Table 1 , whilst the primers and probes used are presented in Table 2 . All reactions were run on a QuantStudio Flex 6 (Applied Biosystems Inc., Waltham, USA), at a reaction volume of 20 μl. Samples were run in duplicate, against a ssRNA (SARS CoV-2 N1 gene fragment, Phi6, Flu A and B), ssDNA (PMMoV, MeV) or plasmid DNA (CrAssphage, AdV and NoVGII) standard curve dilution series of the target sequence in the range of 1–105 copies μl−1 per reaction. The standard curve concentration was determined by a Qubit fluorometer (Invitrogen, Waltham, USA), prior to preparing the 10-fold standard dilution series for N1 and Phi6, as these RNA standards were made in-house as previously described (Kevill et al., 2022). The remaining targets were purchased at 106 copies/μl (Influenza A and B (Twist bioscience, San Francisco, CA, USA)) or 1012 copies/μl (MeV, PMMoV, CrAssphage, AdV and NoV (IDT, Iowa, USA)) from commercial companies. PCR no-template controls (molecular-grade water) determined the absence of contamination during the PCR set-up. For RNA targets (Table 1), the TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems Inc., USA) was used with 4 × Reaction Mix with ROX, 10 pmol of the forward, 20 pmol of the reverse primers and 5 pmol probe, 16 nmol MgSO4, 1 μg bovine serum albumin (BSA), molecular grade water and 4 μl sample/standard/control. For DNA targets (Table 1) the QuantiNova Probe qPCR and QuantiFast SYBR reactions are previously described (Farkas et al., 2018, Farkas et al., 2019).
Table 1.
Viral target, genome structure, kit and qPCR cycling conditions.
| Viral target | Genome | Kit | Cycling conditions |
|---|---|---|---|
| SARS-CoV-2 | RNA | TaqMan Fast Virus 1-Step Master Mix | 50 °C 30 min, 95 °C 20 s, × 45 cycles of 95 °C 0.03 s, 60 °C 3 min |
| Phi6 | |||
| Influenza-A | |||
| Influenza-B | |||
| Norovirus GII | |||
| Measles | |||
| CrAssphage | DNA | QuantiNova | 95 °C 2 min, × 40 cycles of 95 °C 15 s, 60 °C 1 min |
| Adenovirus | DNA | QuantiFast SYBR | 95 °C 5 min, × 40 cycles of 95 °C 15 s, 55 °C 1 min, melt at 95 °C 15 s, 60 °C 1 min, 95 °C 15 s |
Table 2.
Primers and probes used for qPCR viral detection within this study.
| Target | Reference | Primers and probe name | Primer and probe sequence (5′-3′) | Target sequence (5′-3′) |
|---|---|---|---|---|
| SARS-CoV-2 | (Centers for Disease Control and Prevention, 2020) | N1-F | GACCCCAAAATCAGCGAAAT | GTGAAATGGTCATGTGTGGCGGTTCACTATATGTTAA ACCAGGTGGAACCTCATC AGGAGATGCCACAACTGCTTATGCTAATAGTGTTTTTAA CATTTG |
| N1-R | TCTGGTTACTGCCAGTTGAATCTG | |||
| N1-P | ACCCCGCATTACGTTTGGTGGACC | |||
| Norovirus GII | (ISO, 2019) | NoVGII-F | ATGTTCAGRTGGATGAGRTTCTCWGA | ATGTTCAGATGGATGAGAT TCTCAGATCTGAGCACGTGGGAGGGCGATCGCAATCTGGCTCCCAGTTTTGTGAATGAAGATGGCGTCGA |
| NoVGII-R | TCGACGCCATCTTCATTCACA | |||
| NoVGII-P | AGCACGTGGGAGGGCGATCG | |||
| Pepper mild mottle virus | (Haramoto et al., 2013) | PMMoV-F | GAGTGGTTTGACCTTAACGTTTGA | GAGTGGTTTGACCTTAACGT TTGAGAGGCCTACCGAAGCAAATGTCGCACTTGCATTGCAACCGACAA |
| PMMoV-R | TTGTCGGTTGCAATGCAAGT | |||
| PMMoV-P | CCTACCGAAGCAAATG | |||
| CrAssphage | (Stachler et al., 2017) | CrAss-F | CAGAAGTACAAACTCCTAAAAAACGTAGAG | CAGAAGTACAAACTCCTAAAAAACGTAGAGGTAGAGGTATTAATAACGATTTACGTGATGTAACTCGTAAAAAGTTTGATGAACGTACTGATTGTAATAAAGCTAATGGCTTGTTTATTGGTC |
| CrAss-R | GATGACCAATAAACAAGCCATTAGC | |||
| CrAss-P | AATAACGATTTACGTGATGTAAC | |||
| Influenza A | (Shu et al., 2021) | Influ-A-F | CAAGACCAATCYTGTCACCTCTGAC CAAGACCAATYCTGTCACCTYTGAC | AAAGACAAGACCAATCCTGTCACCTCTGACTAAGGGGATTTTAGGATTTGTGTTCACGCTCACCGTGCCCAGTGAGCGAGGACTGCAGCGTAGACGCTTTGTCCAAAATGCCCTAAATGGG |
| Influ-A-R | GCATTYTGGACAAAVCGTCTACG GCATTTTGGATAAAGCGTCTACG | |||
| Influ-A-P | TGCAGTCCTCGCTCACTGGGCACG | |||
| Influenza B | (Shu et al., 2021) | Influ-B-F | TCCTCAAYTCACTCTTCGAGCG | GGATCCTCAACTCACTCTTCGAGCGTTTTGATGAAGGACATTCAAAGCCAATTCGAGCAGCTGAAACTGCGGTGGGAGTCTTATCCCAATTTGGTCAAGAGCACCGATT |
| Influ-B-R | CGGTGCTCTTGACCAAATTGG | |||
| Influ-B-P | CCAATTCGAGCAGCTGAAACTGCGGTG | |||
| Measles virus | (Hummel et al., 2006) | MeV-F | TGGCATCTGAACTCGGTATCAC | TGGCATCTGAACTCGGTATCACTGCTGAGGATGCAAGGCTTGTTTCAGAGATTGCAATGCATACTACTGAGGACA |
| MeV-R | TGTCCTCAGTAGTATGCATTGCAA | |||
| MeV-P | CCGAGGATGCAAGGCTTGTTTCAGA | |||
| Phi6 phage | (Gendron et al., 2010) | Phi6-F | TGGCGGCGGTCAAGAGC | TGGCGGCGGTCAAGAGCAACCCGGTCGTCGCAGGTCTGACACTCGCTCAGATCGGAAGCACCGGTTATGACGCCTATCAGCAGCTTCTGGAGAATCATCC |
| Phi6-R | GGATGATTCTCCAGAAGCTGCTG | |||
| Phi6-P | CGGTCGTCGCAGGTCTGACACTCGC | |||
| Adenovirus | (van Maarseveen et al., 2010) | AdV_R AdV_F |
CCGGCCGAGAAGGGTGTGCGCAGGTA CATGACTTTTGAGGTGGATC |
CATGACTTTTGAGGTGGATCCCATGGATGAGCCCACCCTGCTTTATCTTCTTTTCGAAGTCTTCGACGTGGTCAGAGTGCACCAGCCACACCGCGGCGTCATCGAGGCCGTCTACCTGCGCACACCGTTCTCGGCCGG |
2.6. Data analysis
qPCR data analysis and quality control were performed using the QuantStudio Real-time PCR software v1.7 (Applied Biosystems, Inc., USA). Viral concentrations were expressed as gc/μl nucleic acid extract and were converted to gc/sampler (wastewater or dH2O) by multiplying gc/μl by final nucleic extract elution volume. The data for direct extraction method using tampon passive samplers were multiplied up to one whole tampon, as the PSB-PEG, BE-PEG, and no elution methods recover viruses from one whole tampon; this allowed for direct comparisons between methods. Triplicate 200 μl unconcentrated subsamples also underwent total nucleic acid extraction. These subsamples provided the baseline gene copies spiked per 100 ml of sample and enabled the calculation of viral recoveries. The gc/100 ml were calculated by multiplying gc/μl nucleic extract by volume of eluent (100) and then multiplying by the sample volume extracted (500). Viral recoveries were calculated by taking the average gc/sampler and then dividing by the average baseline viral concentrations. This data was then used for statistical analysis.
Statistical tests were carried out in R; the full script and data are provided in a dedicated repository (https://github.com/CameronPellett/spiked-passive-Bangor).
Passive sample material (tampon and Whatman paper), concentration and extraction method (BE-PEG, direct extraction, no elution PEG, and PBS-PEG), water type (wastewater WW, and dH2O), and virus envelope (enveloped, and non-enveloped) were selected as factors and co-variates of viral recovery. A sequential approach to analysis was adopted, removing data after each test to ensure later results were representative of the expected environmental conditions and best sampling practices. A multiple linear model with interaction effects was considered but deemed unacceptable due to unrepresentative groups of data after initial tests. First, viral recovery was compared between water types to clarify the effect of inhibitors; water type comparisons were made with only spiked samples, due to a lack of naturally present virus in unspiked dH2O. After assessing water type, results for dH2O were removed from later assessment as they would not mimic expected environmental conditions in WBE. Second, the viral recovery using tampon and Whatman paper passive sample materials were compared. Following the material comparison, results using the material with significantly lower recovery were removed so that later comparison would not be skewed by a less effective material. Third, after selecting the best passive sampler material, viral recovery between laboratory processing methods were compared for samples suspended in wastewater. Then finally, the effect of a viral envelope was compared using results from the best passive sampler material suspended in wastewater with results from all processing methods. For statistical tests the recovery percentile was log transformed to meet assumptions of a Gaussian distribution (see Supplementary materials Figs. S1–S5 for quantile-quantile plots). Equality of variances was tested with F tests. Statistical comparisons of features with two levels and non-equal variance were made with Welch two sample t-tests. Comparisons with three or more levels and non-equal variance were made with a Welch ANOVA (one way comparison of means), followed by pairwise two sample t-tests without pooled standard deviations, adjusting p-values with the Holm-Bonferroni method. Paired tests were not selected due to missing data created by removal of undetermined results and sample removal during qPCR quality control.
3. Results
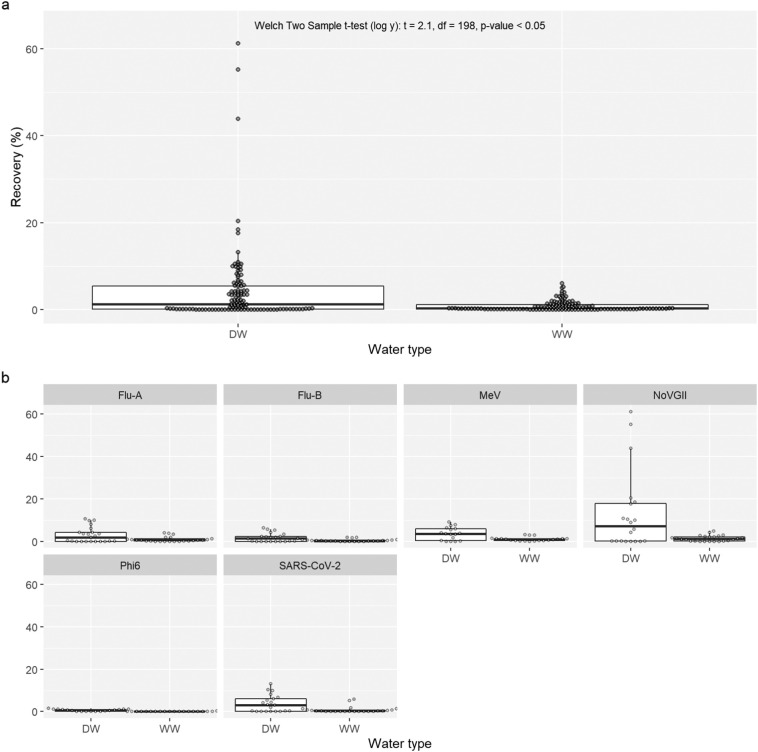
3.1. Wastewater reduces recovery of spiked virus compared to deionised water
To assess the effect of inhibitors, recovery of virus from passive samples suspended in wastewater and dH2O were compared. Passive samples had a greater median recovery of spiked viruses when they were suspended in deionised water (1.26 %; n = 121) compared to suspension in wastewater (0.26 %; n = 136). The difference between water types was found to be significant when comparing log transformed recovery (Fig. 1a; Welch Two Sample t-test (log y): t = 2.1, df = 198, p-value < 0.05); this trend was seen in comparisons between all individual viruses (Fig. 1b). These results suggest chemicals or other materials in wastewater influence adherence of virus on the passive sampler or inhibit later processing and quantification of the viral nucleotides collected in the sample. After identifying the differences in recovery between water types, only data for wastewater suspended samples were taken forward for further analysis, as these better reflected real-world conditions. Details of sample type, sampler type and viral recovery are also individually presented in Supplementary Fig. 1.
Fig. 1.
Comparison of spiked virus recovery using passive samplers in wastewater (WW) and deionised water (DW). Panel ‘a’ combines data for all viruses, whilst panel ‘b’ separates recovery by each virus, such as influenza A and B (Flu-A and Flu-B), measles virus (MeV), SARS-CoV-2 (N1), norovirus GII (NoVGII) and bacteriophage phi6 (Phi6). A Welch two sample t-test was used to compare log transformed recoveries. Biological replicates were not averaged.
3.2. Tampon passive samplers have improved recovery over Whatman paper in wastewater samples
To identify the optimum material of passive samplers, tampon and Whatman paper samplers were compared. The median recovery of Tampon passive samplers (0.59 %; n = 136) were greater than the recoveries observed for Whatman paper (0.16 %; n = 140). This was found to be significant when comparing log transformed recovery (Fig. 2a; Welch Two Sample t-test (log y): t = 11.6, df = 248, p-value < 0.001); the same trend was seen in all individual viruses (Fig. 2b). These results indicate that the Whatman paper is not suitable as a wastewater passive sampler. Therefore, all further data analysis was performed on the tampon passive sampler data.
Fig. 2.
Comparison of viral recovery of tampon (T) and Whatman (W) paper passive samplers suspended in wastewater. Panel ‘a’ combines data for all viruses, whilst panel ‘b’ separates recovery by each virus such as human adenovirus (AdV), crAssphage (CrAss), Flu-A and Flu-B, MeV, SARS-CoV-2, NoVGII, Phi6 and pepper mild mottle virus (PMMoV). A Welch two sample t-test was used to compare log transformed recoveries.
3.3. No elution PEG and direct extraction methods have improved viral recovery
To select the most efficient passive sample viral concentration and extraction method, four processing methods were compared. The no elution PEG concentration method had the highest median viral recovery (1.97 %; n = 36), followed by direct extraction (0.81 %; n = 36), BE-PEG (0.43 %; n = 36), then PBS-PEG methods (0.03 %; n = 28). Significant differences between the log transformed viral recovery of the methods were found (Fig. 3a; Welch ANOVA (log y): F = 28.1, df = 3, p-value < 0.001), though pairwise comparisons found no significant difference between the no elution PEG and direct extraction methods (Fig. 3c). This was likely due to some viruses (AdV and Phi6) having greater recovery with direct extraction compared to no elution PEG (Fig. 3b). These results suggest the no elution PEG method is generally preferred for processing passive samples, but if AdV or Phi6 are the primary targets, the direct extraction method may also be selected.
Fig. 3.
Comparison of viral recovery between processing methods carried out on tampon passive samples suspended in wastewater. Panel ‘a’ combine's data for all viruses, panel ‘b’ separates recovery by each virus, and panel ‘c’ shows p-values (p-value: <0.001 [***]; <0.01 [**]; <0.05 [*]; >0.05 [.]) of pairwise t-tests without pooled standard deviations adjusted with the Holm-Bonferroni method.
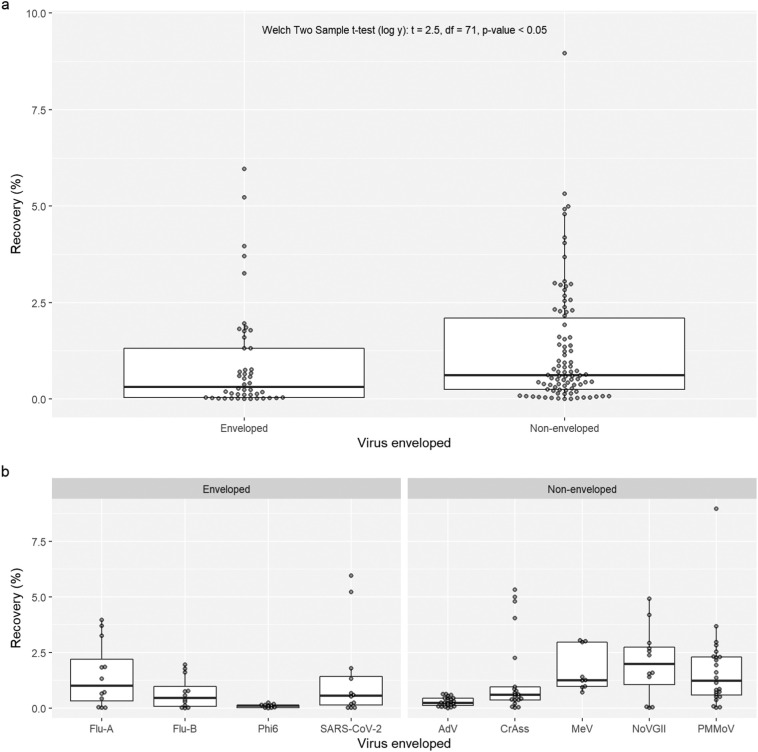
3.4. Enveloped viruses have reduced recovery using passive samplers
The viral envelope was identified as a potentially key virus characteristic that may impact viral recovery. Enveloped and non-enveloped viruses were, grouped and their recovery compared. Non-enveloped viruses had a greater median recovery (0.63 %; n = 90), compared to enveloped viruses (0.32 %; n = 46). The difference in mean log transformed recovery between enveloped and non-enveloped viruses was found to be significant (Welch Two Sample t-test (log y): t = 2.5, df = 71, p-value < 0.05; Fig. 4a). These results indicate the viral envelope may be influencing the adherence of the viral particles to the passive sampler.
Fig. 4.
Comparison of enveloped and non-enveloped virus recovery. Panel ‘a’ combines data from all viruses, whilst panel ‘b’ separates viruses individually. A Welch two sample t-test was used to compare log transformed recoveries.
4. Discussion
Cotton, tampon-based passive sampler devices have been used for wastewater viral infection surveillance in previous studies (Corchis-Scott et al., 2021; Bivins et al., 2022b; Li et al., 2022b) and have proven successful. A study of different passive sampler types show tampons are more effective than some traditional (Moore swab) and novel (cotton-based medical gauze swab) materials for the recovery of SARS-CoV-2 from wastewater in-situ (Lambert-Slosarska et al., 2022, paper in prep). This study compares the two best passive sampler methods; Tampon and Whatman filter paper as per Lambert-Slosarska et al. (2022, paper in prep), for the recovery of eight viruses commonly detected in wastewater and one internal control. This study evidenced that viral recoveries from Whatman paper were poor compared to that of tampons. Both the tampons and Whatman paper were saturated in sample for one hour, showing that short sampling regimes allow viral absorption in tampon passive samplers, yet viruses were barely recovered using the Whatman paper. We recognise that one limitation of this study is that passive sampler materials were submerged in viral contaminated water in a laboratory setting, which does not reflect the field scenario for passive sampling. Therefore, we could not assess the recovery of low abundance viruses or determine the effect of flow rate upon viral capture by the passive sampler material. In this study we placed the passive sampler material in water for one hour, which proved efficient for viral capture. One hour was selected as tampon deployment into the main wastewater stream in field experiments showed that tampons became viral saturated within <6 h (unpublished data). Furthermore, short sampling regimes (<8 h) using tampons have been recommended, whilst other passive sampler types such as electronegative membranes may be better for longer continuous sampling (48 h) (Li et al., 2022b).
A range of viruses commonly detected in wastewater (Rosario et al., 2009; Heijnen and Medema, 2011; Hewitt et al., 2011; Kazama et al., 2016; Eftim et al., 2017; Elmahdy et al., 2019, Elmahdy et al., 2020; Ahmed et al., 2020a; Tiwari et al., 2021), as well as viral process control (phi6) were selected to determine viral affinity for passive sampler type. Understanding viral affinity for passive sampler type is crucial for future experimental design in a world beyond SARS-CoV-2 surveillance where other viral targets begin to be monitored. The method used to recover/precipitate viruses from tampon passive samplers impacted percent recovery, and we found that the no-elution and direct extraction methods were best for viral recoveries from tampons. Viral recoveries were no >10 % when using tampons as passive samplers, whilst other studies evidence far higher viral recoveries of viruses precipitated directly from wastewater (Ahmed et al., 2020b; Brinkman et al., 2013; Farkas et al., 2018; Ikner et al., 2012; Farkas et al., 2022, paper in prep). Passive samplers allow for ease of sampling, are cheaper than using autosamplers, and in the case of the direct extraction method provide a much faster sample-to-data reporting turnaround time. Therefore, whilst recoveries are lower than some liquid sample concentration methods, passive samplers may be preferred when facing constraints, such as short reporting times, lack of funds or expensive equipment. Alternative passive samplers, such as electromagnetic membranes, have also been shown to be comparable to composite sampling (Schang et al., 2021; Habtewold et al., 2022). Their performance should be compared to tampon passive samplers in future studies.
The characteristics of the target virus may influence the choice of method used for viral recovery/precipitation from tampon passive samplers. The no elution PEG method had consistently higher viral recovery than other concentration methods for all viruses except AdV. If AdV is the target virus, then the direct extraction method is preferred for viral recovery from tampon passive samplers. However, in addition to consideration of viral type and method for viral recovery from tampons, it is also worth considering the viral structure (enveloped or unenveloped), as unenveloped viruses had significantly higher mean recovery than enveloped ones. This may be explained by the fact that enveloped viruses are considered more fragile in the environment, as the phospholipid bilayer envelope and its associated proteins are more likely to be affected by changes in temperature, pH, and some disinfectants (Dvorak et al., 2005; Saadatpour and Mohammadipanah, 2020) than non-enveloped viruses (Firquet et al., 2015). Enveloped viruses are also more likely to become inactivated in wastewater than non-enveloped viruses (Casanova et al., 2009; Gundy et al., 2009; Ye et al., 2016); this is particularly true for SARS-CoV-2 (Wang et al., 2005; Ahmed et al., 2020b; Rimoldi et al., 2020; Tran et al., 2021). For this reason, the use of inactivated, enveloped viruses for this study is appropriate. However, it is worth noting that enveloped viruses are diverse and little knowledge is available about their fate and persistence in wastewater environments. The alternative is that non-enveloped viruses have a higher affinity to tampon passive samples than enveloped viruses and hence cannot be eluted efficiently, however, this is yet to be evidenced.
The comparison between viral recoveries from dH2O and wastewater indicate that inhibitors are also present in samples recovered/precipitated from passive samplers, which ultimately affect qPCR and potentially other downstream applications. Inhibitors are naturally found in substances that make up the wastewater matrices such as bile salts in faeces (Lantz et al., 1997), complex polysaccharides found in faeces and plant material (Demeke and Adams, 1992; Monteiro et al., 1997), humic substances found in soils and plant materials (Tsai and Olson, 1992; Watson and Blackwell, 2000), and urea (Khan et al., 1991). Efforts can be made to reduce inhibitor levels, such as precipitating the solids from liquid via the initial centrifugation of the sample and including multiple wash steps during nucleic acid extraction (as per our protocols), however, inhibitor presence in wastewater samples are often unavoidable. Furthermore, the effect of surfactants originating from cleaning products upon passive samplers' ability to retain viral particles and nucleic acids is unknown and may be a potential route for viral loss/degradation; further research into this is suggested.
5. Conclusion and future research
Overall, tampons as passive samplers were more effective than Whatman Si-cellulose ion exchange filter papers for the recovery of viruses from wastewater. Our data suggest that viruses can be recovered from passive samplers by simply draining and concentrating the liquid from the samplers or by extracting viral nucleic acids directly from the passive sampler material. The deployment and process of passive samplers are simple, affordable and can be implemented at any WBE surveillance laboratory. Therefore, we recommend the use of tampons as samplers in areas where composite sampling is not feasible.
Viral structures had a significant effect on viral recoveries, and further work is needed to understand the mechanisms that underpin this. Future studies into the effect of temperature, pH, and disinfectants upon viral recovery from tampon passive samplers are also needed to fully understand the impact of wastewater matrices on viral recovery. In addition, the housing of the passive samplers also requires further consideration as this may also influence the efficiency of viral capture.
CRediT authorship contribution statement
Conceptualisation; KF, JLK. Laboratory work; KLS, NW, IRO, IP, NAS. Data analysis and curation; CP, KLS, JLK. First manuscript draft; JLK, KLS, CP. Final suggested edits; KF, DLJ. Funding acquisition; DLJ, KF.
Funding
The work was supported under the C215.3 Wastewater Based Epidemiology Programme within the UK Government Accelerated Capability Environment (ACE). The Centre for Environmental Biotechnology Project was funded though the European Regional Development Fund (ERDF) by Welsh Government.
Data availability statement
The datasets are available upon request.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank Prof Andrew Weightman (Cardiff University, UK), Dr. Lydia Drumwright, (University of Cambridge, UK) and Eleanor Gaunt (University of Edinburgh. UK) for providing the inactivated SARS-CoV-2, norovirus and influenza virus stocks and Ian Goodfellow (University of Cambridge, UK) for providing the original MNV stock. We also thank Tony Harrington at Dŵr Cymru-Welsh Water alongside staff at the wastewater treatment facility for their support in this project. We acknowledge the support of Daphne Beniston and Malcolm McGeoch at Accelerated Capability Environment (ACE) for project support. Thanks are also due to Andrew Singer at the UK Centre for Ecology and Hydrology and Jasmine Grimsley at the UK Heath Security Agency for their academic input. The work was supported under the C215.3 Wastewater Based Epidemiology Programme within the UK Government Accelerated Capability Environment (ACE). The Centre for Environmental Biotechnology Project was funded though the European Regional Development Fund (ERDF) by Welsh Government.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.156580.
Appendix A. Supplementary data
Supplementary figures
References
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021;11:1–7. doi: 10.1038/s41598-021-84914-2. 2021 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Kaya D., Ahmed W., Brown J., Butler C., Greaves J., et al. 2022. Passive Sampling to Scale Wastewater Surveillance of Infectious Disease: Lessons Learned From COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Lott M., Shaffer M., Wu Z., North D., Lipp E.K., et al. Building-level wastewater surveillance using tampon swabs and RT-LAMP for rapid SARS-CoV-2 RNA detection. Environ. Sci. Water Res. Technol. 2022;8:173–183. doi: 10.1039/d1ew00496d. [DOI] [Google Scholar]
- Blanco A., Abid I., Al-Otaibi N., Pérez-Rodríguez F.J., Fuentes C., Guix S., et al. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019;11:184–192. doi: 10.1007/S12560-019-09378-0/FIGURES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman N.E., Haffler T.D., Cashdollar J.L., Rhodes E.R. Evaluation of methods using celite to concentrate norovirus, adenovirus and enterovirus from wastewater. J. Virol. Methods. 2013;193:140–146. doi: 10.1016/J.JVIROMET.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . Centers Dis. Control Prev; 2020. Real-time RT-PCR Primers and Probes for COVID-19.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html Available at: [Accessed January 28, 2022] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corchis-Scott R., Geng Q., Seth R., Ray R., Beg M., Biswas N., et al. Averting an outbreak of SARS-CoV-2 in a university residence hall through wastewater surveillance. Microbiol. Spectr. 2021;9 doi: 10.1128/SPECTRUM.00792-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeke T., Adams R.P. The effects of plant polysaccharides and buffer additives on PCR. Biotechniques. 1992;12 [PubMed] [Google Scholar]
- Dvorak G., Roth J., Amass S. Disinfection 101. 2005. www.cfsph.iastate.edu/BRM Available at: [Accessed February 23, 2022]
- Eftim S.E., Hong T., Soller J., Boehm A., Warren I., Ichida A., et al. Occurrence of norovirus in raw sewage – a systematic literature review and meta-analysis. Water Res. 2017;111:366–374. doi: 10.1016/J.WATRES.2017.01.017. [DOI] [PubMed] [Google Scholar]
- Elmahdy E.M., Ahmed N.I., Shaheen M.N.F., Mohamed E.C.B., Loutfy S.A. Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health. 2019;17:287–294. doi: 10.2166/WH.2019.303. [DOI] [PubMed] [Google Scholar]
- Elmahdy E.M., Shaheen M.N.F., Rizk N.M., Saad-Hussein A. Quantitative Detection of Human Adenovirus and Human Rotavirus Group A in Wastewater and El-Rahawy Drainage Canal Influencing River Nile in the North of Giza, Egypt. Vol. 12. 2020. pp. 218–225. [DOI] [PubMed] [Google Scholar]
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., de Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- Farkas K., Adriaenssens E.M., Walker D.I., McDonald J.E., Malham S.K., Jones D.L. Critical evaluation of CrAssphage as a molecular marker for human-derived wastewater contamination in the aquatic environment. Food Environ. Virol. 2019;11:113–119. doi: 10.1007/s12560-019-09369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Hillary L.S., Thorpe J., Walker D.I., Lowther J.A., McDonald J.E., et al. Concentration and quantification of SARS-CoV-2 RNA in wastewater using polyethylene glycol-based concentration and qRT-PCR. Methods Protoc. 2021;4:17. doi: 10.3390/mps4010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firquet S., Beaujard S., Lobert P.E., Sané F., Caloone D., Izard D., et al. Survival of enveloped and non-enveloped viruses on inanimate surfaces. Microbes Environ. 2015;30:140. doi: 10.1264/JSME2.ME14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell P.G.H., Hobbs B.C., Allison V.D. An outbreak of water-borne typhoid investigated by bacteriophage typing and ‘selective’ sewage examination. J. Hyg. (Lond) 1945;44:120–128. doi: 10.1017/S0022172400035944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron L., Verreault D., Veillette M., Moineau S., Duchaine C. 2010. Aerosol Science and Technology Evaluation of Filters for the Sampling and Quantification of RNA Phage Aerosols Evaluation of Filters for the Sampling and Quantification of RNA Phage Aerosols. [DOI] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., et al. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood R.(Richard), Mills G.(Graham), Vrana B.(Branislav) Elsevier; 2007. Passive Sampling Techniques in Environmental Monitoring. [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10–14. doi: 10.1007/S12560-008-9001-6. [DOI] [Google Scholar]
- Habtewold J., McCarthy D., McBean E., Law I., Goodridge L., Habash M., et al. Passive sampling, a practical method for wastewater-based surveillance of SARS-CoV-2. Environ. Res. 2022;204 doi: 10.1016/J.ENVRES.2021.112058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., et al. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79:7413. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes E.K., Sweeney C.L., Anderson L.E., Li B., Erjavec G.B., Gouthro M.T., et al. A novel passive sampling approach for SARS-CoV-2 in wastewater in a Canadian province with low prevalence of COVID-19. Environ. Sci. Water Res. Technol. 2021;7:1576–1586. doi: 10.1039/D1EW00207D. [DOI] [Google Scholar]
- Hayes E.K., Sweeney C.L., Fuller M., Erjavec G.B., Stoddart A.K., Gagnon G.A. Operational constraints of detecting SARS-CoV-2 on passive samplers using electronegative filters: a kinetic and equilibrium analysis. ACS Environ. Sci. Technol. Water. 2021 doi: 10.1021/ACSESTWATER.1C00441/ASSET/IMAGES/LARGE/EW1C00441_0004.JPEG. [DOI] [PubMed] [Google Scholar]
- Heijnen L., Medema G. 2011. Surveillance of Influenza A and the Pandemic Influenza A (H1N1) 2009 in Sewage and Surface Water in the Netherlands. [DOI] [PubMed] [Google Scholar]
- Hewitt J., Leonard M., Greening G.E., Lewis G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011;45:6267–6276. doi: 10.1016/J.WATRES.2011.09.029. [DOI] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., et al. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;200 doi: 10.1016/J.WATRES.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Chen S., Yang Z., Guan W., Liu D., Lin Z., et al. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am. J. Respir. Crit. Care Med. 2020;201:1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel K.B., Lowe L., Bellini W.J., Rota P.A. Development of quantitative gene-specific real-time RT-PCR assays for the detection of measles virus in clinical specimens. J. Virol. Methods. 2006;132:166–173. doi: 10.1016/J.JVIROMET.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4:41–67. doi: 10.1007/S12560-012-9080-2/TABLES/6. [DOI] [PubMed] [Google Scholar]
- ISO ISO/TS 15216-1:2013 Microbiology of food and animal feed -- Horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR -. Int. Organ. Stand. 15216–1, 31. 2019. https://www.iso.org/standard/60297.html Available at: [Accessed February 17, 2022]
- Jeong H.W., Kim S.M., Kim H.S., Kim Y.Il, Kim J.H., Cho J.Y., et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang A.Z., Nian F., Chen Han, Mcbean Edward A. Passive samplers, an important tool for continuous monitoring of the COVID-19 pandemic. Environ. Sci. Pollut. Res. 2022;1:1–9. doi: 10.1007/S11356-022-19073-6. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S., Ronquillo N., Belda-Ferre P., Alvarado D., Javidi T., Longhurst C.A., et al. High-throughput wastewater SARS-CoV-2 detection enables forecasting of community infection dynamics in San Diego County. mSystems. 2021;6 doi: 10.1128/MSYSTEMS.00045-21/SUPPL_FILE/MSYSTEMS.00045-21-ST003.DOCX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama S., Masago Y., Tohma K., Souma N., Imagawa T., Suzuki A., et al. Temporal dynamics of norovirus determined through monitoring of municipal wastewater by pyrosequencing and virological surveillance of gastroenteritis cases. Water Res. 2016;92:244–253. doi: 10.1016/J.WATRES.2015.10.024. [DOI] [PubMed] [Google Scholar]
- Kevill J.L., Pellett C., Farkas K., Brown M.R., Bassano I., Denise H., et al. A comparison of precipitation and filtration-based SARS-CoV-2 recovery methods and the influence of temperature, turbidity, and surfactant load in urban wastewater. Sci. Total Environ. 2022;808 doi: 10.1016/J.SCITOTENV.2021.151916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan G., Kangro H.O., Coates P.J., Heath R.B. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J. Clin. Pathol. 1991;44:360–365. doi: 10.1136/JCP.44.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronbichler A., Kresse D., Yoon S., Lee K.H., Effenberger M., Shin J.Il. Asymptomatic patients as a source of COVID-19 infections: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;98:180–186. doi: 10.1016/J.IJID.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Joshi M., Patel A.K., Joshi C.G. Unravelling the early warning capability of wastewater surveillance for COVID-19: a temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environ. Res. 2021;196 doi: 10.1016/j.envres.2021.110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini E., Spencer S.K., Bertz P.D., Loge F.J., Kieke B.A., Borchardt M.A. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 2008;74:2990–2996. doi: 10.1128/AEM.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz P.G., Matsson M., Wadström T., Rådström P. Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J. Microbiol. Methods. 1997;28:159–167. doi: 10.1016/S0167-7012(97)00979-2. [DOI] [Google Scholar]
- Li J., Ahmed W., Metcalfe S., Smith W.J.M., Tscharke B., Lynch P., et al. Monitoring of SARS-CoV-2 in sewersheds with low COVID-19 cases using a passive sampling technique. Water Res. 2022;218 doi: 10.1016/J.WATRES.2022.118481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Verhagen R., Ahmed W., Metcalfe S., Thai P.K., Kaserzon S.L., et al. In situ calibration of passive samplers for viruses in wastewater. ACS ES&T Water. 2022 doi: 10.1021/ACSESTWATER.1C00406. [DOI] [Google Scholar]
- Liu P., Ibaraki M., VanTassell J., Geith K., Cavallo M., Kann R., et al. A sensitive, simple, and low-cost method for COVID-19 wastewater surveillance at an institutional level. Sci. Total Environ. 2022;807 doi: 10.1016/J.SCITOTENV.2021.151047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X., et al. Evaluation of sars-cov-2 rna shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maarseveen N.M., Wessels E., de Brouwer C.S., Vossen A.C.T.M., Claas E.C.J. Diagnosis of viral gastroenteritis by simultaneous detection of Adenovirus group F, Astrovirus,Rotavirus group A, Norovirus genogroups I and II, and Sapovirus in two internally controlled multiplex real-time PCR assays. J. Clin. Virol. 2010;49:205–210. doi: 10.1016/J.JCV.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Monteiro L., Bonnemaison D., Vekris A., Petry K.G., Bonnet J., Vidal R., et al. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 1997;35:995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. The detection of enteric carriers in towns by means of sewage examination. J. R. Sanit. Inst. 1951;71:57–60. doi: 10.1177/146642405107100109. [DOI] [PubMed] [Google Scholar]
- Rafiee M., Isazadeh S., Mohseni-Bandpei A., Mohebbi S.R., Jahangiri-rad M., Eslami A., et al. Moore swab performs equal to composite and outperforms grab sampling for SARS-CoV-2 monitoring in wastewater. Sci. Total Environ. 2021;790 doi: 10.1016/J.SCITOTENV.2021.148205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/J.SCITOTENV.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M., Petersburg S. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009;75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadatpour F., Mohammadipanah F. 2020. Physicochemical Susceptibility of SARS-CoV-2 to Disinfection and Physical Approach of Prophylaxis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguti F., Magnil E., Enache L., Churqui M.P., Johansson A., Lumley D., et al. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 2021;189 doi: 10.1016/j.watres.2020.116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schang C., Crosbie N.D., Nolan M., Poon R., Wang M., Jex A., et al. Passive sampling of SARS-CoV-2 for wastewater surveillance. Environ. Sci. Technol. 2021;55:10432–10441. doi: 10.1021/ACS.EST.1C01530/SUPPL_FILE/ES1C01530_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- Shah S., Gwee S.X.W., Ng J.Q.X., Lau N., Koh J., Pang J. Wastewater surveillance to infer COVID-19 transmission: a systematic review. Sci. Total Environ. 2022;804 doi: 10.1016/J.SCITOTENV.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., et al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B., Kirby M.K., Davis W.G., Warnes C., Liddell J., Liu J., et al. Multiplex real-time reverse transcription PCR for influenza A virus, influenza B virus, and severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2021;27:1821. doi: 10.3201/EID2707.210462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski M.J., Levine M.M. Reviving the “Moore Swab”: a Classic Environmental Surveillance Tool Involving Filtration of Flowing Surface Water and Sewage Water To Recover Typhoidal Salmonella Bacteria. 2020. https://journals.asm.org/journal/aem Available at: [Accessed February 23, 2022] [DOI] [PMC free article] [PubMed]
- Sinclair R.G., Choi C.Y., Riley M.R., Gerba C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008;65:249–269. doi: 10.1016/S0065-2164(08)00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51:9146. doi: 10.1021/ACS.EST.7B02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Gahlot P., Tyagi V.K., Zhang L., Zhou Y., Kazmi A.A., et al. Surveillance of wastewater for early epidemic prediction (SWEEP): environmental and health security perspectives in the post COVID-19 anthropocene. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.N., Le G.T., Nguyen D.T., Juang R.S., Rinklebe J., Bhatnagar A., et al. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ. Res. 2021;193 doi: 10.1016/J.ENVRES.2020.110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.L., Olson B.H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent-Hubert F., Morga B., Renault T., Le Guyader F.S. Adsorption of norovirus and ostreid herpesvirus type 1 to polymer membranes for the development of passive samplers. J. Appl. Microbiol. 2017;122:1039–1047. doi: 10.1111/JAM.13394. [DOI] [PubMed] [Google Scholar]
- Vincent-Hubert F., Wacrenier C., Morga B., Lozach S., Quenot E., Mège M., et al. Passive samplers, a powerful tool to detect viruses and bacteria in marine coastal areas. Front. Microbiol. 2021;12:333. doi: 10.3389/FMICB.2021.631174/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M.J., Lo Jacomo A., Armenise E., Brown M.R., Bunce J.T., Cameron G.J., et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022;424 doi: 10.1016/J.JHAZMAT.2021.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Guo T.K., Zhen B., Kong Q.X., Yi B., et al. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital. J. Virol. Methods. 2005;128:156. doi: 10.1016/J.JVIROMET.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu P., Zhang H., Ibaraki M., Vantassell J., Geith K., et al. 2022. Early Warning of a COVID-19 Surge on a University Campus Based on Wastewater Surveillance for SARS-CoV-2 at Residence Halls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R.J., Blackwell B. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 2000;46:633–642. doi: 10.1139/w00-043. [DOI] [PubMed] [Google Scholar]
- Wei X., Li L., Zhang F. The impact of the COVID-19 pandemic on socio-economic and sustainability. Environ. Sci. Pollut. Res. 2021;28:68251–68260. doi: 10.1007/S11356-021-14986-0/TABLES/6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- World Health Organization . WHO Coronavirus (COVID-19) Dashboard. WHO Coronavirus (COVID-19) Dashboard With Vaccination Data. WHO; 2021. pp. 1–5.https://covid19.who.int/ Available at: [Accessed February 23, 2022] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. 2016. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. [DOI] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/BMJ.M1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Oishi W., Maruo C., Saito M., Chen R., Kitajima M., et al. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks. Sci. Total Environ. 2021;767 doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Data Availability Statement
The datasets are available upon request.