Abstract
Prior to gene transfer experiments performed with nonsterile soil, plasmid pJP4 was introduced into a donor microorganism, Escherichia coli ATCC 15224, by plate mating with Ralstonia eutropha JMP134. Genes on this plasmid encode mercury resistance and partial 2,4-dichlorophenoxyacetic acid (2,4-D) degradation. The E. coli donor lacks the chromosomal genes necessary for mineralization of 2,4-D, and this fact allows presumptive transconjugants obtained in gene transfer studies to be selected by plating on media containing 2,4-D as the carbon source. Use of this donor counterselection approach enabled detection of plasmid pJP4 transfer to indigenous populations in soils and under conditions where it had previously not been detected. In Madera Canyon soil, the sizes of the populations of presumptive indigenous transconjugants were 107 and 108 transconjugants g of dry soil−1 for samples supplemented with 500 and 1,000 μg of 2,4-D g of dry soil−1, respectively. Enterobacterial repetitive intergenic consensus PCR analysis of transconjugants resulted in diverse molecular fingerprints. Biolog analysis showed that all of the transconjugants were members of the genus Burkholderia or the genus Pseudomonas. No mercury-resistant, 2,4-D-degrading microorganisms containing large plasmids or the tfdB gene were found in 2,4-D-amended uninoculated control microcosms. Thus, all of the 2,4-D-degrading isolates that contained a plasmid whose size was similar to the size of pJP4, contained the tfdB gene, and exhibited mercury resistance were considered transconjugants. In addition, slightly enhanced rates of 2,4-D degradation were observed at distinct times in soil that supported transconjugant populations compared to controls in which no gene transfer was detected.
Genes for metal resistance or contaminant degradation are often plasmid encoded (5, 6, 11, 12, 14, 22, 24). Accordingly, microbial inocula for bioaugmentation often contain such plasmids. Unfortunately, due to biotic and abiotic stresses, a rapid decline in inoculum size or cell death is frequently observed following addition to the environment. This suggests that transfer of a catabolic plasmid from an introduced microorganism to indigenous soil microorganisms may enhance bioaugmentation by providing an environmentally stable host for the plasmid. Limited study workers evaluated the potential for transfer of large catabolic plasmids from an introduced donor to indigenous microbial recipients (3, 5, 7, 9, 23). Furthermore, it has been shown that remediation of contaminated soils may be enhanced as a result of such transfers (5, 7, 23).
Of pertinence to this study is the 80-kb, broad-host-range, self-transmissible, IncP1 group, catabolic plasmid pJP4 (8). This plasmid encodes resistance to mercuric ions and phenyl mercury acetate and partial catabolism of 2,4-dichlorophenoxyacetic acid (2,4-D), 2-methyl-4-chlorophenoxyacetic acid, and 3-chlorobenzoate (8). Genes carried on plasmid pJP4 are responsible for transformation of 2,4-D to 2-chloromaleylacetate (21). Transfer of pJP4 in soil has been observed previously (3, 7, 10, 15). It has been found that this plasmid is transferred from Ralstonia eutropha JMP134 to indigenous Madera Canyon soil recipients in soil amended with 1,000 μg of 2,4-D g of dry soil−1 but not in soil amended with 500 μg of 2,4-D g of dry soil−1 (7). No pJP4 transfer with this donor at either level of 2,4-D was detected in studies performed with other soils in which a significant population of R. eutropha JMP134 remained culturable after inoculation.
Low-frequency transfer events are difficult to observe if the level of the donor that survives is at or above the order of magnitude of the level of any transconjugant due to the high soil dilution that must be plated in order to obtain distinct colonies. Donor counterselection can enhance gene transfer detection and quantification by eliminating donor interference. The following two main approaches for donor counterselection have been described: (i) using a donor-specific bacteriophage (16, 20) and (ii) plating preparations onto selective media that do not allow growth of the donor microorganism (10). Both of these approaches have been shown to be useful in facilitating the detection of indigenous transconjugants under various environmental conditions.
It has been shown that microorganisms that harbor pJP4 but lack the chromosomally encoded maleylacetate reductase required for utilization of 2,4-D are not detected on selective media in which 2,4-D is the sole carbon source (13). Little information regarding the occurrence of maleylacetate reductase genes in indigenous populations is available; however, the ubiquity of soil microbial populations capable of mineralizing 2,4-D suggests that these genes are probably relatively common. In pure-culture experiments, Don and Pemberton (8) identified several microorganisms, including an Escherichia coli strain, that were not able to degrade 2,4-D despite the fact that they harbored pJP4. An E. coli strain was chosen to be the novel pJP4 host and donor microorganism for soil gene transfer studies for the following reasons: (i) this strain lacks the chromosomal component necessary for mineralization of 2,4-D; (ii) it has a rapid doubling time; and (iii) its background level in soil is low. Furthermore, an E. coli strain has been used previously as a pJP4 donor in plate mating experiments in which there was subsequent transconjugant selection on 2,4-D-containing media (10), which showed that the approach was feasible.
The four main objectives of this study were (i) to develop a system in which donor counterselection is used to facilitate detection of low-frequency gene transfer in soil, (ii) to evaluate pJP4 transfer to indigenous soil microbial populations, (iii) to assess the diversity of presumptive transconjugants, and (iv) to assess the effects of augmentation of the soil degradative gene pool on biodegradation of 2,4-D.
MATERIALS AND METHODS
Donor generation and maintenance.
To generate the E. coli donor, plasmid pJP4 was introduced into E. coli ATCC 15224 by plate mating with R. eutropha JMP134(pJP4). The latter microorganism is considered the natural host of pJP4 and until recently was classified as Alcaligenes eutrophus JMP134. Plate matings were conducted on nonselective peptone-yeast extract plates by using late-exponential-phase cultures. Plate mating patches were suspended in sterile saline and then plated onto peptone-yeast extract plates amended with 25 ppm of mercury (PH plates) (see below). Isolates from the PH plates were then screened for the ability to utilize 2,4-D as a soil carbon source. Enterobacterial repetitive intergenic consensus (ERIC) PCR (25) and tfdB PCR (15) analyses were performed with mercury-resistant non-2,4-D-degrading isolates to confirm that the isolates were E. coli pJP4 hosts. Confirmed E. coli transconjugant isolate 11 was used as the donor in subsequent studies and was designated E. coli D11.
Soils.
All soils were collected from sites that were not previously exposed to 2,4-D. Surface soils were sieved (pore size, 2 mm) and, if they were not used within 1 week of collection, were stored at 4°C. Stored soils were incubated at 28°C for 1 week before amendments were added, which allowed the microbial populations to acclimate. Madera Canyon soil was collected from the Madera Canyon Recreational Area of the Coronado National Forest near Tucson, Ariz. Rose Canyon and Bear Canyon soils were collected from Mt. Lemmon, which is located in the Catalina Mountains National Forest, Tucson, Ariz. Brazito soil was collected from the University of Arizona's Campus Agricultural Center in Tucson, Ariz. Table 1 shows chemical and physical properties of each soil.
TABLE 1.
Characteristics of the soils used
| Soil | pHa | Textureb | Composition (%)
|
|||
|---|---|---|---|---|---|---|
| Sand | Silt | Clay | Organic matterc | |||
| Brazito | 8.5 | Sandy loam | 76 | 12 | 12 | 1.1 |
| Bear Canyon | 6.1 | Sandy loam | 68 | 22 | 10 | 11.4 |
| Rose Canyon | 5.4 | Sandy loam | 74 | 18 | 8 | 17.1 |
| Madera Canyon | 6.5 | Sandy loam | 78 | 16 | 6 | 3.3 |
Determined by using a soil-water (1:1) extract.
Determined by the hydrometer method.
Determined by the Walkley-Black method (2).
Gene transfer studies in soil.
The soil microcosms used consisted of 100-g (dry weight) portions of nonsterile soil in 0.5-liter polypropylene wide-mouth screw-cap jars. Enough 1% 2,4-D stock solution was added to each treated microcosm to obtain a 2,4-D concentration of either 500 or 1,000 μg g of dry soil−1. The 2,4-D stock solution was prepared by adding 10 g of 2,4-D (Sigma Chemical Co., St. Louis, Mo.) to 900 ml of distilled water. Ten milliliters of 5 N NaOH was added to facilitate dissolution of the 2,4-D. The pH was then adjusted to 7.0 with concentrated HCl, the total volume was adjusted to 1.0 liter, and the solution was filter sterilized. Microcosms that were not contaminated with 2,4-D received sterile water so that the moisture contents were comparable to the moisture contents of microcosms contaminated with 2,4-D. A late-exponential-phase culture of the donor, E. coli D11, was harvested from PH broth, washed, and resuspended in 0.85% sterile saline. This inoculum was added to soil microcosms so that the inoculum densities were approximately 106 CFU g of dry soil−1. All of the microcosms were maintained at gravimetric moisture contents of 20 to 25% depending on the soil type. These moisture contents corresponded to 65 to 75% of the water-holding capacities of the soils. Thus, the soils were moist but not saturated. The corresponding control microcosms contained the same soils amended with the same concentrations of 2,4-D and had the same moisture contents but lacked the donor inoculum. In a preliminary study designed to screen for the potential for plasmid transfer to indigenous populations, single control and treatment microcosms were prepared for each set of conditions. For subsequent studies performed with the Madera Canyon soil, triplicate nonsterile control and treated microcosms were prepared.
Isolation and characterization of presumptive transconjugants.
Bacterial recipients of plasmid pJP4, referred to as transconjugants, were isolated and then characterized by performing the following phenotypic and molecular analyses. The microbial extraction process involved adding 1.2 g of moist soil to a 9.5-ml extraction solution blank (6 μM Zwittergent detergent, 0.2% sodium hexametaphosphate [1]) and then vortexing the preparation for 2 min. Bacterial populations extracted from the soil were plated onto 2,4-D indicator plates. The 2,4-D indicator plates contained (per liter of distilled water) 112 mg of MgSO4 · 7H2O, 5 mg of ZnSO4 · 7H2O, 2.5 mg of Na2MoO4 · 2H2O, 218 mg of K2HPO4, 14 mg of CaCl2 · 2H2O, 0.22 mg of FeCl3 · 6H2O, 0.5 g of NH4Cl, 500 mg of 2,4-D, 80 mg of eosin B, 13 mg of methylene blue, and 20 g of purified agar. Cells that could mineralize the 2,4-D in this medium formed dark purple colonies on the plates due to a change in pH.
Individual colonies were selected from the indicator plates, streaked and isolated on PH agar plates (which contained [per liter of distilled water] 5.0 g of peptone, 3.0 g of yeast extract, 1.1 g of CaCl2, 25 mg of Hg [added as HgCl2], and 15 g of agar), and then used as inocula for PH broth (which contained all of the compounds in PH agar except agar but contained only 5 mg of Hg [added as HgCl2]). Overnight PH broth cultures of each isolate were centrifuged at 5,220 × g for 5 min, and the resulting pellets were resuspended in saline. Aliquots (500 μl) of each suspension were transferred to 3 ml of 2,4-D indicator broth, which contained (per liter of distilled water) 112 mg of MgSO4 · 7H2O, 5 mg of ZnSO4 · 7H2O, 2.5 mg of Na2MoO4 · 2H2O, 340 mg of KH2PO4, 305 mg of Na2HPO4, 14 mg of CaCl2 · 2H2O, 0.22 mg of FeCl3 · 6H2O, 0.5 g of NH4Cl, 500 mg of 2,4-D, and 0.004% bromthymol blue (pH 7.0). In addition, 100 μl of each cell suspension was lysed by boiling it at 98°C for 10 min and was used as a template in two PCR-based analyses, and another 500 μl was stored at 4°C and used for plasmid analysis. An ERIC PCR (25) analysis was performed with each sample in order to generate a molecular fingerprint of each isolate. ERIC PCR was performed as described by Versalovic et al. (26), with minor modifications to confirm that isolates were unique. The primers used were primers ERIC IR and ERIC 2 (4). We confirmed that the pJP4 plasmid-borne tfdB gene was present in unique isolates by performing PCR amplification of a 205-bp portion of the gene (15). A modified miniscreening procedure for large plasmids (18) was used to assess the presence of an 80-kb plasmid. All DNA was visualized with an AlphaImager 2000 gel imager (Alpha Innotech Corp., San Leandro, Calif.) following gel electrophoresis and ethidium bromide staining. In addition, the Biolog procedure (Biolog, Inc., Hayward, Calif.) was used to identify a subset of the confirmed transconjugants.
Heterotrophic plate counts.
R2A (Difco Laboratories, Detroit, Mich.) plates were used to enumerate heterotrophic microorganisms extracted from soil as described above. The plates were incubated at 28°C for 6 days.
Quantitation of 2,4-D biodegradation.
The level of 2,4-D in triplicate microcosms was measured spectrophotometrically by using a procedure modified from the procedure of DiGiovanni et al. (7). A 1.0-ml aliquot of a vortexed soil extraction solution was placed in a 1.2-ml microcentrifuge tube and centrifuged at 16,000 × g for 10 min. The absorbance of the supernatant was measured with a model Spectronic Genesys 2 spectrophotometer (Spectronic Instruments, Inc., Rochester, N.Y.) at a wavelength of 230 nm. Any necessary dilutions were made with extracting solution. A blank microcosm (containing soil that had the same moisture content, was not inoculated, and was not amended with 2,4-D) was analyzed in duplicate on each sampling day in order to evaluate the natural soil components that absorbed at this wavelength. The average blank value was subtracted from the corresponding 2,4-D values obtained on each sampling day.
RESULTS
Screening to assess the potential for transconjugant formation.
Single inoculated and control microcosms for each treatment were used to qualitatively assess plasmid pJP4 transfer from E. coli 15224(pJP4) to indigenous populations in four sandy loam soils containing different concentrations of 2,4-D. Unamended soil samples and soil samples amended with 500 or 1,000 μg of 2,4-D g of dry soil−1 were examined. Soil microcosm samples were obtained 3, 7, 11, and 15 days after inoculation. The numbers of presumptive transconjugants detected varied with time, soil, and the level of 2,4-D. Transconjugants were detected in unamended soil and in soil amended with 500 and 1,000 μg of 2,4-D g of dry soil−1 when the Madera Canyon soil was used. When the Bear Canyon soil was used, transconjugants were detected in the samples amended with 500 or 1,000 μg of 2,4-D g of dry soil−1, and when the Rose Canyon soil was used, transconjugants were found only in samples amended with 500 μg of 2,4-D g of dry soil−1. The earliest time that transconjugants were detected in each soil ranged from 3 to 11 days. Despite the lack of replicates, information concerning the conduciveness of the soils used to transfers was obtained by examining the numbers of transconjugants detected. In all of the soil samples except the unamended Madera Canyon soil sample, the levels of presumptive transconjugants reached 106 to 107 CFU g of dry soil−1. In unamended Madera Canyon soil, only 102 presumptive transconjugants g of dry soil−1 were detected. ERIC PCR and tfdB PCR of several presumptive transconjugants revealed that gene transfer had occurred.
Enumeration and characterization of presumptive transconjugants in Madera Canyon soil.
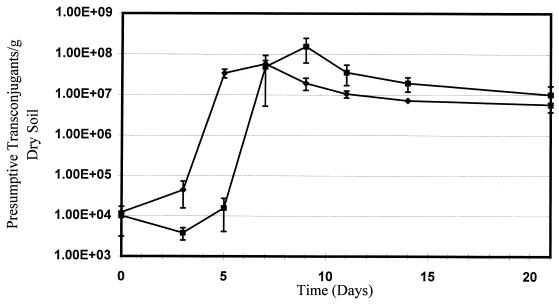
An indigenous population of 2,4-D degraders was present in the Madera Canyon soil, as shown by the growth of light purple pinpoint colonies on the 2,4-D indicator plates by day 7 when either control or treated soil was used. In contrast, presumptive transconjugants formed deep purple colonies that often had a metallic sheen. The number of indigenous 2,4-D-degrading colonies increased with time from approximately 104 colonies g of dry soil−1 on day 7 to 106 colonies g of dry soil−1 on day 21. Most indigenous degraders resisted subculturing, even when the 2,4-D indicator media on which they were initially isolated was used, which made further analysis difficult. However, no randomly selected indigenous degraders were found to be mercury resistant or to contain an 80-kb plasmid. Accordingly, light purple pinpoint colonies were not included in the plate counts when presumptive transconjugants were enumerated. Transconjugants not only persisted but increased in number throughout the 21-day incubation period, reaching maximal levels of 107 and 108 presumptive transconjugants g of dry soil−1 for samples amended with 500 and 1,000 μg of 2,4-D g of dry soil−1, respectively (Fig. 1). Based on heterotrophic plate counts, which remained fairly constant at approximately 2 × 108 transconjugants g of dry soil−1, the maximal presumptive transconjugant levels represented approximately 10% of the carrying capacity (culturable microorganisms) of the soil. Similar trends in the number of transconjugants with time were observed for samples amended with both levels of 2,4-D, and there was a slight lag in the increase in the number of transconjugants in the soil amended with 1,000 μg of 2,4-D g of dry soil−1.
FIG. 1.
Enumeration of presumptive transconjugants in Madera Canyon soil microcosms. The data points and error bars show the means and standard deviations based on data from three replicate microcosms. No presumptive transconjugants were detected in control microcosms. Symbols: ⧫, soil amended with 500 μg of 2,4-D g of dry soil−1 and E. coli D11; ■, soil amended with 1,000 μg of 2,4-D g of dry soil−1 and E. coli D11.
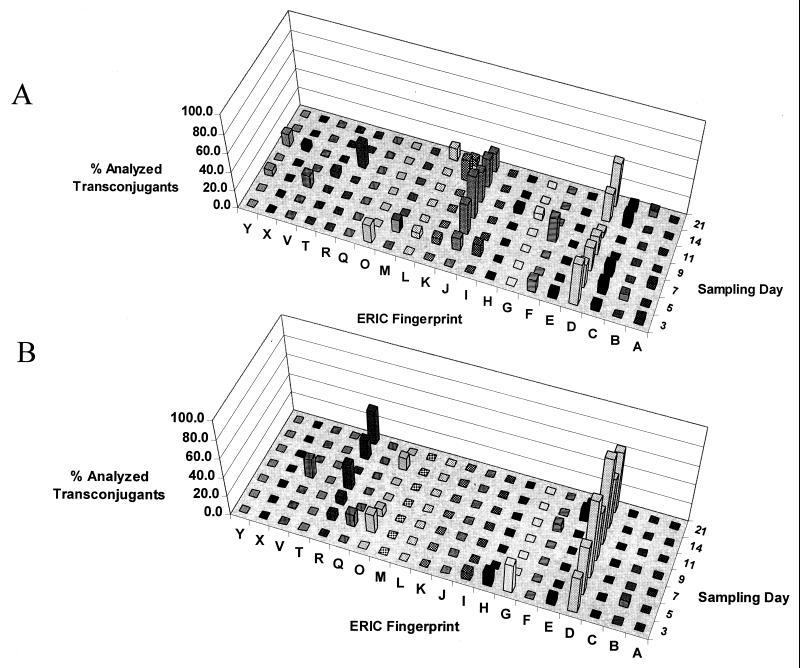
Several presumptive transconjugants obtained on each sampling day were streaked and isolated on PH agar plates and were characterized further by performing ERIC PCR (Fig. 2), tfdB PCR, and plasmid profile size (Fig. 3) analyses. The ERIC PCR analysis of presumptive transconjugants confirmed that none of the transconjugants were E. coli D11. In all, 20 different ERIC fingerprints of presumptive transconjugants were obtained. Representative isolates that produced each ERIC fingerprint were subjected to additional analyses. Figure 4 shows semiquantitatively the distribution over time of ERIC fingerprints of randomly selected presumptive transconjugants. For the samples amended with 500 and 1,000 μg of 2,4-D g of dry soil−1, 103 and 96 presumptive transconjugants, respectively, were characterized by ERIC PCR. Transconjugants could only be identified to the genus level by Biolog analysis due to the limited database available for environmental isolates and the low similarity scores obtained for the environmental isolates. All 199 transconjugants analyzed were identified as members of one of two closely related genera, the genera Pseudomonas and Burkholderia (levels of similarity, 0.39 to 0.72). For each level of 2,4-D, two dominant recipient populations were detected in the study; populations J and D were detected in samples amended with 500 μg of 2,4-D g of dry soil−1, and populations R and D were detected in samples amended with 1,000 μg of 2,4-D g of dry soil−1. For each sampling day, presumptive transconjugants were randomly selected for analysis from the highest-dilution 2,4-D indicator plates. Using the highest-dilution plates ensured that dominant transconjugant populations were selected and reduced the likelihood of plate matings.
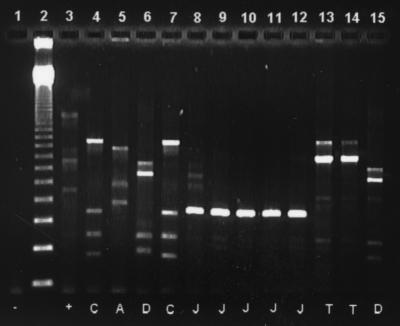
FIG. 2.
Representative day 7 presumptive transconjugant fingerprints generated by ERIC PCR. Lane 1, negative control; lane 2, 123-bp ladder; lane 3, E. coli donor; lanes 4 to 15, presumptive transconjugants. The letters at the bottom are ERIC fingerprint designations.
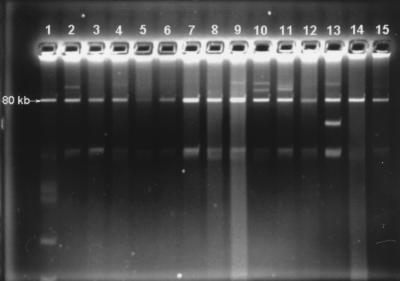
FIG. 3.
Plasmid profiles of selected isolates. Lane 1, 80-kb plasmid isolated from E. coli donor as a size marker (positive control); lanes 2 to 15, presumptive transconjugants. A negative control from the same plasmid preparation was electrophoresed on another gel.
FIG. 4.
Semiquantitative analysis of presumptive transconjugant diversity over time. (A) Soil amended with 500 μg of 2,4-D g of dry soil−1. (B) Soil amended with 1,000 μg of 2,4-D g of dry soil−1. ERIC fingerprints, expressed as percentages of presumptive transconjugants analyzed per sample day, are plotted against sampling day.
Degradation of 2,4-D.
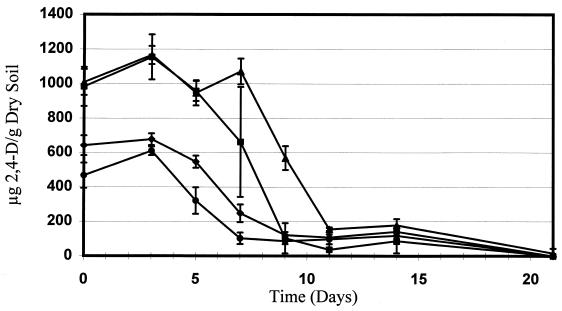
At distinct times, particularly in microcosms amended with 1,000 μg of 2,4-D g of dry soil−1, an increase in the rate of 2,4-D degradation was observed in microcosms inoculated with E. coli D11 compared to the corresponding controls (Fig. 5). However, after 21 days, 2,4-D was completely degraded in all of the microcosms, which showed that the indigenous microorganisms were able to degrade 2,4-D in the soil at the contaminant levels used.
FIG. 5.
Biodegradation of 2,4-D in Madera Canyon soil microcosms. The data points and error bars show the means and standard deviations based on data from three replicate microcosms. Symbols: ⧫, soil amended with 500 μg of 2,4-D g of dry soil−1 (control); ▴, soil amended with 1,000 μg of 2,4-D g of dry soil−1 (control); ●, soil amended with 500 μg of 2,4-D g of dry soil−1 and E. coli D11; ■, soil amended with 1,000 μg of 2,4-D g of dry soil−1 and E. coli D11.
DISCUSSION
The frequency of gene transfer in nonsterile soil systems may vary with time, soil type, and level of 2,4-D, as suggested by the results of screening experiments conducted to assess the potential for transconjugant formation in four soils contaminated with different levels of 2,4-D. Inoculation with E. coli D11 followed by donor counterselection facilitated detection of gene transfer not only in Madera Canyon soil but also in Rose Canyon and Bear Canyon soils. In similar studies in which R. eutropha JMP134 was used as the donor, plasmid pJP4 transfer to indigenous Madera Canyon soil recipients was detected in soil amended with 1,000 μg of 2,4-D g of dry soil−1 but not in soil amended with 500 μg of 2,4-D g of dry soil−1 (4). Use of the E. coli D11 donor facilitated detection of plasmid transfer to Madera Canyon soil recipients at both levels of 2,4-D. Furthermore, use of the E. coli D11 donor allowed detection of plasmid transfer in Rose Canyon and Bear Canyon soils. No gene transfer was detected in these soils in studies performed with either level of 2,4-D when R. eutropha JMP134, which remained culturable at significant levels, was used as the donor (unpublished data). Using E. coli D11 and subsequent plating of extracted populations on 2,4-D indicator media circumvented this problem by eliminating donor interference.
Although all four soils examined were very similar in terms of texture, differences in pH and soil organic matter content (Table 1), along with presumed differences in other biological and physicochemical properties, may explain in part the observed differences in detection of presumptive transconjugants. For example, bioavailability and thus presumptive toxicity of 2,4-D may be influenced by sorption to the organic matter of the soil. A threshold 2,4-D stress may be necessary in certain soils for presumptive transconjugant detection via selection for plasmid-bearing microorganisms and/or increasing transfer frequencies. Although the scenario is plausible, it should be noted that strictly speaking there is no direct evidence which supports the latter hypothesis. As suggested by the Rose Canyon microcosm results, high concentrations of 2,4-D may inhibit presumptive transconjugant detection by stressing the indigenous populations to such an extent that gene transfer does not occur at a detectable level.
Figure 1 shows that the trends in transconjugant numbers with time for the two levels of 2,4-D used were similar. Presumptive transconjugants detected at time zero may have been the result, at least in part, of plate matings due to the low soil dilutions used. One potential problem with gene transfer studies is the occurrence of gene transfer on agar surfaces following extraction from the soil, which results in overestimation of the true frequency of gene transfer in the soil (15, 19, 27). It has been suggested that when the number of donor cells is low (<104 cells per plate), plate mating is a minimal concern (19). Taking into consideration the dilutions counted when presumptive transconjugants were assessed and the heterotrophic plate counts obtained in the same sampling experiment, it was unlikely that plate mating was a significant concern except perhaps at time zero in the Madera Canyon study. However, it should be noted that no transconjugants were detected at time zero in Bear Canyon or Rose Canyon soils, suggesting that the time zero transconjugants in the Madera Canyon study were not the result of plate mating. The possibility that gene transfer occurred in the soil at time zero cannot be completely ruled out since approximately 0.5 h elapsed between inoculation and plating of each microcosm. In addition, when presumptive transconjugant numbers are evaluated, it is important to note that only the culturable transconjugants are detected by plating on 2,4-D indicator media. Viable but nonculturable microorganisms, in addition to any transconjugants that lack the chromosomally encoded genes necessary for complete mineralization, escape detection. Accordingly, the absolute numbers of transconjugants are probably higher. There are several plausible explanations for the observed increase in the number of transconjugants with time: ongoing transfer from E. coli D11 to indigenous populations, successive gene transfer between indigenous populations, and growth of the initial transconjugants themselves. However, the diversity of the ERIC fingerprints indicates that the presumptive transconjugants did not arise simply from a single transfer event followed by growth but that numerous gene transfer events occurred at some point. The longer lag phase observed when the soil was amended with 1,000 μg of 2,4-D g of dry soil−1 may have been due to increased toxicity of the herbicide to indigenous populations compared to the soil amended with 500 μg of 2,4-D g of dry soil−1. Presumably for similar reasons, a decrease in the diversity of transconjugants at the higher level of 2,4-D was observed, as indicated by ERIC PCR results (Fig. 4). The ERIC fingerprint analysis also verified that the presumptive transconjugants isolated were not mutant, 2,4-D-degrading donors. Transconjugants were identified by the Biolog method only to the genus level due to the limited database available for environmental isolates.
At distinct times, microcosms inoculated with E. coli D11 exhibited slightly increased rates of 2,4-D degradation (Fig. 5). Although no presumptive-transconjugant colonies were detected on the 2,4-D indicator plates prepared by using control microcosms, the rapid decreases in the 2,4-D concentration in control microcosms suggested that indigenous 2,4-D degraders or consortia of microorganisms capable of carrying out degradation were present. Degradation of 2,4-D by indigenous soil populations, which blurs the action of introduced genes, is fairly common and expected. The hypothesis that such populations were present was also supported by the growth of light purple pinpoint colonies on the 2,4-D indicator plates. These indigenous degraders, in conjunction with the fact that the donor itself is not capable of completely mineralizing 2,4-D, may explain the only moderately increased rate of 2,4-D degradation at distinct times after E. coli D11 was inoculated.
Taken together, our results suggest that using the E. coli donor counterselection system improved detection of plasmid pJP4 transfer to indigenous soil populations. In addition, these results indicate that gene transfer from an introduced donor to indigenous soil populations occurs in a variety of soils and at different contaminant levels. Inoculating E. coli D11 generated a variety of transconjugants, and this technique has potential for increasing the ability of soil to biodegrade 2,4-D, particularly in soils that lack an adequate intrinsic ability to degrade this herbicide (17). Furthermore, based on this model system for studying plasmid transfer to indigenous populations, similar transfers of other plasmids may facilitate degradation of more recalcitrant organic compounds and/or perhaps organic compounds that are present at sites which are cocontaminated with a metal. The ability to counterselect the donor may prove to be useful for assessing the potential for gene release, whether intentional or not, into the environment.
ACKNOWLEDGMENT
This work was supported by grant 5 P42 ESO4940-09 from the NIEHS Basic Research Superfund.
REFERENCES
- 1.Brendecke J W, Axelson R D, Pepper I L. Soil microbial activity as an indicator of soil fertility; the long-term effects of municipal sewage sludge on an arid soil. Soil Biol Biochem. 1993;25:751–758. [Google Scholar]
- 2.Chapman H, Pratt P. Methods of analysis for soils, plants and waters. Riverside: University of California; 1961. pp. 44–46. [Google Scholar]
- 3.Daane L L, Molina J A E, Berry E C, Sadowsky M J. Influence of earthworm activity on gene transfer from Pseudomonas fluorescens to indigenous soil bacteria. Appl Environ Microbiol. 1996;62:515–521. doi: 10.1128/aem.62.2.515-521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Rore H, Demolder K, De Wilde K, Top E, Houwne F, Verstraete W. Transfer of the catabolic plasmid RP4::Tn4371 to indigenous soil bacteria and its effect on respiration and biphenyl breakdown. FEMS Microbiol Ecol. 1994;15:71–77. [Google Scholar]
- 6.De Rore H, Top E, Houwen F, Mergeay M, Verstraete W. Evolution of heavy metal resistant transconjugants in a soil environment with a concomitant selective pressure. FEMS Microbiol Ecol. 1994;14:263–273. [Google Scholar]
- 7.DiGiovanni G D, Neilson J W, Pepper I L, Sinclair N A. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl Environ Microbiol. 1996;62:2521–2526. doi: 10.1128/aem.62.7.2521-2526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan K. Fitnesses of a conjugative plasmid and its host bacteria in soil microcosms. Mol Biol Evol. 1995;12:1012–1021. [Google Scholar]
- 10.Friedrich B, Meyer M, Schlegel H G. Transfer and expression of the herbicide-degrading plasmid pJP4 in aerobic autotrophic bacteria. Arch Microbiol. 1983;134:92–97. doi: 10.1007/BF00407938. [DOI] [PubMed] [Google Scholar]
- 11.Ghosal D, You I S. Microbial degradation of halogenated compounds. Science. 1985;228:135–142. doi: 10.1126/science.228.4696.135. [DOI] [PubMed] [Google Scholar]
- 12.Ghosal D, You I S, Chatterjee D K, Chakrabarty A M. Plasmids in the degradation of chlorinated aromatic compounds. In: Helinski D R, editor. Plasmids in bacteria. New York, N.Y: Plenum Press; 1985. pp. 667–686. [DOI] [PubMed] [Google Scholar]
- 13.Kinkle B K, Sadowsky M J, Schmidt E L, Koskinen W C. Plasmids pJP4 and r68.45 can be transferred between populations of bradyrhizobia in nonsterile soil. Appl Environ Microbiol. 1993;59:1762–1766. doi: 10.1128/aem.59.6.1762-1766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozdroj J. Effect of copper(II) on survival of Pseudomonas fluorescens and transfer of plasmid RP4 in soil. World J Microbiol Biotechnol. 1994;10:175–177. doi: 10.1007/BF00360881. [DOI] [PubMed] [Google Scholar]
- 15.Neilson J W, Josephson K L, Pepper I L, Arnold R B, Di Giovanni G D, Sinclair N A. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl Environ Microbiol. 1994;60:4053–4058. doi: 10.1128/aem.60.11.4053-4058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richaume A, Smit E, Faurie G, van Elsas J D. Influence of soil type on the transfer of plasmid RP4p from Pseudomonas fluorescens to introduced recipient and to indigenous bacteria. FEMS Microbiol Ecol. 1992;101:281–292. [Google Scholar]
- 17.Roane T M. Bioaugmentation with metal-resistant microorganisms in the remediation of metal and organic contaminated soils. Ph.D. dissertation. Tucson: University of Arizona; 1999. [Google Scholar]
- 18.Rodriguez R L, Tait R C. Recombinant DNA techniques: an introduction. Menlo Park, Calif: The Benjamin-Cummings Publishing Co., Inc.; 1983. pp. 160–162. [Google Scholar]
- 19.Smit E, van Elsas J D. Determination of plasmid transfer frequency in soil: consequences of bacterial mating on selective agar media. Curr Microbiol. 1990;21:151–157. [Google Scholar]
- 20.Smit E, van Elsas J D, van Veen J A, de Vos W M. Detection of plasmid transfer from Pseudomonas fluorescens to indigenous bacteria in soil by using bacteriophage φR2F for donor counterselection. Appl Environ Microbiol. 1991;57:3482–3488. doi: 10.1128/aem.57.12.3482-3488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Top E M, Holben W E, Forney L J. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl Environ Microbiol. 1995;61:1691–1698. doi: 10.1128/aem.61.5.1691-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Top E M, Van Daele P, De Saeyer N, Forney L J. Enhancement of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation in soil by dissemination of catabolic plasmids. Antonie Leeuwenhoek. 1998;73:87–94. doi: 10.1023/a:1000663619522. [DOI] [PubMed] [Google Scholar]
- 24.Trevors J T, Berg G. Conjugal RP4 transfer between pseudomonads in soil and recovery of RP4 plasmid DNA from soil. Syst Appl Microbiol. 1989;11:223–227. [Google Scholar]
- 25.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Versalovic J, Schneider M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 27.Walter M V, Porteous L A, Seidler R J. Evaluation of a method to measure conjugal transfer of recombinant DNA in soil slurries. Curr Microbiol. 1989;19:365–370. [Google Scholar]