Abstract
Denaturing gradient gel electrophoresis (DGGE) of DNA fragments obtained by PCR amplification of the V2-V3 region of the 16S rRNA gene was used to detect the presence of Lactobacillus species in the stomach contents of mice. Lactobacillus isolates cultured from human and porcine gastrointestinal samples were identified to the species level by using a combination of DGGE and species-specific PCR primers that targeted 16S-23S rRNA intergenic spacer region or 16S rRNA gene sequences. The identifications obtained by this approach were confirmed by sequencing the V2-V3 region of the 16S rRNA gene and by a BLAST search of the GenBank database.
The gastrointestinal tracts of animals, including humans, harbor complex microbial communities comprised of possibly hundreds of bacterial species (13). Members of the genus Lactobacillus are commonly present as members of these communities and have received considerable attention with respect to their putative health-conferring properties (as probiotics [6]).
Analysis of gastrointestinal communities has relied traditionally on bacteriological culture methods and microscopy. There is doubtless a bias present in the results of these studies, however, because not all of the members of the communities can be cultured (11). The results of monitoring the composition of gastrointestinal communities may be more reliable if molecular methods were used in addition to traditional approaches (5, 16). Microbiological investigations of the compositions of terrestrial and aquatic microbial communities have shown the versatility of denaturing gradient gel electrophoresis (DGGE) combined with PCR as a molecular analytical method (4, 10). In this technique, DNA is extracted from cells of all of the species represented in the habitat of interest, and a hypervariable sequence region of the 16S rRNA gene is amplified by PCR. The mixture of 16S fragments is subjected to DGGE in order to separate the fragments and thus obtain a profile of the community. This profile is generated because of differences in the chemical stability, and hence the distance into the gradient where denaturation occurs and migration ceases, of the 16S fragments that have different nucleotide base compositions (10). We have for many years maintained a mouse colony devoid of lactobacilli as gastrointestinal inhabitants (14). They provided a suitable system with which to test the efficacy of PCR-DGGE as an analytical method in studies of the gastrointestinal microflora.
Even when they can be cultured reliably, gastrointestinal species of bacteria can be difficult to identify. The identification of Lactobacillus isolates by phenotypic methods is difficult because it requires, in several cases, determination of bacterial properties beyond that of the common fermentation tests (for example, cell wall analysis and electrophoretic mobilities of lactate dehydrogenase [8]). In general, about 17 phenotypic tests are required to identify a Lactobacillus isolate accurately to the species level (7). The logistics, regardless of the doubtful accuracy of phenotypic identification methods, are daunting when large-scale investigations of the intestinal microflora are pursued. We reasoned that, since modern bacterial classification is greatly influenced by knowledge of 16S rRNA gene sequences, PCR-DGGE could provide a more practical approach to the identification of lactobacilli.
We demonstrate here that addition of Lactobacillus species to the stomach microflora of mice could be detected by PCR-DGGE analysis. Further, we demonstrate that Lactobacillus isolates cultured from gastrointestinal samples obtained from humans and pigs could be identified, or at least grouped, by PCR-DGGE. The species identities of the isolates were then further investigated with species-specific PCR primers that targeted 16S-23S intergenic spacer region or 16S rRNA gene sequences. These identifications were subsequently confirmed by obtaining 16S rRNA gene sequences that were compared to those in GenBank.
MATERIALS AND METHODS
Bacterial cultures.
The lactobacilli used in this study are listed in Table 1. The bacteria were cultured with Lactobacilli MRS medium (Difco Laboratories, Detroit, Mich.) incubated under anaerobic conditions at 37°C. Thirty-six of the strains used in this study were unidentified isolates (“unknowns”) originating in gastrointestinal samples (Table 1).
TABLE 1.
Lactobacillus strains
| Strain | Sourcea | Identification by:
|
GenBank accession no. for sequence | ||
|---|---|---|---|---|---|
| DGGE | Species-specific primers | 16S V2-V3 sequence | |||
| L. acidophilus ATCC 4356T | ATCC | NAb | L. acidophilus | NA | NA |
| L. agilis DSM 20509T | DSM | NAb | NA | NA | NA |
| L. brevis ATCC 14869T | ATCC | NAb | NA | NA | NA |
| L. casei ATCC 334 | ATCC | NAb | L. casei | NA | NA |
| L. casei/L. zeae ATCC 393T | ATCC | NAb | L. zeae | NA | NA |
| L. casei ATCC 4684 | ATCC | NAb | L. casei | NA | NA |
| L. crispatus ATCC 33820T | ATCC | NAb | L. crispatus | NA | NA |
| L. delbrueckii subsp. bulgaricus ATCC 11842T | ATCC | NAb | NA | NA | NA |
| L. fermentum ATCC 14869T | ATCC | NAb | L. fermentum | NA | NA |
| L. gasseri ATCC 33323T | ATCC | NAb | L. gasseri | NA | NA |
| L. helveticus ATCC 15009T | ATCC | NAb | NA | NA | NA |
| L. johnsonii ATCC 33200T | ATCC | NAb | L. johnsonii | NA | NA |
| L. paracasei subsp. paracasei ATCC 25302 | ATCC | NAb | L. casei | NA | NA |
| L. plantarum ATCC 1988 | ATCC | NAb | L. plantarum | NA | NA |
| L. plantarum ATCC 14917T | ATCC | NAb | L. plantarum | NA | NA |
| L. rhamnosus ATCC 8530 | ATCC | NAb | L. rhamnosus | NA | NA |
| L. rhamnosus DSM 20021T | DSM | NAb | L. rhamnosus | NA | NA |
| L. reuteri DSM 20016T | DSM | NAb | L. reuteri | NA | NA |
| L. ruminis ATCC 27780T | ATCC | NAb | NA | NA | NA |
| L. salivarius subsp. salicinius ATCC 11742T | ATCC | NAb | NA | NA | NA |
| L. salivarius subsp. salivarius ATCC 11741T | ATCC | NAb | NA | NA | NA |
| L. sharpeae DSM 20505T | DSM | NAb | L. sharpeae | NA | NA |
| L. vitulinus ATCC 27783T | ATCC | NAb | NA | NA | NA |
| L. zeae | |||||
| DSM 20178T | DSM | NAb | L. zeae | NA | NA |
| GTP5 | Pig | L. acidophilus/L. crispatus/L. helveticus | L. crispatus | L. crispatus | AF157035 |
| GT3C1 | Pig | L. acidophilus/L. crispatus/L. helveticus | L. crispatus | L. crispatus | AF158588 |
| L8 | Human | L. acidophilus/L. crispatus/L. helveticus | L. acidophilus | L. acidophilus | AF158988 |
| L35 | Human | L. acidophilus/L. crispatus/L. helveticus | L. acidophilus | L. acidophilus | AF159014 |
| L43 | Human | L. acidophilus/L. crispatus/L. helveticus | L. acidophilus | L. acidophilus | AF159021 |
| L10 | Human | L. plantarum | L. plantarum | L. plantarum group | AF158990 |
| L12 | Human | L. plantarum | L. plantarum | L. plantarum group | AF158992 |
| L13 | Human | L. plantarum | L. plantarum | L. plantarum group | AF158993 |
| L33 | Human | L. plantarum | L. plantarum | L. plantarum group | AF159012 |
| L45 | Human | L. plantarum | L. plantarum | L. plantarum group | AF159023 |
| L44 | Human | L. gasseri/L. johnsonii | L. gasseri | L. gasseri | AF159022 |
| GTH5 | Human | L. gasseri/L. johnsonii | L. gasseri | L. gasseri | AF157044 |
| L. johnsonii | |||||
| NCK800 | Culture collection of T. K. Klaenhammer | L. gasseri/L. johnsonii | L. johnsonii | NA | NA |
| GTH10 | Human | L. fermentum | L. fermentum | L. fermentum | AF158581 |
| GT3S3 | Pig | L. reuteri | L. reuteri | L. reuteri | AF158590 |
| GTH18 | Human | L. brevis/L. sharpeae | L. brevis (because primer pair 11 negative and aerogenic) | L. brevis | AF157038 |
| L4 | Human | L. reuteri | L. reuteri | L. reuteri | AF158983 |
| L38 | Human | L. fermentum | L. fermentum | L. fermentum | AF159016 |
| L3 | Human | L. casei group | L. casei | L. casei group | AF158982 |
| L6 | Human | L. casei group | L. casei | L. casei group | AF158986 |
| L9 | Human | L. casei group | L. casei | L. casei group | AF158989 |
| L14 | Human | L. casei group | L. casei | L. casei group | AF158994 |
| L19 | Human | L. casei group | L. casei | L. casei group | AF158998 |
| L20 | Human | L. casei group | L. casei | L. casei group | AF158999 |
| L22 | Human | L. casei group | L. rhamnosus | L. casei group | AF159001 |
| L25 | Human | L. casei group | L. casei | L. casei group | AF159004 |
| L27 | Human | L. casei group | L. rhamnosus | L. casei group | AF159006 |
| L28 | Human | L. casei group | L. rhamnosus | L. casei group | AF159007 |
| L29 | Human | L. casei group | L. rhamnosus | L. casei group | AF159008 |
| L30 | Human | L. casei group | L. casei | L. casei group | AF159009 |
| L32 | Human | L. casei group | L. rhamnosus | L. casei group | AF159011 |
| L37 | Human | L. casei group | L. rhamnosus | L. casei group | AF159015 |
| L39 | Human | L. casei group | L. casei | L. casei group | AF159017 |
| L41 | Human | L. casei group | L. rhamnosus | L. casei group | AF159019 |
| L42 | Human | L. casei group | L. casei | L. casei group | AF159020 |
| L. rhamnosus | |||||
| DR20 | Culture collection, NZDRI | L. casei group | L. rhamnosus | NA | NA |
| GTH1 | Human | L. salivarius | NA | L. salivarius subsp. salivarius | AF158557 |
| L5 | Human | L. ruminis | NA | L. ruminis | AF158984 |
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen; NZDRI, New Zealand Dairy Research Institute.
NA, not applicable (these were reference strains).
Mouse experiments.
Lactobacillus-free mice were maintained in isolators by gnotobiotic technology. The animals harbored a microbial community in their gastrointestinal tracts equivalent to that of conventional mice except that lactobacilli were absent (14). To test the ability of PCR-DGGE to detect changes in the microbial community of the murine stomach, we maintained a group of five lactobacillus-free mice in an isolator. At day 0, we removed one of the mice (mouse 1) as a control. Then we inoculated the remainder of the animals, by mouth, with a culture of strain 100-23. Four days later, we removed one of the mice (mouse 2) and inoculated the remainder with Lactobacillus sp. strain 100-5. Then 4 days later, we removed another mouse (mouse 3) and inoculated the remaining mice with Lactobacillus sp. strain 21. Another mouse (mouse 4) was removed at day 12, and the remaining animal (mouse 5) was inoculated with strain 20 and examined 4 days later. Upon removal from the isolator, the mice were killed by carbon dioxide anesthesia and cervical dislocation, the stomach was removed from each mouse, and the stomach contents were retained and stored at −20°C. To extract bacterial DNA, the stomach contents were homogenized in 1 ml of TN150 buffer (10 mM Tris-HCl, 150 mM NaCl [pH 8]) and centrifuged at 14,600 × g for 5 min (5°C). DNA was extracted from the resulting pellet with a FastDNA kit (Bio 101, Vista, Calif.) by using CLS-TC (cell lysis solution for animal tissues and bacteria) and a 1/4-in. sphere plus garnet matrix (see the kit data sheet) according to the manufacturer's instructions.
Extraction of DNA from Lactobacillus cultures.
Growth from pure cultures on Lactobacilli MRS agar plates was used to prepare a heavy suspension of cells in 1 ml of sterile deionized water. The suspensions were centrifuged at 14,600 × g (3 min, 5°C) and washed with 1 ml of TN150 buffer. The pellets were resuspended in 1 ml of TN150 buffer and transferred to sterile tubes containing 0.3 g of sterile zirconium beads (diameter, 0.1 mm). The tubes were placed in a mini-bead beater (Biospec Products, Bartlesville, Okla.), shaken at 5,000 rpm for 3 min, and then stored on ice. Five hundred microliters of this crude DNA solution was extracted sequentially with 500 μl of TE (10 mM Tris, 1 mM EDTA [pH 8.5])-saturated phenol and with chloroform-isoamyl alcohol (24:1). The DNA was precipitated overnight by the addition of 2 volumes of cold ethanol and a 0.1 volume of 3 M sodium acetate at −20°C. The preparations were centrifuged at 14,600 × g (20 min, −5°C) and the pellets were dried at 37°C. The pellets were then dissolved in 500 μl of TE buffer (pH 8.5), and 25 μl of DNase-free RNase (2 mg/ml) was added. After incubation at 37°C for 1 h, the phenol-chloroform extraction, precipitation, and drying steps were repeated, and the DNA was dissolved in 20 μl of TE buffer (pH 7.5). The amount of DNA per microliter was measured by spectrophotometry.
Amplification of the DNA target sequence for DGGE.
The V2-V3 region of the 16S ribosomal DNA (rDNA) gene (position 339 to 539 in the Escherichia coli gene) of bacteria in the stomach contents or from pure cultures of lactobacilli was amplified with primers HDA1-GC (CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′; the GC clamp is in boldface) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′). The HDA primers were originally described by Turner and colleagues (S. J. Turner, G. D. Lewis, D. J. Saul, C. S. Baker, and A. G. Rodrigo, New Zealand Microbiol. Soc. Ann. Meet., poster paper, 1998). PCR was performed in 0.2-ml tubes in a PCR Express thermal cycler (Hybaid, Teddington, United Kingdom). For the amplification of the target sequence from DNA extracted from pure cultures, the reaction mixture (50 μl) consisted of reaction buffer (final concentrations, 10 mM Tris-HCl, 2.5 mM MgCl2, and 50 mM KCl [pH 8.3]), a 200 μM concentration of each deoxynucleoside triphosphate, 20 pmol of each primer, 500 ng of bacterial DNA, and 2.5 U of Taq DNA polymerase (Boehringer Mannheim, Mannheim, Germany). The amplification program was 94°C for 4 min; 30 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 60 s; and finally 68°C for 7 min. PCR amplifications from DNA extracted from the stomach contents were carried out with the Expand high-fidelity PCR system (Boehringer Mannheim) or Taq polymerase as described above, but the reaction mixture (50μl) contained 1 μl of DNA solution. The amplification program was 93°C for 2 min and 30 cycles of 93°C for 30 s, 57°C for 30 s, and 72°C for 30 s, followed by 2 min at 72°C.
Lactobacillus identification ladder.
An identification ladder containing 16S V2-V3 region sequences of 19 Lactobacillus reference strains was prepared. The sequences were obtained by PCR from individual pure cultures of the reference strains. The PCR products were then mixed to obtain the identification ladder. It was possible to generate further supplies of the ladder by PCR with the V2-V3 primers and 1 μl of mixture as the template, in the presence of 2.5 mM MgCl2.
DGGE.
DGGE was performed with a DCode universal mutation detection system (Bio-Rad, Hercules, Calif.) utilizing 16-cm by 16-cm by 1-mm gels. Eight percent polyacrylamide gels were prepared and run with 1× TAE buffer diluted from 50× TAE buffer (2 M Tris base, 1 M glacial acetic acid, and 50 mM EDTA). The denaturing gradient was formed with two 8% acrylamide (acrylamide-bis, 37.5:1) stock solutions (Bio-Rad). The gels contained a 30 to 50% gradient of urea and formamide increasing in the direction of electrophoresis. A 100% denaturing solution contained 40% (vol/vol) formamide and 7.0 M urea. The electrophoresis was conducted with a constant voltage of 130 V at 60°C for about 4 h 30 min. The run was stopped when a xylene cyanol dye marker reached the bottom of the gel. Gels were stained with ethidium bromide solution (5 μg/ml; 20 min), washed with deionized water, and viewed by UV transillumination.
Species-specific primers.
The 11 species-specific primer pairs that we derived were based on the 16S rRNA gene or the 16S-23S rRNA intergenic spacer region (Table 2). A reaction mixture (25 μl) consisted of reaction buffer (10 mM Tris-HCl [final concentration], a variable MgCl2 concentration [Table 2], and 50 mM KCl [pH 8.3]), a 200 μM concentration of each deoxynucleoside triphosphate, 10 pmol of each primer, 50 ng of bacterial DNA (extracted from pure cultures as described above), and 1.75 U of Taq DNA polymerase (Boehringer Mannheim). The amplification program was 92°C for 2 min, followed by 30 cycles of 95°C for 30 s, 30 s at the appropriate annealing temperature (Table 2), and 72°C for 30 s. A cycle of 72°C for 1 min concluded the program. Amplification products were detected by agarose gel electrophoresis (5 μl of PCR mixture, 2% agarose gel), ethidium bromide staining, and UV transillumination.
TABLE 2.
Species-specific primers
| Primer pair | Species | Primers | Target | Sequence (5′–3′) | PCR annealing temp (°C) | MgCl2 concn (mM) |
|---|---|---|---|---|---|---|
| 1 | L. acidophilus | Aci 16SI | 16S gene | AGCTGAACCAACAGATTCAC | 62 | 1.5 |
| 16SII | ACTACCAGGGTATCTAATCC | |||||
| 2 | L. crispatus | Cri 16SI | 16S gene | GTAATGACGTTAGGAAAGCG | 60 | 1.5 |
| 16SII | ACTACCAGGGTATCTAATCC | |||||
| 3 | L. gasseri | GasI | 16S-23S spacer | GAGTGCGAGAGCACTAAAG | 55 | 2.5 |
| GasII | CTATTTCAAGTTGAGTTTCTCT | |||||
| 4 | L. johnsonii | Joh 16SI | 16S gene | GAGCTTGCCTAGATGATTTTA | 57 | 1.5 |
| 16SII | ACTACCAGGGTATCTAATCC | |||||
| 5 | L. plantarum | Lfpr | 16S-23S spacer | GCCGCCTAAGGTGGGACAGAT | 55 | 2.0 |
| PlanII | TTACCTAACGGTAAATGCGA | |||||
| 6 | L. casei | PrI | 16S-23S spacer | CAGACTGAAAGTCTGACGG | 55 | 2.0 |
| CasII | GCGATGCGAATTTCTTTTTC | |||||
| 7 | L. zeae | ZeaI | 16S-23S spacer | TGTTTAGTTTTGAGGGGACG | 58 | 2.0 |
| ZeaII | ATGCGATGCGAATTTCTAAATT | |||||
| 8 | L. rhamnosus | PrI | 16S-23S spacer | CAGACTGAAAGTCTGACGG | 58 | 2.0 |
| RhaII | GCGATGCGAATTTCTATTATT | |||||
| 9 | L. reuteri | Lfpr | 16S-23S spacer | GCCGCCTAAGGTGGGACAGAT | 55 | 2.0 |
| Reu | AACACTCAAGGATTGTCTGA | |||||
| 10 | L. fermentum | Lfpr | 16S-23S spacer | GCCGCCTAAGGTGGGACAGAT | 55 | 3.0 |
| FermII | CTGATCGTAGATCAGTCAAG | |||||
| 11 | L. sharpeae | ShaI | 16S-23S spacer | GATAATCATGTAAGAAACCGC | 58 | 1.5 |
| ShaII | ATATTGTTGGTCGCGATTCG |
Amplification and sequencing of the 16S V2-V3 region.
To confirm the identification of the gastrointestinal lactobacilli examined by using DGGE and species-specific primers, we amplified and sequenced (one polynucleotide strand only) the V2-V3 region of the 16S rRNA gene of each isolate and conducted a search of sequences deposited in the GenBank DNA database by using the BLAST algorithm (1). The identities of the isolates were determined on the basis of highest score. Amplification of the V2-V3 region was accomplished with primers HDA1 (lacking the GC clamp) and HDA2 and the same thermal cycler program as described above for DGGE. Sequencing was carried out by the Centre for Gene Research, University of Otago, by the dideoxy method of Sanger et al. (12) by using the PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems Inc., Foster City, Calif.) in combination with an Applied Biosystems model 377A automated sequencing system. Analysis of nucleotide sequence data was carried out by using the SeqEd program, version 1.0.3 (Applied Biosystems Inc.).
RESULTS
DGGE detection of Lactobacillus populations in mouse stomach contents.
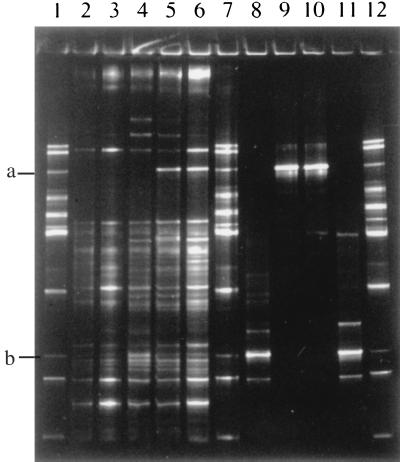
Comparison of the 16S V2-V3 rDNA profiles obtained from murine stomach contents showed that this method could detect changes in the microbial community, in this case the intentional addition of lactobacilli (Fig. 1). Multiple DNA fragments were present in the profiles, even from the stomach contents of mouse 1 (Fig. 1, lane 2). These fragments probably represent either fecal bacteria present in the stomach contents of these coprophagous animals or bacterial species not detected previously by culture methods in the stomachs of mice. Inoculation of the mice with Lactobacillus strains 100-23 and 20 led to the appearance of a new 16S fragment in the profiles of samples from mice 2, 3, 4, and 5 (fragment b in Fig. 1, lanes 3, 4, 5, and 6). This fragment migrates to the same position as the 16S V2-V3 fragment obtained from pure cultures of strains 100-23 and 20 (lanes 8 and 11). It also coincides with the 16S fragment generated from Lactobacillus reuteri DSM 20016T (fragment b in Fig. 1, lanes 1, 7, and 12). Similarly, inoculation of the mice with Lactobacillus strains 100-5 and 21 resulted in their detection in the stomach contents of animals 4 and 5 (fragment a in Fig. 1, lanes 5 and 6). A faint band at this location was also present in the preparation from mouse 3, but it is not visible in Fig. 1 (lane 4). The 16S fragments of these strains (fragment a in Fig. 1, lanes 9 and 10) coincide with that of Lactobacillus fermentum ATCC 14931T (fragment a in Fig. 1, lanes 1, 7, and 12).
FIG. 1.
Detection of Lactobacillus strains in stomach contents of mice. Lane 1, Lactobacillus identification ladder (from top to bottom, L. plantarum, L. johnsonii/L. gasseri, L. fermentum, L. agilis, L. sharpeae/L. brevis, L. acidophilus/L. crispatus/L. helveticus, L. delbrueckii subsp. bulgaricus, L. salivarius, L. ruminis, L. reuteri, L. casei group, L. vitulinus). The 16S V2-V3 rDNA fragment of L. fermentum ATCC 14931T is fragment a; that of L. reuteri DSM 20016T is fragment b. Lanes 2 to 6, 16S V2-V3 rDNA profiles from stomach contents from mouse 1 (lactobacilli absent) (lane 2); mouse 2, inoculated with strain 100-23 (lane 3); mouse 3, inoculated with strains 100-23 and 100-5 (lane 4); mouse 4, inoculated with strains 100-23, 100-5, and 21 (lane 5); and mouse 5, inoculated with strains 100-23, 100-5, 21, and 20 (lane 6). Lane 7, Lactobacillus identification ladder. Lanes 8 to 11, 16S V2-V3 rDNA fragments from pure cultures of strains 100-23, 100-5, 21, and 20, respectively. Lane 12, Lactobacillus identification ladder.
Identification ladder for lactobacilli.
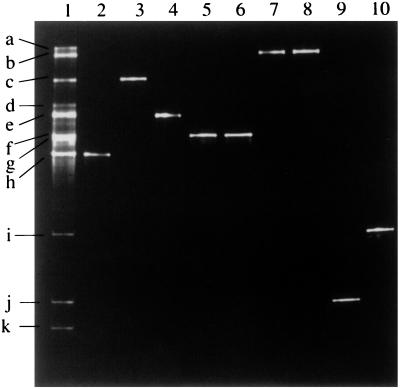
It was possible to at least group the Lactobacillus reference strains according to the migration of their 16S V2-V3 regions in a denaturing gradient gel (Fig. 2). Thus, the fragments from Lactobacillus johnsonii and Lactobacillus gasseri migrated the same distance in the gel (Fig. 2, lanes 2 and 3). DNA fragments from Lactobacillus acidophilus, Lactobacillus helveticus, and Lactobacillus crispatus (not shown in Fig. 2) had similar migration distances (Fig. 2, lanes 8 and 9), as did those of Lactobacillus salivarius subsp. salivarius and Lactobacillus salivarius subsp. salicinius (Fig. 2, lanes 11 and 12). The fragments from Lactobacillus casei, Lactobacillus casei/L. zeae, and Lactobacillus rhamnosus (Fig. 2, lanes 15, 16, and 17) and L. paracasei (not shown in Fig. 2) migrated the same distance in the gel. Fragments from Lactobacillus brevis and Lactobacillus sharpeae had almost identical migration properties (Fig. 2, lanes 6 and 7). DNA fragments from the remaining seven reference strains that we tested could be distinguished individually by DGGE (Fig. 2).
FIG. 2.
DGGE of 16S V2-V3 rDNA sequences from Lactobacillus reference strains with an 8% polyacrylamide 30 to 50% denaturing gradient gel. Lanes: 1, L. plantarum ATCC 14917T; 2, L. johnsonii ATCC 33200T; 3, L. gasseri ATCC 33323T; 4, L. fermentum ATCC 14931T; 5, L. agilis DSM 20509T; 6, L. sharpeae DSM 20505T; 7, L. brevis ATCC 14869T; 8, L. acidophilus ATCC 4356T; 9, L. helveticus ATCC 15009T; 10, L. delbrueckii subsp. bulgaricus ATCC 11842T; 11, L. salivarius subsp. salivarius ATCC 11741T; 12, L. salivarius subsp. salicinius ATCC 11741T; 13, L. ruminis ATCC 27780T; 14, L. reuteri DSM 20016T; 15, L. casei/L. zeae ATCC 393T; 16, L. rhamnosus ATCC 8530; 17, L. casei ATCC 4684; 18, L. vitulinus ATCC 27783T. The DNA fragment from L. crispatus ATCC 33820T (not run in this gel) migrated the same distance as those of L. acidophilus and L. helveticus. The L. paracasei subsp. paracasei ATCC 25302 fragment (not run in this gel) migrated the same distance as the fragments of L. casei, L. rhamnosus, and L. casei/L. zeae.
Identification of unknowns by DGGE.
Migration distances of 16S V2-V3 fragments obtained from unidentified isolates of lactobacilli were compared to those of reference strains in the identification ladder. Such comparisons enabled the unknowns to at least be placed in a Lactobacillus group, if not identified to the species level (examples are shown in Fig. 3; results are in Table 1).
FIG. 3.
DGGE of 16S V2-V3 rDNA sequences from gastrointestinal isolates of Lactobacillus with an 8% polyacrylamide 30 to 50% denaturing gradient gel. Lane 1, ladder of sequences from reference strains (a, L. plantarum; b, L. johnsonii/L. gasseri; c, L. fermentum; d, L. agilis; e, L. sharpeae/L. brevis; f, L. acidophilus/L. crispatus/L. helveticus; g, L. delbrueckii subsp. bulgaricus; h, L. salivarius; i, L. ruminis; j, L. reuteri; k, L. casei group); lane 2, GTH1; lane 3, GTH10; lane 4, GTH18; lane 5, GT3C1; lane 6, GTP5; lane 7, GTH5; lane 8, NCK800; lane 9, GT3S3; lane 10, L5.
Identification of unknowns by species-specific primers.
Our strategy was to first group or identify each isolate by DGGE and then specifically identify (or confirm) the species to which the isolate belonged by using species-specific primers. This meant that each isolate needed to be tested with only a few primer pairs and that extensive testing of the primers across all known Lactobacillus species was unnecessary (Table 3). Thus, isolates grouped as L. acidophilus, L. crispatus, or L. helveticus by DGGE were tested with primer pairs 1 and 2. L. gasseri and L. johnsonii isolates were tested with primer pairs 3 and 4. Isolates classified as belonging to the L. casei group were tested with primer pairs 6, 7, and 8. Isolates identified as L. reuteri or L. fermentum were tested with primer pairs 9 and 10. L. brevis and L. sharpeae isolates were tested with primer 11. The results obtained by using these primer pairs with reference strains are shown in Table 3, and identification results for unknowns are given in Table 1.
TABLE 3.
Specificity of primer pairs
| Species | PCR product obtained with primer paira
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| L. acidophilus ATCC 4356T | + | − | − | − | − | ||||||
| L. crispatus ATCC 33820T | − | + | − | − | − | ||||||
| L. gasseri ATCC 33323T | − | − | + | − | − | ||||||
| L. johnsonii ATCC 33200T | − | − | − | + | − | ||||||
| L. plantarum ATCC 14917T | − | − | − | − | + | ||||||
| L. casei ATCC 334 | + | − | − | ||||||||
| L. casei ATCC 4684 | + | − | − | ||||||||
| L. paracasei subsp. paracasei ATCC 25302 | + | − | − | ||||||||
| L. rhamnosus ATCC 8530 | − | − | + | ||||||||
| L. rhamnosus DSM20021T | − | − | + | ||||||||
| L. casei/L. zeae ATCC 393T | − | + | − | ||||||||
| L. zeae DSM 20178T | − | + | − | ||||||||
| L. brevis ATCC 14869T | − | − | − | ||||||||
| L. sharpeae DSM 20505T | − | − | + | ||||||||
| L. fermentum ATCC 14931T | − | + | |||||||||
| L. reuteri DSM 20016T | + | − | |||||||||
See Table 2 for primer details.
Comparison of V2-V3 sequences with those of lactobacilli in the GenBank database.
Identifications obtained by a BLAST search of the GenBank database with V2-V3 region sequences correlated with those derived by DGGE and species-specific PCR primers (Table 1). Thus, for each isolate, the group or species identification achieved by DGGE or species-specific PCR primers was confirmed by analysis of the V2-V3 sequence. The species-specific primers, however, gave definitive identifications of isolates belonging to L. rhamnosus or L. casei, whereas V2-V3 sequences did not differentiate between these species but indicated the group (L. casei/L. paracasei/L. rhamnosus/L. zeae) to which they belonged.
DISCUSSION
PCR-DGGE clearly has potential in the analysis of gastrointestinal communities. Using the defined system consisting of our unique lactobacillus-free mice, we were able to detect the addition of Lactobacillus species to the gastric microflora of the animals. The sensitivity of the method was limited to detection of different Lactobacillus species, rather than strains, as expected of a method based on 16S rRNA gene sequences. It will be interesting to apply this method to more complex communities, such as those inhabiting large-bowel ecosystems.
PCR-DGGE also proved a practical addition to available identification methods for lactobacilli of gastrointestinal origin. Multiple fragments of different sizes were sometimes present in PCR products from pure cultures (see Fig. 1, lanes 8 and 11, for examples). There was, however, always one major fragment (most dense in the DGGE gel) which indicated the identity of the isolate. The minor (less dense fragments) were probably PCR artifacts resulting from the highly folded (loops and stems) nature of the V2-V3 region (Turner et al., New Zealand Microbiol. Soc. Ann. Meet.). Lactobacillus ruminis, L. reuteri, L. fermentum, Lactobacillus vitulinus, and Lactobacillus agilis could be identified directly on the basis of the migration of their V2-V3 sequences. DNA fragments from Lactobacillus plantarum and Lactobacillus delbrueckii subsp. bulgaricus had characteristic migration behaviors, but we have not tested representative strains of all of the members of the L. plantarum and L. delbrueckii taxa.
DGGE results provided a useful initial screen for the remaining gastrointestinal species because it narrowed the possible identities of the isolates. These grouped isolates could then be identified by application of the species-specific PCR primers. This meant that an isolate needed to be tested only with two or three primer pairs to obtain a specific identification. We do not yet have species-specific primers for all of the gastrointestinal species of lactobacilli, but this would be a desirable goal for future research.
The species-specific PCR primers based on the 16S-23S rRNA intergenic spacer regions were particularly valuable in the identification of the L. casei group isolates. They permitted discrimination to be made between L. casei/L. paracasei, L. rhamnosus, and L. zeae. These species cannot be differentiated by DGGE or BLAST comparisons of V2-V3 sequences. The information that we have obtained using these species-specific primers may be helpful in unraveling the somewhat confused situation regarding the taxonomy of the L. casei group. Dellaglio and colleagues (2), on the basis of DNA-DNA homology studies, requested in 1991 that strain ATCC 334 replace ATCC 393 as the neotype strain of Lactobacillus casei subsp. casei. They rejected the species name L. paracasei. In 1996, Dicks et al. (3) proposed that strain ATCC 393 be reclassified as L. zeae. Subsequently, Mori and colleagues (9) proposed that the L. casei group be reclassified to include three species: L. zeae containing strains ATCC 15820T (DSM 20178) and ATCC 393, a species containing L. paracasei and ATCC 334, and L. rhamnosus. Our PCR primer pair 7, based on the 16S-23S spacer region sequences of ATCC 393 (which we have considered on the basis of the work of Dicks et al. [3] to be L. zeae), produced a PCR product with DNA from both ATCC 393 and ATCC 15820 (DSM 20178), but not with DNA from ATCC 334, L. paracasei ATCC 25302, or L. rhamnosus strains. PCR primer pair 6, based on the intergenic spacer region of ATCC 334, produced products only from L. casei and L. paracasei cultures. Primer pair 8, based on the intergenic spacer region of L. rhamnosus (15), produced PCR products only from L. rhamnosus cultures. The application of our primers to a collection of strains belonging to L. casei may therefore assist in future taxonomic considerations of this group. Additionally, it may be possible to differentiate between the members of this group by DGGE if primers that targeted another region of the 16S rRNA gene are used. A potentially useful region is located between nucleotides 73 and 111 (L. casei numbering), where variation in sequences has been observed among the members of the L. casei group (9).
Identifications obtained by a BLAST search of the GenBank database with V2-V3 region sequences correlated with those obtained by DGGE and species-specific PCR primers. We are therefore confident that the approaches to the detection and identification of Lactobacillus species that we have described in this report will contribute to future studies of the composition of the intestinal microflora and to a better understanding of Lactobacillus taxonomy.
ACKNOWLEDGMENTS
S. Rodtong was supported by a Teaching and Research Observation Fellowship from the Ministry of University Affairs, Thailand. Work conducted in Finland was aided by the Technology Development Centre of Finland, and A. Tilsala-Timisjarvi was the recipient of a grant from the Finnish Cultural Foundation. The support of the University of Otago Research Committee and the New Zealand Lottery Board is gratefully acknowledged. The mouse experiments were kindly supported by the Yakult Central Institute for Microbiological Research, Tokyo, Japan.
G. Tannock thanks the Akkerman group, University of Wageningen, Wageningen, The Netherlands, for the introduction to gradient gel electrophoresis. J. Walter thanks W. Hammes, University of Hohenheim, Stuttgart, Germany, for his support and encouragement.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Dellaglio F, Dicks L M T, du Toit M, Torriani S. Designation of ATCC 334 in place of ATCC 393 (NCDO 161) as the neotype strain of Lactobacillus casei subsp. casei and rejection of the name Lactobacillus paracasei (Collins et al., 1989). Request for an opinion. Int J Syst Bacteriol. 1991;41:340–342. [Google Scholar]
- 3.Dicks L M T, Du Plessis E M, Dellaglio F, Lauer E. Reclassification of Lactobacillus casei subsp. casei ATCC 393 and Lactobacillus rhamnosus ATCC 15820 as Lactobacillus zeae nom. rev., designation of ATCC 334 as the neotype of L. casei subsp. casei, and rejection of the name Lactobacillus paracasei. Int J Syst Bacteriol. 1996;46:337–340. doi: 10.1099/00207713-46-1-337. [DOI] [PubMed] [Google Scholar]
- 4.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franks A H, Harmsen H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldin B R, Gorbach S L. Probiotics for humans. In: Fuller R, editor. Probiotics. The scientific basis. London, United Kingdom: Chapman and Hall; 1992. pp. 355–376. [Google Scholar]
- 7.Hammes W P, Vogel R F. The genus Lactobacillus. In: Wood B J B, Holzapfel W H, editors. The lactic acid bacteria. 2. The genera of lactic acid bacteria. London, United Kingdom: Blackie Academic and Professional; 1995. pp. 19–54. [Google Scholar]
- 8.Kandler O, Weiss N. Regular, nonsporing gram-positive rods. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams and Wilkins; 1986. pp. 1208–1234. [Google Scholar]
- 9.Mori K, Yamazaki K, Ishiyama T, Katsumata M, Kobayashi K, Kawai Y, Inoue N, Shinano H. Comparative sequence analyses of the genes coding for 16S rRNA of Lactobacillus casei-related taxa. Int J Syst Bacteriol. 1997;47:54–57. doi: 10.1099/00207713-47-1-54. [DOI] [PubMed] [Google Scholar]
- 10.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 11.O'Sullivan D J. Methods of analysis of the intestinal microflora. In: Tannock G W, editor. Probiotics: a critical review. Wymondham, United Kingdom: Horizon Scientific Press; 1999. pp. 23–44. [Google Scholar]
- 12.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tannock G W. Normal microflora. An introduction to microbes inhabiting the human body. London, United Kingdom: Chapman and Hall; 1995. [Google Scholar]
- 14.Tannock G W, Crichton C, Welling G W, Koopman J P, Midtvedt T. Reconstitution of the gastrointestinal microflora of lactobacillus-free mice. Appl Environ Microbiol. 1988;54:2971–2975. doi: 10.1128/aem.54.12.2971-2975.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilsala-Timisjarvi A, Alatossava T. Development of oligonucleotide primers for the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int J Food Microbiol. 1997;35:49–56. doi: 10.1016/s0168-1605(97)88066-x. [DOI] [PubMed] [Google Scholar]
- 16.Zoetendal E, Akkermans A D, De Vos W M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]