Abstract
The catabolite control protein CcpA is a central regulator in low-G+C-content gram-positive bacteria. It confers carbon catabolite repression to numerous genes required for carbon utilization. It also operates as a transcriptional activator of genes involved in diverse phenomena, such as glycolysis and ammonium fixation. We have cloned the ccpA region of Lactobacillus pentosus. ccpA encodes a protein of 336 amino acids exhibiting similarity to CcpA proteins of other bacteria and to proteins of the LacI/GalR family of transcriptional regulators. Upstream of ccpA was found an open reading frame with similarity to the pepQ gene, encoding a prolidase. Primer extension experiments revealed two start sites of transcription for ccpA. In wild-type cells grown on glucose, mRNA synthesis occurred only from the promoter proximal to ccpA. In a ccpA mutant strain, both promoters were used, with increased transcription from the distant promoter, which overlaps a presumptive CcpA binding site called cre (for catabolite responsive element). This suggests that expression of ccpA is autoregulated. Determination of the expression levels of CcpA in cells grown on repressing and nonrepressing carbon sources revealed that the amounts of CcpA produced did not change significantly, leading to the conclusion that the arrangement of two promoters may ensure constant expression of ccpA under various environmental conditions. A comparison of the genetic structures of ccpA regions revealed that lactic acid bacteria possess the gene order pepQ-ccpA-variable while the genetic structure in bacilli and Staphylococcus xylosus is aroA-ccpA-variable-acuC.
The catabolite control protein CcpA is a global regulator controlling carbon catabolite repression (CCR), glycolysis, fermentative metabolism, and fixation of ammonium in low-G+C-content gram-positive bacteria (42). Thus, bacteria carrying a defect in ccpA exhibit deregulated CCR and reduced growth rates (20). The molecular mechanism of CcpA function in bacilli is well understood (for a review, see reference 14). When Bacillus subtilis is grown on a preferred carbon source, such as glucose or fructose, the metabolite-activated HPr kinase/phosphatase PtsK phosphorylates the proteins HPr and Crh at a seryl residue via ATP (9, 19, 32). Both seryl-phosphorylated proteins activate CcpA by forming a complex with it, thereby enabling CcpA to bind to catabolite responsive elements (cre) found within promoter or coding regions of catabolite-controlled genes (5). This results in a repression of gene expression at the level of mRNA synthesis, as has been demonstrated, for example, for the gnt operon of B. subtilis and the xyl operon of Bacillus megaterium (8, 10). Besides its repressor function, CcpA operates as a pleiotropic activator, as has been reported for the B. subtilis genes encoding acetate kinase, α-acetolactate synthase, phosphotransacetylase, and glutamate synthase and for the Lactococcus lactis las operon, encoding phosphofructokinase, pyruvate kinase, and l-lactate dehydrogenase (7, 11, 26, 33, 36).
Lactic acid bacteria, which are of central importance for the food industry, apparently control utilization of carbon sources via CCR. An internal fragment of the ccpA gene of Lactobacillus pentosus, an organism involved in fermentation of cucumbers, olives, and cabbage (the latter for sauerkraut production), has been cloned and used to construct a ccpA mutant (22). Glucose repression of the xylAB operon, encoding d-xylose isomerase and d-xylulose kinase, which are required for xylose fermentation, was relieved in the ccpA mutant strain (22). This led to the conclusion that the mechanism of CCR in L. pentosus is similar to that found in bacilli. Data supporting this conclusion have been reported for the CcpA proteins of Lactobacillus casei and Lactococcus lactis and for the CcpA homologue PepR1 of Lactobacillus delbrueckii subsp. lactis (25, 26, 30, 35). Analysis of the CcpA homologue RegM of Streptococcus mutans revealed an opposite effect on CCR (37). When regM was inactivated, the mutant strain showed an increase in glucose repression. As in the case of pepR1 of L. delbrueckii subsp. lactis and the ccpA genes of L. casei and Lactococcus lactis, regM is linked to the gene pepQ, encoding a prolidase, suggesting that these genes have similar functions (3, 37, 41; C. Esteban and G. Pérez-Martínez, unpublished data, 1999). Information on the genetic context of L. pentosus ccpA is lacking.
In this communication, we report the completion of the determination of the L. pentosus ccpA sequence and that of the surrounding genes. We use this information to assess the regulation of ccpA itself, showing that it has features distinctly different from those of L. casei and Staphylococcus xylosus. We inspect CcpA-specific residues among CcpAs and the closely related proteins RegM and PepR1, and we compare the ccpA region of L. pentosus with ccpA regions of other bacteria, providing new insights into the genetic organization of ccpA among lactic acid bacteria and other low-G+C-content gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains.
L. pentosus MD363 (wild type) was used for isolation of the ccpA genomic region (22). L. pentosus MD363 and the isogenic ccpA mutant L. pentosus LPE4 (ccpA::erm) were used for primer extension and Western blot analyses (22). Escherichia coli DH5α was used for standard cloning procedures (34).
Southern blot analysis.
Chromosomal DNA of L. pentosus MD363 was isolated as described previously (24). Ten-microgram quantities of chromosomal DNA were digested with SalI, PstI, SpeI, DraI, or EcoRV, and restriction fragments were separated on a 1% agarose gel. Plasmid pEI2 (1 μg) was labelled by nick translation, using a biotin-7-dATP labelling kit (BioNick labelling system; Gibco BRL) according to the recommendations of the manufacturer. DNA fragments from agarose gels were transferred to a nylon membrane (Gibco BRL) and fixed by UV irradiation (UV Stratalinker; Stratagene). Standard aqueous conditions were employed for hybridization and washing of membranes (34). Hybridized DNA fragments on the membrane were visualized by using the PhotoGENE nucleic acid detection system (Gibco BRL). Single ccpA-hybridizing DNA fragments of 6.4, 14.0, 10.0, 8.0, and 4.3 kb were obtained after restriction with SalI, PstI, SpeI, DraI, and EcoRV, respectively.
Inverse PCR.
L. pentosus MD363 chromosomal DNA (200 ng) was digested with SalI and religated overnight at 14°C in a 200-μl volume of T4 DNA ligase buffer containing 10 U of T4 DNA ligase (Boehringer). The religated DNA was precipitated by addition of 700 μl of ice-cold ethanol (96%), 20 μl of 3 M sodium acetate (pH 4.8), and glycogen (1 μg ml−1) and incubation for 2 h at −70°C. The precipitated DNA was resuspended in 20 μl of Tris-EDTA buffer, pH 8.0. For inverse PCR, the Tth-Taq enzyme mixture included in the Sawady-Long PCR Kit from Peqlab was used. The reaction mixture consisted of 3 μl of religated DNA (about 30 ng), 1.75 mM MgCl2, 350 μM each deoxyribonucleoside triphosphate, 2.5 U of the Tth-Taq enzyme mixture, and 15 pmol of each primer (LPE1 [5′-CCATTAACCACCCGTGAAACCGTTGCC-3′] and LPE2 [5′-TCCAGCCATCACCTCAATCACGCAACC-3′]) in a volume of 50 μl. PCR was conducted as follows: 2 min at 93°C; 10 cycles of 10 s at 93°C, 30 s at 55°C, and 5 min at 68°C; 20 cycles of 10 s at 93°C, 30 s at 55°C, and 5 min plus 20 s at 68°C; and a final incubation for 18 min at 68°C. DNA sequencing of inverse-PCR products was performed on an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer) with fluorescence-labelled dideoxyribonucleoside triphosphates provided in the BigDye Terminator Mix (Perkin-Elmer).
Plasmid construction.
The ccpA gene was cloned in two steps. First, a DNA fragment of 1,260 bp was amplified by PCR from L. pentosus MD363 chromosomal DNA, using oligonucleotides LPE9 (5′-AAAAATACAATCTCCGTGTGG-3′) and LPE10 (5′-GATTCCAAACCTAGTATACCGC-3′). This fragment was used as a template for a second PCR with oligonucleotides LPE11 (5′-GGGCTATTTTCATATGGAAAAGC-3′) and LPE12 (5′-ACTTCCCGGATCCGGCGTCTCATTAG-3′) to create an NdeI and a BamHI restriction site, respectively (underlined). The NdeI-BamHI fragment was cloned into plasmid pET15b (Novagen) which had been cut with the same restriction endonucleases, giving plasmid pWH154 (ccpA). The pepQ-ccpA intergenic region was amplified by PCR with oligonucleotides LPE17 (5′-CACCGAGGTCGAACAAGACC-3′) and LPE26 (5′-GCCACATCATAAATTGTTACTG-3′). The PCR amplification product of 978 bp, containing 689 bp of pepQ, 254 bp of the pepQ-ccpA intergenic region, and 35 bp of ccpA, was cloned into the SmaI restriction site of plasmid pSU2718, giving plasmid pWH156 (27). Nucleotide sequences of inserts were determined by DNA sequencing as described above.
Isolation of RNA.
Cells of L. pentosus MD363 and LPE4 (ccpA::erm) were grown overnight at 37°C under static conditions on minimal (M) medium supplemented with 50 mM glucose (23). Cells were harvested by centrifugation and washed twice with 10 ml of M medium. A 50-ml volume of M medium supplemented with 50 mM glucose was inoculated with cells to an optical density at 600 nm (OD600) of 0.1. Cells were grown at 37°C to an OD600 of 0.8. A 10-ml volume of the culture was harvested, and the resulting cell pellet was resuspended in 100 μl of Tris-EDTA buffer, pH 8.0. Lysozyme was added to a final concentration of 10 mg ml−1, and the suspension was incubated at 37°C for 1 h. Lysis of the cells and preparation of total RNA were performed with an RNeasy Minikit from Qiagen according to the recommendations of the manufacturer.
Primer extension analysis.
Total RNA of L. pentosus MD363 and LPE4 was isolated as described above. Primer extension experiments were performed with avian myeloblastosis virus reverse transcriptase (Stratagene) and oligonucleotide LPE27 (5′-AATTGTTACTGTTTGCTTTTCC-3′), which is complementary to positions 24 to 3 of the ccpA coding sequence (see Fig. 3A). Oligonucleotides were 5′ labelled by the use of T4 polynucleotide kinase (New England Biolabs). In primer extension reactions, 500 fmol of labelled primer was used with 20 μg of cellular RNA. Reverse transcripts were resolved on 6% urea–polyacrylamide gels. Standard DNA sequencing reactions with Sequenase (U.S. Biochemical Corp.), using the same oligonucleotides, were performed for sizing of the primer extension products. The positions of transcriptional start sites were confirmed by using a second oligonucleotide, LPE26 (5′-GCCACATCATAAATTG-TTACTG-3′), hybridizing to positions 35 to 14 of the ccpA coding sequence.
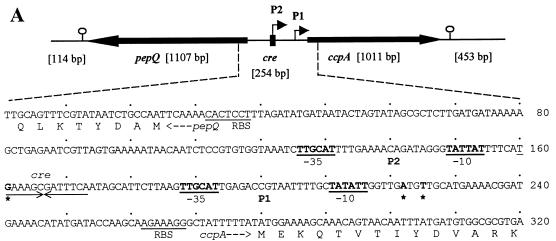
FIG. 3.
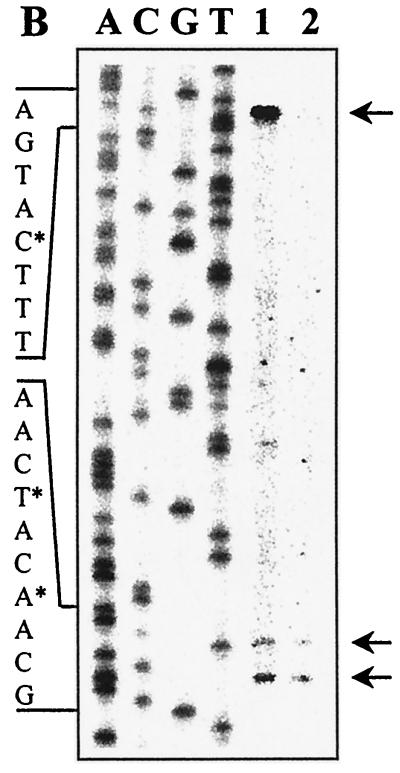
Genetic organization and transcriptional regulation of L. pentosus ccpA. (A) Genetic organization of the ccpA region. The size and orientation of the ORFs were deduced from the nucleotide sequence. The ccpA promoter region is depicted at the DNA sequence level. ccpA transcriptional start sites are in boldface and are marked by asterisks. Potential RBSs and cre motifs are underlined. Putative RNA polymerase binding sites (−10 region and −35 region) in the sequence are in boldface letters and underlined and are indicated with P1 and P2. The N-terminal protein sequences of pepQ and ccpA are shown. (B) Primer extension analysis of ccpA gene transcription of L. pentosus wild-type and ccpA mutant strains. Total RNA was prepared from cells grown on M medium supplemented with 50 mM glucose. Reverse transcription was carried out with end-labelled oligonucleotide LPE27. DNA sequencing reactions were performed with the same oligonucleotide and with pWH156 as template DNA. Primer extension products were analyzed on 6% polyacrylamide–urea gels. Lane 1, RNA (20 μg) from L. pentosus LPE4 (ccpA mutant); lane 2, RNA (20 μg) from L. pentosus MD363 (wild type). The sequence interpretations around the +1 sites (asterisks and arrows) of the two ccpA promoters are shown.
Western blot analysis.
L. pentosus MD363 and LPE4 (ccpA::erm) were grown individually overnight at 37°C under static conditions in 10 ml of M medium supplemented with 50 mM glucose (23). Cells were harvested by centrifugation and washed twice with 10 ml of M medium. A 50-ml volume of M medium supplemented with 50 mM glucose or xylose was inoculated with culture to an OD600 of 0.1. Cells were grown at 37°C to an OD600 of 0.8. After the cells were harvested, the cell pellet was resuspended in 1 ml of starting buffer (20 mM Tris-HCl [pH 7.5], 3 mM dithiothreitol). Crude extracts were prepared by sonification at 45 W (Labsonic U [Braun]; twice, for 20 s each time) and subsequent removal of cell debris by centrifugation. Proteins of cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 7.5% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Fluorotrans) by electroblotting. CcpA was detected with a rabbit polyclonal antiserum raised against CcpA of B. megaterium (21). CcpA antibodies on the polyvinylidene difluoride membrane were visualized by using the ECL Western blot analysis system (Amersham).
Computer analyses.
DNA and protein data bank searches were performed with the BLAST server of the National Center for Biotechnology Information at the National Institutes of Health, Bethesda, Md. (URL http://www.ncbi.nlm.nih.gov). The LaserGene workstation software (DNASTAR, Inc.) was used to process DNA and protein sequence data.
Nucleotide sequence accession number.
The pepQ-ccpA DNA sequence has been submitted to GenBank under accession no. AF176799.
RESULTS AND DISCUSSION
Isolation of the ccpA region of L. pentosus.
The isolation of an internal ccpA gene fragment from L. pentosus MD353 has been described in a previous publication (22). This fragment was used to construct a ccpA mutant in the more highly transformable strain MD363. Thus, we chose MD363 to determine the sequences of the complete ccpA gene and flanking genes.
A Southern blot analysis of chromosomal DNA of MD363 was performed with plasmid pEI2, which contains the internal ccpA fragment ′ccpA′, as a probe. Chromosomal DNA restricted with SalI gave a signal of 6.4 kb, indicating the presence of the ccpA gene on the chromosomal SalI DNA fragment (data not shown). The 6.4-kb fragment was amplified by inverse PCR with oligonucleotides LPE1 and LPE2, which hybridized to the 5′ end and to the 3′ end of the known ′ccpA′ fragment, respectively. As a result, an amplification product of 5.6 kb was generated (Fig. 1). The PCR fragment was directly subjected to sequencing with the same oligonucleotides. The determined nucleotide sequence comprised the missing ends of ccpA and adjacent intergenic sequences, as revealed by analysis of similarity to the ccpA gene of L. casei. Further sequencing of the PCR product yielded 1,379 bp upstream and 453 bp downstream of ccpA (see the section on genetic organization below).
FIG. 1.
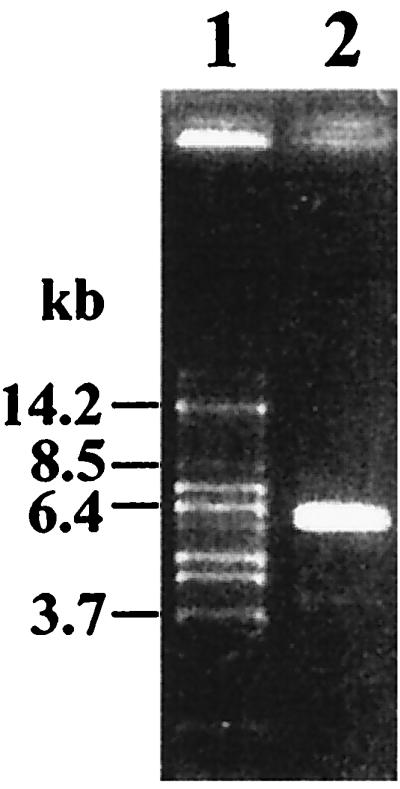
Amplification product of 5.6 kb of the ccpA region of L. pentosus on a 1% agarose gel. Sizes of DNA fragments are indicated in kilobase pairs. Lane 1, DNA marker fragments; lane 2, 3 μl of a 50-μl-volume inverse PCR using oligonucleotides LPE1 and LPE2 and religated L. pentosus MD363 chromosomal DNA.
Analysis of CcpA.
The L. pentosus ccpA gene was amplified by PCR, using chromosomal DNA as a template, as described in Materials and Methods. For further analysis, the fragment was cloned in plasmid pET15b by insertion into NdeI and BamHI sites, giving plasmid pWH154. Sequencing of L. pentosus MD363 ccpA revealed an open reading frame (ORF) of 1,011 bp encoding 336 amino acids with a calculated molecular mass of 36,316 Da. A potential ribosome binding site (RBS; AGAAAGG) is located at positions −12 to −18. At the DNA level, ccpA of L. pentosus MD363 showed 97% identity to the ′ccpA′ fragment of L. pentosus MD353 and 76% identity to the next related ccpA gene of L. casei. The 20 differences in nucleotides found in the ccpA genes of the two L. pentosus strains do not affect the amino acid sequence; thus, the gene products are identical.
At the protein level, CcpA of L. pentosus shows 66 to 39% identity to CcpA and CcpA-like proteins of L. casei (66%), Enterococcus faecalis (62%; accession no. AJ011113), Listeria monocytogenes (60%), Streptococcus mutans (RegM; 59%), B. subtilis (56%), B. megaterium (55%), Lactococcus lactis (53%), L. delbrueckii (PepR1; 50%), Thermoactinomyces sp. strain E79 (50%; accession no. AF055979), Staphylococcus xylosus (48%), and Clostridium acetobutylicum (39%) (1, 13, 16, 26, 37, 41).
The CcpA proteins comprise a subgroup of the LacI/GalR family of bacterial transcriptional regulators, to which they exhibit about 30% protein sequence identity (43). The CcpA subgroup has been defined by physiological functionality (18). On this basis, 67 CcpA-specific residues have been proposed to be present in all CcpA proteins and predominantly absent from the other members of the family (18). The CcpA-specific residues are clustered in three blocks throughout the amino acid sequence. One block covers the N-terminal helix-turn-helix motif, while most other conserved residues form a continuous patch on the protein surface (18). The contents of LacI/GalR-specific residues among all proteins of the family are similar, ranging between 63 and 83 LacI/GalR-specific positions (Table 1). CcpA of L. pentosus possesses 61 of the 67 CcpA-specific amino acid positions (Table 1 and Fig. 2). Thus, CcpA of L. pentosus contains the same number of CcpA-specific residues as the well-characterized CcpA of B. megaterium (Table 1). An increased number of CcpA-specific residues compared to that of non-CcpA proteins of the LacI/GalR family can be found in the regulator protein RegA of C. acetobutylicum (24 of 67). This protein was shown to complement a B. subtilis ccpA mutant and must therefore have features necessary for CcpA function (4). The CcpA homologues RegM of Streptococcus mutans and PepR1 of L. delbrueckii comprise 60 and 47 CcpA-specific residues, respectively, a good indication of a CcpA-like function in these organisms.
TABLE 1.
Comparison of contents of CcpA- and LacI/GalR-specific residues
| Organism or sequence | Protein | No. of:
|
% Identitya | Accession no. | |
|---|---|---|---|---|---|
| CcpA-specific residues | LacI/GalR-specific residues | ||||
| Consensus | 67 | 94 | |||
| L. pentosus | CcpA | 61 | 79 | 100 | AF176799 |
| L. casei | CcpA | 64 | 79 | 66 | U28137 |
| E. faecalis | CcpA | 63 | 75 | 62 | AJ011113 |
| Listeria monocytogenes | CcpA | 62 | 80 | 60 | AF076520 |
| Streptococcus mutans | RegM | 60 | 78 | 59 | O07329 |
| B. subtilis | CcpA | 63 | 80 | 56 | P25144 |
| B. megaterium | CcpA | 61 | 83 | 55 | P46828 |
| Lactococcus lactis | CcpA | 51 | 75 | 53 | AF106673 |
| L. delbrueckii subsp. lactis | PepR1 | 47 | 71 | 50 | Q48544 |
| Staphylococcus xylosus | CcpA | 55 | 82 | 48 | Q56194 |
| C. acetobutylicum | RegA | 24 | 71 | 39 | Q45831 |
| E. coli | LacI | 0 | 63 | 30 | P03023 |
| GalR | 0 | 69 | 29 | P03024 | |
Percent amino acid sequence identity with respect to CcpA of L. pentosus.
FIG. 2.
Protein sequence of L. pentosus CcpA and comparison to a consensus sequence. Lowercase letters of the consensus sequence designate conserved amino acid residues specific for proteins of the LacI/GalR family of regulators, whereas uppercase letters designate amino acid residues specific for CcpA proteins (18).
Identification of the genes flanking ccpA.
To determine the genes flanking ccpA, parts of the 5.6-kb amplification product were sequenced by primer walking. Upstream of ccpA, an ORF of 1,107 bp whose orientation was opposite that of ccpA and which exhibited a high degree of similarity to pepQ genes of other lactic acid bacteria was identified and found to encode a dipeptidase which specifically cleaves X-prolyl peptide bonds (40). The gene was therefore designated pepQ. Multiple-alignment analysis revealed a possible start codon, TTG, which is preceded by a potential RBS (AGGAGTG) located at positions −8 to −14. A potential transcriptional termination site (AATCCGCCCCATCCTGATAAGCACGATGGGGCGGATT; the inverted repeat is underlined) was found 33 bp downstream of pepQ. PepQ of L. pentosus showed 57% identity to PepQ of Lactobacillus helveticus (accession no. AK012084) and L. delbrueckii subsp. lactis, 50% identity to PepQ of Streptococcus mutans, and 42% identity to L. delbrueckii subsp. bulgaricus (31, 37, 40). A catalytical center comprising a zinc-binding motif which is typical of zinc-dependent metallopeptidases has been described (17). PepQ of L. pentosus contains this unique signature at amino acid positions 294 to 304.
ccpA and pepQ have similar G+C contents (49 and 50%, respectively). The 453-bp sequence downstream of ccpA (48% G+C) exhibited no similarity to any known proteins of the data bank. A potential stem-loop structure (underlined), AGGTTTGGAATCTGATTCCAAACCT, could be identified 58 bp downstream of ccpA; this might serve as a transcriptional termination site. The pepQ-ccpA intergenic region (254 bp) exhibits a decreased G+C content of 32%, typical of promoter-containing regions (39). A fully conserved cre site (TGAAAGCGATTTCA) was found to be located at positions −107 to −120 with respect to the ccpA gene, suggesting autoregulation of ccpA.
Transcriptional regulation of ccpA.
To determine the transcriptional start point and to examine the potential of ccpA for autoregulation, primer extension analyses were performed on RNA extracted from cells of wild-type strain L. pentosus MD363 and from cells of ccpA mutant strain L. pentosus LPE4, respectively, grown on M medium supplemented with 50 mM glucose. In both strains, a transcription start site was found (at positions −55 and −58, respectively), as indicated by a double band in Fig. 3B. Such a double signal can be explained by an alternative use of start sites through the same promoter (P1). Interestingly, the primer extension analysis of mRNA of the ccpA mutant strain revealed a second transcription initiation site, indicated by a stronger primer extension signal. It could be assigned to a G at position −119, which represents the second base of the potential cre site (P2) (Fig. 3A). Both promoters P1 (TTGCAT-17 bp-TATATT) and P2 (TTGCAT-17 bp-TATTAT) are well conserved compared to the ςA-dependent promoters of B. subtilis (12). The finding that the P2 transcript could be formed only when functional CcpA protein was missing suggests autoregulation of ccpA.
CcpA expression levels on various carbon sources.
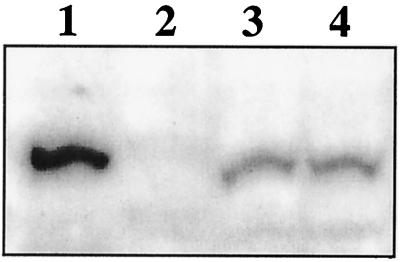
To further determine the effect of transcriptional regulation on the amount of CcpA protein synthesized under different growth conditions, Western blotting experiments were performed (Fig. 4). L. pentosus MD363 wild-type cells were grown on M medium under repressing and nonrepressing conditions, using 50 mM glucose and 50 mM xylose, respectively. As can be seen in Fig. 4, CcpA signals of equal intensities were found in cell extracts grown under repressing and nonrepressing conditions (lanes 3 and 4). This finding suggests that nearly constant amounts of cellular CcpA are present independently of the presence or absence of glucose.
FIG. 4.
Western blot analysis of L. pentosus grown on various carbon sources. A Western blot of a sodium dodecyl sulfate–7.5% polyacrylamide gel is shown after incubation with polyclonal antibodies derived against CcpA of B. megaterium. Lane 1, 50 ng of purified CcpA of B. megaterium; lane 2, 0.2 OD600 equivalents of protein extract of L. pentosus LPE4 (ccpA mutant); lanes 3 and 4, 0.2 OD600 equivalents of protein extract of L. pentosus MD363 grown on M medium with 50 mM glucose and 50 mM xylose, respectively.
Regulation of ccpA has been studied in only a few organisms, namely, B. subtilis, B. megaterium, Staphylococcus xylosus, and L. casei (6, 16, 29, 30). Western blot analysis and ccpA::lacZ fusion measurements led to the conclusion that CcpA is constitutively expressed in B. subtilis and B. megaterium (16, 29). However, in neither case has the promoter been mapped. Constitutive expression of ccpA of L. casei was also revealed by primer extension and Northern blot analyses (30). The ccpA gene of L. casei is expressed via only one promoter, which apparently is lacking a cre site and is therefore not subject to autoregulation. As in L. pentosus, transcription of ccpA of Staphylococcus xylosus is driven by a tandem promoter and triggered through autoregulation via a cre site. However, here the cre site overlaps the transcription start site of the promoter proximal to ccpA (6). The authors (6) reported that in Staphylococcus xylosus there exists a carbon source-dependent autoregulation of ccpA mediated by cre and resulting in a slight decrease of CcpA expression when cells are grown in the presence of glucose.
In conclusion, we suggest that in all organisms investigated to date, CcpA is produced in more or less constant amounts regardless of growth conditions. Transcriptional regulation of ccpA expression may be a means of reaching this goal in Staphylococcus xylosus and L. pentosus.
Genetic organization.
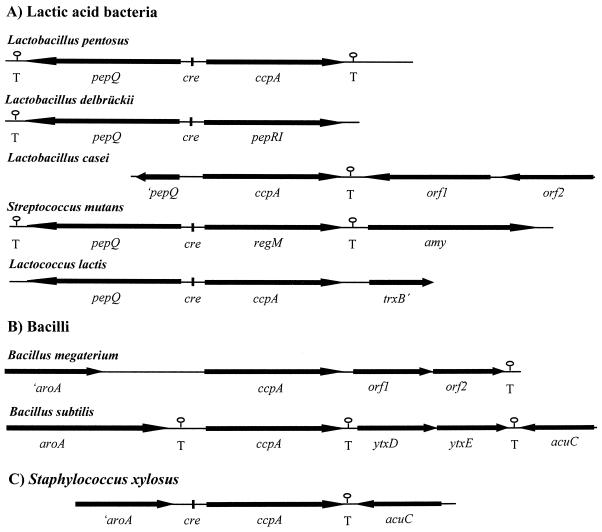
The finding that pepQ genes are also located divergently from ccpA genes in other lactic acid bacteria led us to compare the genetic organizations of all published ccpA regions. As depicted in Fig. 5, the gene order pepQ-ccpA (or pepQ-ccpA homologue) is found in all lactic acid bacteria, namely L. pentosus, L. delbrueckii, L. casei, Streptococcus mutans, and Lactococcus lactis (3, 37, 41; C. Esteban and G. Pérez-Martínez, unpublished data, 1999). In contrast, the sequences downstream of ccpA differ throughout the lactic acid bacteria. While there are no sequence similarities to other known genes in L. pentosus, orf1 of L. casei is homologous to genes encoding transposases of IS30 family insertion elements (30). The genes downstream of ccpA in Lactococcus lactis and Streptococcus mutans encode homologues of thioredoxin reductase (accession no. AF106673) and α-amylase, respectively (38). Taken together, these findings indicate that pepQ is always associated with ccpA or a ccpA homologue in lactic acid bacteria.
FIG. 5.
Comparison of ccpA regions of L. pentosus, L. delbrueckii subsp. lactis (40, 41), L. casei (30; C. Esteban and G. Pérez-Martínez, unpublished data), Streptococcus mutans (37), Lactococcus lactis (accession no. AF106673) (3) (A); B. megaterium (15), B. subtilis (2, 11, 13) (B); and Staphylococcus xylosus (6) (C). Orientations of genes are indicated by arrows. Potential transcriptional termination structures (T) and cre motifs are indicated.
Here, the questions of whether pepQ is regulated by CcpA and whether PepR1 and RegM are truly functional equivalents of CcpA of L. pentosus, L. casei, and Lactococcus lactis arise. The contents of CcpA-specific residues support this assumption (Table 1). While the involvement of CcpA in CCR is apparently established for L. pentosus, L. casei, and Lactococcus lactis, recent data suggest that this is also the case for PepR1 of L. delbrueckii (35). In the case of Streptococcus mutans, inactivation of regM resulted in an opposite CCR phenotype; i.e., CCR was enhanced (37). The picture concerning regulation of pepQ by CcpA is not clear. While a regulatory effect has been described for L. delbrueckii, this was not the case in Streptococcus mutans (35, 37).
A very different gene arrangement is found in non-lactic acid gram-positive bacteria. Upstream regions of ccpA in B. megaterium, B. subtilis, and Staphylococcus xylosus comprising the aroA gene, whose product in B. subtilis has been shown to possess chorismate mutase activity, are conserved (2). In B. megaterium and B. subtilis, two ORFs are found downstream of ccpA (orf1 and orf2 in B. megaterium and ytxD and ytxE in B. subtilis) that are homologous to the motA and motB genes of B. subtilis (15). MotA and MotB are integral membrane proteins involved in flagellar movement (28). In Staphylococcus xylosus, no motA or motB homologues are linked to ccpA; here acuC is the gene downstream of ccpA encoding a protein involved in acetoin and butanediol metabolism (11). The acuC gene of B. subtilis is located downstream of ytxE, showing that the order of the genes in the ccpA region of B. subtilis is similar to that of Staphylococcus xylosus.
The overall genetic context of ccpA and the fact that in most cases a terminator-like structure downstream of ccpA is predicted suggest a monocistronic operon organization. This is supported by transcript size mapping of the ccpA mRNAs of L. casei and L. pentosus (22, 30). However, in L. pentosus, a second transcript, of 10 kb, has been detected (22). While in RNA of glucose-grown cells the short transcript was the primary product, the 10-kb transcript was predominantly present in RNA of xylose-grown cells. This suggests that transcription of ccpA and the genes downstream might be coordinately regulated.
Conclusions.
Analysis of the ccpA region of L. pentosus revealed the gene order pepQ-ccpA-variable. This genetic organization is found in all lactic acid bacteria described to date, which may indicate that the ccpA genes of L. pentosus, L. casei, and Lactococcus lactis, the pepR1 gene of L. delbrueckii, and the regM gene of Streptococcus mutans are functional equivalents. The fact that no common gene is linked to ccpA throughout the low-G+C-content gram-positive bacteria indicates that none of the flanking genes play a role in CCR. The reported data on the regulation of ccpA in L. pentosus show that CcpA levels are constant under different environmental conditions. This is a common feature also found in L. casei, bacilli, and Staphylococcus xylosus. Yet, the mechanisms of ccpA transcription differ among these organisms. In L. pentosus, ccpA transcription is realized by a tandem promoter, the more distant component of which is apparently subject to autoregulation. Further studies are required to elucidate whether CcpA of L. pentosus is involved in the regulation of other catabolite-controlled genes; whether CcpA regulates pepQ, thereby linking carbon utilization to proteolysis; and whether CcpA triggers gene activation as well.
ACKNOWLEDGMENTS
We thank Elke Küster-Schöck for gifts of protein and antibodies and for helpful discussions. We thank Peter Pouwels and Stephane Chaillou for gifts of L. pentosus strains and plasmid pEI2 and for many suggestions and discussions. We are grateful to Joachim Schick, Carlos Esteban, and Gaspar Pérez-Martínez for making data available prior to publication. Stephan Parche is acknowledged for critical reading of the manuscript.
This work was financed by the EU project BIO4-CT96-0380.
REFERENCES
- 1.Behari J, Youngman P. A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes. J Bacteriol. 1998;180:6316–6324. doi: 10.1128/jb.180.23.6316-6324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin A, Khazak V, Stoynova N, Ratmanova K, Yomantas Y, Kozlov Y. Identical amino acid sequence of the aroA(G) gene products of Bacillus subtilis 168 and B. subtilis Marburg strain. Microbiology. 1995;141:2219–2222. doi: 10.1099/13500872-141-9-2219. [DOI] [PubMed] [Google Scholar]
- 3.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 4.Davison S P, Santangelo J D, Reid S J, Woods D R. A Clostridium acetobutylicum regulator gene (regA) affecting amylase production in Bacillus subtilis. Microbiology. 1995;141:989–996. doi: 10.1099/13500872-141-4-989. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 6.Egeter O, Brückner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 7.Faires N, Tobisch S, Bachem S, Martin-Verstraete I, Hecker M, Stülke J. The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis. J Mol Microbiol Biotechnol. 1999;1:141–148. [PubMed] [Google Scholar]
- 8.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 9.Galinier A, Haiech J, Kilhofer M-C, Jaquinod M, Stülke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes an HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 11.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 12.Helmann J D. Compilation and analysis of Bacillus subtilis ςA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 14.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 15.Hueck C J, Kraus A, Hillen W. Sequences of ccpA and two downstream Bacillus megaterium genes with homology to the motAB operon from Bacillus subtilis. Gene. 1994;143:147–148. doi: 10.1016/0378-1119(94)90621-1. [DOI] [PubMed] [Google Scholar]
- 16.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning, expression and functional analyses of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 17.Jongeneel C V, Bouvier J, Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- 18.Kraus A, Küster E, Wagner A, Hoffmann K, Hillen W. Identification of a co-repressor binding site in catabolite control protein CcpA. Mol Microbiol. 1998;30:955–963. doi: 10.1046/j.1365-2958.1998.01123.x. [DOI] [PubMed] [Google Scholar]
- 19.Kravanja M, Engelmann R, Dossonnet V, Bluggel M, Meyer H E, Frank R, Galinier A, Deutscher J, Schnell N, Hengstenberg W. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol Microbiol. 1999;31:59–66. doi: 10.1046/j.1365-2958.1999.01146.x. [DOI] [PubMed] [Google Scholar]
- 20.Küster E, Hilbich T, Dahl M K, Hillen W. Mutations in catabolite control protein CcpA separating growth effects from catabolite repression. J Bacteriol. 1999;181:4125–4128. doi: 10.1128/jb.181.13.4125-4128.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Küster E, Luesink E J, de Vos W M, Hillen W. Immunological crossreactivity to catabolite control protein CcpA from Bacillus megaterium is found in many gram-positive bacteria. FEMS Microbiol Lett. 1996;139:109–115. doi: 10.1111/j.1574-6968.1996.tb08188.x. [DOI] [PubMed] [Google Scholar]
- 22.Lokman B C, Heerikhuisen M, Leer R J, van den Broek A, Borsboom Y, Chaillou S, Postma P W, Pouwels P H. Regulation of expression of the Lactobacillus pentosus xylAB operon. J Bacteriol. 1997;179:5391–5397. doi: 10.1128/jb.179.17.5391-5397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lokman B C, Leer R J, van Sorge R, Pouwels P H. Promoter analysis and transcriptional regulation of Lactobacillus pentosus genes involved in xylose catabolism. Mol Gen Genet. 1994;245:117–125. doi: 10.1007/BF00279757. [DOI] [PubMed] [Google Scholar]
- 24.Lokman B C, van Santen P, Verdoes J C, Kruse J, Leer R J, Posno M, Pouwels P H. Organization and characterization of three genes involved in d-xylose catabolism in Lactobacillus pentosus. Mol Gen Genet. 1991;230:161–169. doi: 10.1007/BF00290664. [DOI] [PubMed] [Google Scholar]
- 25.Luesink E J, Beumer C M A, Kuipers O P, De Vos W M. Molecular characterization of the Lactococcus lactis ptsHI operon and analysis of the regulatory role of HPr. J Bacteriol. 1999;181:764–771. doi: 10.1128/jb.181.3.764-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luesink E J, van Herpen R E, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 27.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 28.Mirel D B, Lustre V M, Chamberlin M J. An operon of Bacillus subtilis motility genes transcribed by the ςD form of RNA polymerase. J Bacteriol. 1992;174:4197–4204. doi: 10.1128/jb.174.13.4197-4204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miwa Y, Saikawa M, Fujita Y. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology. 1994;140:2567–2575. doi: 10.1099/00221287-140-10-2567. [DOI] [PubMed] [Google Scholar]
- 30.Monedero V, Gosalbes M J, Pérez-Martínez G. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morel F, Frot-Coutaz J, Aubel D, Portalier R, Atlan D. Characterization of a prolidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397 with an unusual regulation of biosynthesis. Microbiology. 1999;145:437–446. doi: 10.1099/13500872-145-2-437. [DOI] [PubMed] [Google Scholar]
- 32.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Saier M H J, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 33.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schick J, Weber B, Klein J R, Henrich B. PepR1, a CcpA-like transcription regulator of Lactobacillus delbrückii ssp. lactis. Microbiology. 1999;145:3147–3154. doi: 10.1099/00221287-145-11-3147. [DOI] [PubMed] [Google Scholar]
- 36.Shin B S, Choi S K, Park S H. Regulation of the Bacillus subtilis phosphotransacetylase gene. J Biochem (Tokyo) 1999;126:333–339. doi: 10.1093/oxfordjournals.jbchem.a022454. [DOI] [PubMed] [Google Scholar]
- 37.Simpson C L, Russell R R B. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson C L, Russell R R B. Intracellular α-amylase of Streptococcus mutans. J Bacteriol. 1998;180:4711–4717. doi: 10.1128/jb.180.17.4711-4717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stucky K, Klein J R, Schuller A, Matern H, Henrich B, Plapp R. Cloning and DNA sequence analysis of pepQ, a prolidase gene from Lactobacillus delbrueckii subsp. lactis DSM7290, and partial characterization of its product. Mol Gen Genet. 1995;247:494–500. doi: 10.1007/BF00293152. [DOI] [PubMed] [Google Scholar]
- 41.Stucky K, Schick J, Klein J, Henrich B, Plapp R. Characterization of pepR1, a gene coding for a potential transcriptional regulator of Lactobacillus delbrueckii subsp. lactis DSM7290. FEMS Microbiol Lett. 1996;136:63–69. doi: 10.1016/0378-1097(95)00494-7. [DOI] [PubMed] [Google Scholar]
- 42.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 43.Weickert M J, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]