Abstract
Cannabinoids are synthesized in glandular stalked trichomes on the female flowers of Cannabis sativa (cannabis). The regulation of glandular trichome development has not been characterized in cannabis. We recently identified an R2R3-MYB transcription factor, CsMIXTA, which could be involved in trichome morphogenesis in cannabis. Some homologous genes of CsMIXTA are known to function in glandular trichome initiation in other plant species. CsMIXTA is highly expressed in flower tissue compared to vegetative tissues. Interestingly, CsMIXTA is also highly expressed in trichomes isolated from female flower tissue. In addition, CsMIXTA is upregulated during the peak stages of female flower maturation in correlation with some cannabinoid biosynthetic genes. Transient expression in Nicotiana benthamiana showed that CsMIXTA is localized in the nucleus. Furthermore, yeast transcriptional activation assay demonstrated that CsMIXTA has transactivation activity. Overexpression of CsMIXTA in Nicotiana tabacum resulted in higher trichome density, larger trichome size, and more branching on stalked glandular trichomes. The results indicate that CsMIXTA not only promotes glandular trichome initiation in epidermal cells, but also regulates trichome development in tobacco leaves. In this report, we characterized the novel function of the first cannabis transcription factor that may be critical for glandular trichome morphogenesis.
Keywords: Cannabis sativa, glandular trichome morphogenesis, transcription factor, cannabinoid biosynthesis, female flower development
1. Introduction
Glandular trichomes are epidermal outgrowths characterized by the presence of a secretory cavity or “gland”, which houses secretory gland cells capable of synthesizing secondary metabolites and exporting them into a secretory reservoir [1,2]. Glandular stalked trichomes (GSTs) on cannabis female flowers emerge from an epidermal cell and develop multicellular stalks which are usually less than 500 µm long [3,4,5,6]. In cannabis, glandular trichomes contain a rosette of secretory cells that synthesize and export biosynthetic enzymes into the secretory reservoir [7,8]. The direct precursor of cannabinoids, cannabigerol (CBG), is synthesized in the plastid of the secretory gland cell, and then exported (by an unknown mechanism) into the secretory reservoir, where it is converted to tetrahydrocannabinolic acid (THCA), cannabidiolic acid (CBDA), or cannabichromenic acid (CBCA) by their respective synthases [7].
Current models indicate that the regulation of secondary metabolite production is associated with the regulation of flowering and trichome development [9,10]. After an epidermal cell receives a signal to initiate trichome morphogenesis, it develops into a sessile glandular trichome [1]. In concert with the maturation of the flower, sessile trichomes will develop into the bulbous phase and then eventually reach the capitate-stalked glandular (CSG) phase [11]. This is the mature phase at which cannabinoids are predominantly produced [10].
A complex of transcription factors (TFs) in the R2R3-MYB and HD-ZIP IV families has been shown to regulate the development of glandular trichomes in other plant species [11,12,13]. This complex includes MIXTA-like proteins, which interact with other HD-ZIP IV TFs to coordinate trichome development in concert with flower development [13]. Understanding the genetic regulation of the development of a glandular trichome from sessile to stalked is of particular importance for cannabis biology because it has been shown that cannabinoids are not produced in any considerable amount until the trichome reaches the CSG phase [10].
In cannabis, the genetic regulation of glandular trichome initiation and development is still unknown. In this study, we identified a novel R2R3-MYB TF in cannabis, CsMIXTA, which could positively regulate cannabis CSG development. We showed that CsMIXTA was highly expressed in flowers; CsMIXTA also showed higher expression in trichome cells isolated from female flower tissue. Ectopic expression of CsMIXTA in tobacco not only significantly increased glandular trichome size and density but also promoted trichome branching on tobacco leaves. Our study suggests that CsMIXTA is a TF that could potentially control glandular trichome initiation and development in female cannabis flowers.
2. Results
2.1. CsMIXTA Is Upregulated during Female Flower Development
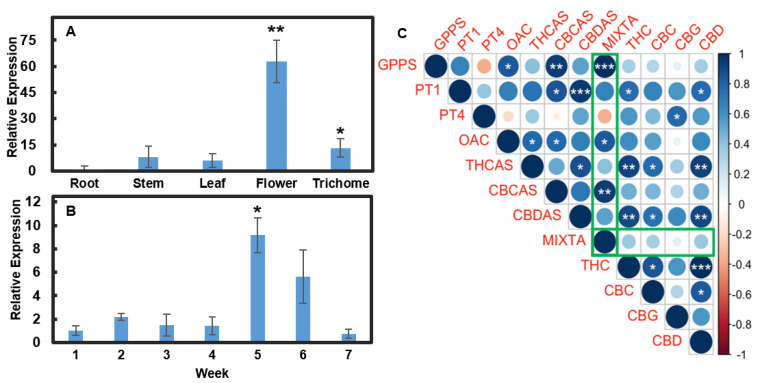
MIXTA has been reported to be involved in glandular trichome formation in various plant species, such as snapdragon and artemisia [12,14]. We searched the NCBI database using the known MIXTA sequences from snapdragon and artemisia to identify a homolog in Cannabis sativa. We named the cannabis gene CsMIXTA. We first determined CsMIXTA expression in different tissues and during female flower development in a hemp variety, Cherry Wine (CW). CsMIXTA is highly expressed in flower tissues compared to the vegetative tissues (Figure 1A). CsMIXTA is also expressed in stem and leaf tissues, where trichomes are also present. Interestingly, CsMIXTA was detected in isolated trichomes, suggesting that CsMIXTA may have distinct functions in processes other than trichome initiation. We further examined CsMIXTA expression during a period of 7-week flower development. The results showed that CsMIXTA was significantly upregulated in week 5 and reduced thereafter (Figure 1B). CsMIXTA showed a similar expression pattern to that of the enzymes in the cannabinoid biosynthetic pathway [15]. We used a Pearson’s correlation matrix test to evaluate the relationship (positive and negative) between expression of MIXTA and other genes involved in cannabinoid synthesis with expression of cannabinoids across the full seven weeks of flower development (Figure 1C). This analysis shows that CsMIXTA has a significant positive correlation with the expression of the two genes (Geranyl Pyrophosphate Synthase (GPPS) and Olivetolic Acid Cyclase (OAC)) required to generate the substrates (GPP and OA, respectively) necessary for the synthesis of the CBG, the first cannabinoid in the biosynthetic pathway. This analysis indicated that during flower development, CsMIXTA expression was also positively correlated with cannabinoid levels and expression of Cannabichromenic Acid Synthase (CBCAS) which generates the end product cannabinoid CBCa (Figure 1C). This analysis indicates that through coordinating trichome development, CsMIXTA may play an important role in cannabinoid biosynthesis.
Figure 1.
Expression analysis of CsMIXTA. (A) Relative expression of CsMIXTA in 5 different tissue types harvested from Cherry Wine. (B) Relative Expression of CsMIXTA over 7 weeks of flowering. Results ((A,B)) are shown as means ± SE (n = 4). Means separation between expression in various tissues compared to the level in roots (A) and at various times during flower development as compared to expression at week one (B) was evaluated using Student’s t-test; * indicates p < 0.05, and ** indicates p < 0.01. (C) Integrated expression analyses of CsMIXTA and genes encoding cannabinoid biosynthetic enzymes along with cannabinoid levels monitored over the 7-week flowering period were evaluated using Pearson’s correlation coefficient analyses. Green boxes highlight CsMIXTA. Blue indicates a positive correlation, while red indicates a negative correlation. Size and shade of circles represent strength of correlation. Figures were generated using the R corrplot package (https://github.com/taiyun/corrplot). (*, p < 0.05; **, p < 0.01; ***, p < 0.001.) Percentage data was arcsine transformed prior to statistical analysis.
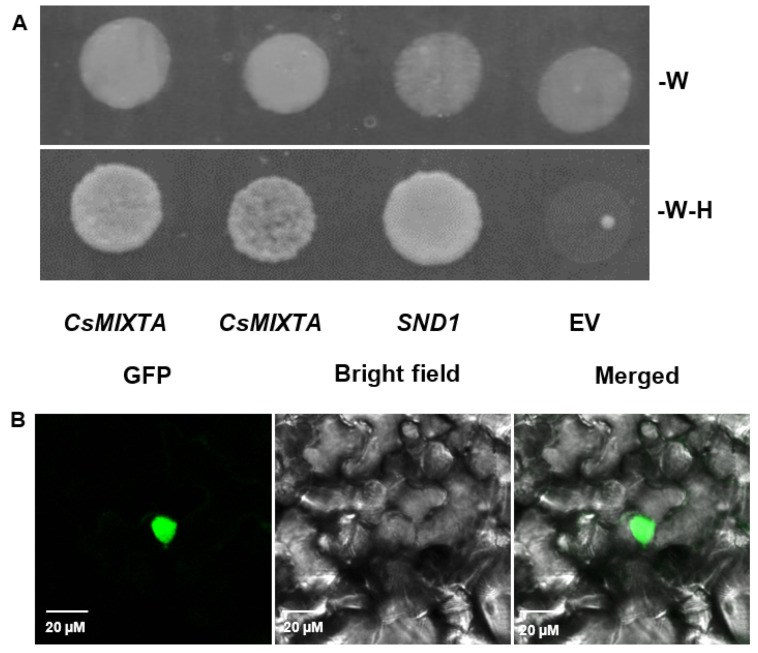
CsMIXTA belongs to the R2R3-MYB TF family and showed homology with the known R2R3-MYB and MIXTA-like TFs from other plant species. Using a yeast activation assay, we demonstrated that CsMIXTA is capable of transcriptional activation (Figure 2A). Transient expression of CsMIXTA in N. benthamiana leaves showed nuclear localization (Figure 2B). In some cases, we also observed plasma membrane localization of CsMIXTA (Figure S2).
Figure 2.
Yeast transactivation assay and subcellular localization of CsMIXTA. (A) Top: yeast cells grown on SD medium lacking W. All cells grew normally, including cells transformed with the empty vector (EV). Bottom: yeast cells grown on SD medium lacking tryptophan (W) and histidine (H). Cells expressing CsMIXTA produced healthy cultures, while pAS2 empty vector transformed culture did not. (B) N. benthamiana epidermal cells expressing a CsMIXTA-YFP fusion protein, observed using Nikon A1R Confocal microscope with excitation at 488 nm. YFP signal clearly indicates nuclear localization.
2.2. CsMIXTA Promotes Glandular Trichome Formation in Tobacco Leaves
Our work monitoring CsMIXTA expression in cannabis shows evidence of an associative relationship between CsMIXTA expression during the female flower development and cannabinoid synthesis, presumably mediated by CsMIXTA-mediated control of trichome development. We undertook further work to examine if there is a causal relationship between CsMIXTA expression and trichome development in a heterologous system. Cannabis is recalcitrant to genetic transformation; a few reports have been published recently [16,17,18], but their transformation efficiencies are not robust. In lieu of a reliable cannabis transformation protocol, we used a heterologous system to determine if there is a causal effect of CsMIXTA expression on trichome morphogenesis. Tobacco (Nicotiana tabacum), like cannabis, has glandular stalked trichomes but in the case of tobacco, GSTs are abundant on the leaves [19]. We ectopically expressed CsMIXTA (controlled by a constitutive (35S CaMV) promoter) in tobacco plants to evaluate if this cannabis TF could regulate glandular trichome initiation/development on tobacco leaves.
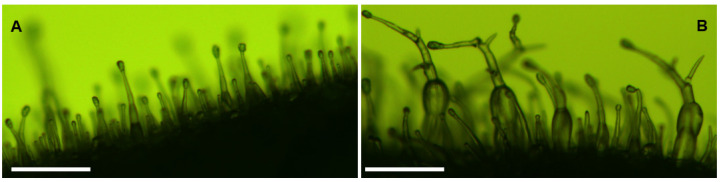
Figure 3 displays a side view of leaves from wt and a transgenic tobacco line, L1. An increase in trichome size and branching is clearly observed on the transgenic tobacco leaf. In addition, the upper leaf surface of the transgenic tobacco L1 is noticeably “hairier” than that of the wt, indicating a greater density of leaf trichomes produced in the CsMIXTA overexpressing tobacco line (Figure S3).
Figure 3.
Trichomes on wt (A) and transgenic (B) tobacco leaf edges visualized using a dissection microscope. Samples were viewed at 38× magnification. It is noticeable that the base of some trichomes has become enlarged and trichomes are exhibiting novel branching in the transgenic line (B). Scale bars: 200 µm.
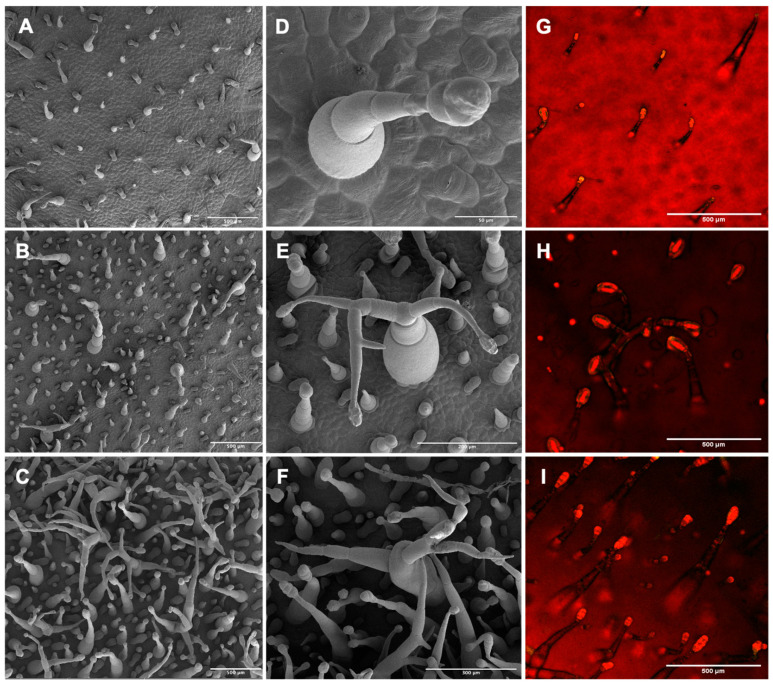
We further examined trichome morphology on tobacco leaves using SEM. Two transgenic lines (L1 and L3) were evaluated in this experiment (Figure 4). Both lines (Figure 4B,C) showed substantially higher trichome density compared to wt (Figure 4A). Typical wt trichomes possess only one gland and rarely branch [19]. However, we observed highly branched glandular stalked trichomes in both transgenic lines. In addition, trichomes in L1 showed higher density, size, and branching than L3, which could be due to the higher CsMIXTA expression in L1 (Figure S4). Close-up micrographs of representative highly branched glandular trichomes on the transgenic tobacco leaves (Figure 4E,F) and the regular wt glandular trichome are shown (Figure 4D). Floral organs of the transgenic tobacco plants were also investigated. Glandular trichomes are also formed on the abaxial side of the petal lobe (Figure S5). Anecdotal results showed that there were slightly more trichomes formed on the abaxial side of the petal lobe in the transgenic lines than those in wt, but no obvious structural difference was observed compared to what was seen in leaf glandular trichomes (Figure 3 and Figure 4).
Figure 4.
Scanning Electron Microscopy and Fluorescence Microscopy micrographs of wt and transgenic tobacco leaves. (A–C), SEM images of adaxial leaf surface of wt (A) and two transgenic lines L3 (B) and L1 (C). (D–F) Higher resolution micrographs of representative glandular trichomes from wt (D) and two transgenic lines L3 (E) and L1 (F). (G–I) Fluorescence Microscopy images of wt (G) and two transgenic lines L3 (H) and L1 (I) captured with a Nikon A1R confocal microscope. All samples were taken from tissue adjacent to the midrib of the third leaf from the apical meristem, and 2 cm from the petiole. In all cases, the images shown for each genotype are representative of other biological replicates. Fluorescence signals indicating glandular trichomes were counted from each tissue sample (n = 3). Scale bars in (A–C): 500 µm; in (D–F): 200 µm; in (G–H): 500 µm.
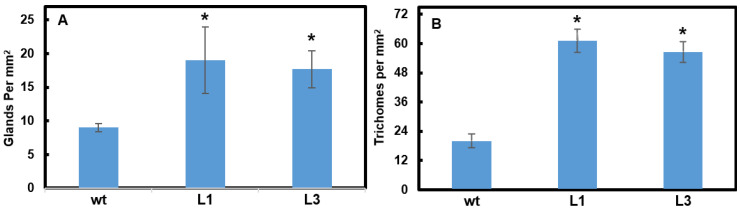
Fluorescence Microscopy was used to produce intrinsic fluorescence images of the trichomes at the leaf surface (Figure 4G–I). Glandular trichome heads filled with secondary metabolites show strong fluorescence: multiple studies have used this method to quantify trichome gland number [10,12,13]. It is clear that the trichome heads were much larger in the two transgenic lines than those in the wt. Trichome counting based on the images obtained from Fluorescence Microscopy and SEM further showed that trichome glands (those that emitted yellow fluorescence in Figure 4G–I) in transgenic tobacco significantly increased nearly twofold compared to wt (Figure 5A); the total trichomes on the leaves of the transgenic tobacco lines significantly increased threefold compared to the wt (Figure 5B). The results indicate that CsMIXTA plays an essential role in glandular trichome initiation and development. Our findings provide evidence that CsMIXTA also functions in CSG cells to modulate trichome growth and development, consistent with the higher CsMIXTA expression detected in isolated female flower glandular trichomes (Figure 1A).
Figure 5.
Tobacco transgenic lines overexpressing CsMIXTA generated more trichomes than wt. (A) Average number of glands on the glandular trichomes per mm2, observed by intrinsic fluorescence as shown in Figure 4G–I. (B) Average trichome number per mm2, obtained from SEM images in Figure 4A–C. Averages taken from 3 biological reps (n = 3) for each transgenic line and wt. * indicates p < 0.05, which was determined using Student t-test by comparing the two transgenic lines to wt.
3. Discussion
In this study we identified CsMIXTA, the first characterized cannabis TF that could potentially influence cannabis CSG development. CsMIXTA is highly expressed in the female cannabis flowers, with expression patterns that are coordinated with cannabinoid biosynthesis and the development of inflorescences. Phylogenetic analysis indicated that CsMIXTA is closely related to Arabidopsis thaliana AtMYB16 and Artemisia annua AaMIXTA (Figure S1). MIXTA-like proteins have been shown to coordinate cuticle deposition in tomato and Arabidopsis thaliana [20,21]. Modification of AaMIXTA expression altered the biosynthesis of wax and cutin monomers, components of the cuticle [12], and MIXTA-like proteins identified in liverworts have implicated MIXTA in the early evolution of cuticle formation and therefore the colonization of land by plants [22]. Because cutin is an important structural component of the trichome, CsMIXTA may coordinate the biosynthesis of this compound in the trichome.
Our previous study demonstrated that expression of cannabinoid biosynthetic genes peaked in weeks 4 or 5 [15]; CsMIXTA expression showed a similar pattern. It has been shown that trichomes produce most cannabinoids during the latter capitate-stalked glandular phase. Therefore, it is reasonable to propose that the development of the stalk cells also occurs around week 4 of floral development, in correlation with the expression of CsMIXTA.
There is a strong positive correlation between CsMIXTA expression and the expression of both GPPS and OAC, enzymes which synthesize the necessary precursors (GPP and olivetolic acid, respectively) [15] for both cannabinoids and, in the case of GPPS, monoterpenes. Sessile (non-stalked) glandular trichomes produce minimal monoterpenes or cannabinoids before maturation, possessing a sesquiterpene-dominant chemotype. However, after the sessile trichomes develop into CSGs, cannabinoid and monoterpene biosynthesis takes place. Previous studies have shown that GPPS was more highly expressed in stalked trichomes, but OAC expression was similar among different glandular trichome types [10].
The heterologous overexpression of CsMIXTA in tobacco has shown a significant impact on trichome development with increased density, enlarged size and more branching (Figure 4). The overexpression of CsMIXTA in tobacco showed more distinct trichome phenotypes than that of AmMIXTA overexpression [14,23]. The overexpression of known MIXTA or MIXTA-like genes did not show a similar glandular trichome phenotype caused by overexpression of CsMIXTA, indicating that CsMIXTA may have unique functions in glandular trichome morphogenesis in cannabis. Figure 5 quantifies a modest (twofold) increase in trichome glands which can be seen in the Fluorescence Microscopy images (Figure 4G–I). What cannot be seen in the fluorescence images, however, is the forest of trichomes and trichome branches, which becomes evident in the SEM photos (Figure 4A–C). The increase in trichome density (threefold) as well as trichome branching indicates that CsMIXTA may also function to regulate the initiation and development of trichome types other than glandular stalked trichomes. According to these findings, genetic engineering of CsMIXTA may have the potential to promote cannabinoid production by increasing glandular trichome formation.
Our data provide evidence associating CsMIXTA with trichome development in cannabis. However, a definitive causal relationship between CsMIXTA expression, glandular trichome development, and cannabinoid production in cannabis is, at this point, beyond experimental verification due to the recalcitrance of cannabis concerning stable transformation. We also note that our work does not delineate the specific molecular steps in a CsMIXTA signaling cascade in cannabis. Future research is needed that might focus on the cell-to-cell signaling pathways by which glandular trichome development is generally coordinated, and on proteins and promoter elements which interact with CsMIXTA in C. sativa.
4. Materials and Methods
4.1. Cannabis Plant Growth
A hemp variety, Cherry Wine, was used in this study. Plants were acquired using cuttings from mother plants in our research greenhouses. Cuttings were treated with Hormodin powder and stuck in rockwool cubes soaked with 20 mL/L Clonex Nutrient Solution (Growth Technologies Ltd., Taunton, UK). Cuttings were allowed to root for 3 weeks before transplant. All plants were grown in #600 standard nursery pots filled with Promix-BX25 (Premier Tech Horticulture, Quakertown, PA, USA) soilless medium and Osmocote 15-9-12 (Scott’s Miracle-Gro, Marysville, OH, USA). Plants were grown under 16 h light/8 h dark conditions with supplemental lighting from high-pressure sodium lighting. The light intensity from the high-pressure sodium lights is 568 µmol m–2 s–1 nm−1. Photoperiod was changed to 12 h light/12 h dark using blackout curtains. Jack’s nutrient solution fed via drip fertigation.
4.2. Isolation of RNA and cDNA Synthesis
Different tissue samples were collected in the eighth week of flowering. Amounts totaling 100 mg of plant tissues were collected and immediately frozen in liquid nitrogen. The NucleoSpin Plant and Fungi RNA Isolation Kit (Macherey-Nagel) was used for RNA isolation according to manufacturer’s manual. cDNA was synthesized from 1 µg RNA using the iScript Reverse Transcriptase Master Mix (BioRad).
4.3. Isolation of Cannabis Trichomes
Trichome isolation was performed following the protocol developed by Livingston et al. (2020) [10] with slight modifications. Isolation buffer was made as described by Livingston et al. but excluding Amberlite. Approximately 3 g of fresh inflorescence tissue was used for trichome isolation. After isolation, the tube containing approximately 100–300 mg of trichome tissue enriched with glands was then frozen at −80 °C until RNA isolation.
4.4. Analysis of CsMIXTA Expression Using qPCR
qPCR analysis was performed using Bio-Rad CFX. iTaq Universal Sybr Green Master Mix was used (Bio-Rad). For all qPCR reactions, CsUbiquitin was used as the internal reference [24]. The CsMIXTA Primers were forward 5′-TCCATGCTTTACTAGGCAACAG-3′, reverse 5′-CCACCGTCTTGTTGAGAGAG-3′. Experiments were performed with four biological replicates.
4.5. Molecular Cloning of CsMIXTA
CsMIXTA was cloned out of cDNA using iProof HF Master Mix (Bio-Rad). The primers for cloning were forward 5′-CAGTCGACTGGATCCGGTACCATGGGTCGGTCACCATGCTG-3′, reverse 5′-GAAAGCTGGGTCTAGATATCTCGAGAACATAGGAGAATCTG-3′. NEBuilder HiFi DNA Assembly kit (New England Biolabs) was used for the cloning of CsMIXTA into KpnI and XhoI digested pENTR-3C vector (Invitrogen, Waltham, MA, USA). LR reaction was then performed to clone CsMIXTA into the pB7YWG2 binary vector with a 35S CaMV promoter using LR Clonase II (Invitrogen), as well as the pAS2 and pUBC vectors.
4.6. Agrobacterium Mediated Transformation of Nicotiana tabacum
N. tabacum (tobacco) seeds were acquired from Dr. Yi Li at the University of Connecticut. Seeds were sterilized using 3% bleach and 70% ethanol and planted in sterile magenta boxes containing 50 mL full strength Murashige and Skoog (MS) salts (Caisson Labs, Smithfield, UT, USA), MES (adjusted to pH 5.7 with Tris), 1% sucrose, and 1% agar. Seedlings were grown under fluorescent lighting in a growth chamber at 28 °C with a 16-h photoperiod. Leaves from 3-week-old sterile tobacco plants were used for transformation by Agrobacterium tumefacience GV3101 harboring pB7YWG2-CsMIXTA. Tobacco leaf disc transformation was performed as described previously (Zheng et al., 2007). Rooted transformants were transferred to Promix-BX amended with 12 g/gal Osmocote 15-9-12 and maintained in the greenhouse with 16 h photoperiod.
4.7. Yeast Transcriptional Activation Assay
CsMIXTA was fused with the GAL4-BD domain in the pAS2 vector using LR Clonase II (Invitrogen). The resulting plasmid was transformed into yeast strain AH109 (Clontech, Mountain View, CA, USA) using the Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA, USA). A known Arabidopsis TF, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 1 (SND1) (provided by Dr. Huanzhong Wang), was used as a positive control and empty vector as a negative control. The transformed cells were plated on synthetic defined (SD) media to select positive transformants. A 4 μL amount of yeast culture harboring corresponding constructs was dropped onto a solid SD medium with or without histidine to check for activation of reporter genes. Yeast growth was observed and documented after cultivation at 28 °C for 2 days.
4.8. Transient Expression of CsMIXTA in N. benthamiana Leaves
CsMIXTA was fused to YFP using a ubiquitin promoter-containing plasmid (pUBC-YFP) and LR Clonase II (Invitrogen), and the resulting construct was transformed into Agrobacterium tumefacience GV3101. Transient transfection protocol was performed as described by Espinoza-Patharkar et al., modified from [25].
4.9. Scanning Electron Microscopy
Squares of tobacco leaf tissue 3 mm in size were excised with midribs and the veins were removed. These squares were fixed for 24 h in formaldehyde-acetic acid-ethanol (10% formaldehyde, 5% acetic acid, 50% ethanol, 35% MilliQ H2O). These samples were then transferred to a 70% ethanol solution twice, and then dehydrated through a graded ethanol series. Samples were collected and placed into stainless steel containers for critical point drying while completely submerged in 100% ethanol. Dehydrated and dried samples were mounted onto SEM stubs using double-sided carbon tape, and sputter coated with gold nanoparticles. The samples were mounted into a Nova Nanosem 450 and imaged.
4.10. Fluorescence and Confocal Microscopy
Samples were mounted between two cover slips and viewed using a Nikon A1R confocal microscope through a 10× Plan Apo lens. Both channels were excited at 488 nm. Emissions were collected with a GFP filter (488 nm) as well as a red filter (700 nm). Z-stacks were collected at a step size of 27 microns. Composite channel/stack images/scale bars were produced in ImageJ. Trichomes were counted in photographs using “cell counter” in ImageJ.
5. Conclusions
Overexpression of the cannabis TF CsMIXTA in tobacco resulted in an increase in trichome density, size, and branching on the leaves of multiple transgenic lines, demonstrating that CsMIXTA is involved in trichome initiation and development. In cannabis, CsMIXTA is strongly expressed in inflorescence tissue and has an expression pattern which coordinates with key cannabinoid biosynthesis genes during female flower development. Through analyzing cannabis gene expression data, we established an association between the expression of CsMIXTA and cannabinoid biosynthetic genes as well as flower/trichome development. CsMIXTA is also highly expressed in the isolated trichome, suggesting a function within the trichome. CsMIXTA expression has a significantly positive correlation with the expression of GPPS and OAC: two key enzymes in the cannabinoid biosynthesis pathway. These data suggest that CsMIXTA not only regulates glandular trichome morphogenesis, but also cannabinoid and/or cutin biosynthesis. Together, these data create a strong case to support the hypothesis that CsMIXTA is a TF involved in the coordination of CSG development in female cannabis flowers; increasing CsMIXTA expression in cannabis could potentially enhance cannabinoid production in female flowers.
6. Patents
A provisional patent application has been submitted based on the findings in this report.
Acknowledgments
We thank Frederick Pettit and Shelly Durocher for their assistant in maintaining cannabis and tobacco plants in the greenhouse. We thank Yi Li at the University of Connecticut for providing us tobacco seeds. We thank Huan-zhong Wang at the University of Connecticut for providing us the pGBKT7-SND1 vector and the AH109 yeast strain.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11111519/s1, Figure S1: Phylogenetic tree of MIXTA and MIXTA-like genes; Figure S2: Confocal microscopy showing CsMIXTA localization; Figure S3: photographic comparison of upper leaf surface in WT and transgenic tobacco (line 1); Figure S4: qPCR results showing CsMIXTA expression in Transgenic lines 1, 3, and 6 vs. WT; Figure S5: SEM micrographs of trichomes on the abaxial side of the petal lobe in WT and transgenic lines 1 and 3.
Author Contributions
S.R.H., Y.M. and G.A.B. designed the research; S.R.H. performed experiments and analyzed data; P.V.A. generated Pearson’s matrix; S.R.H., Y.M. and G.A.B. wrote the manuscript; G.A.B. and Y.M. supervised the research. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Accession Numbers
The accession numbers for proteins in the phylogenetic tree are: CsMIXTA, XP_030485058.1; CsMIXTA2, XP_030481714.1; AaMIXTA1, ALD84252.1; AtMYB16, NP_197035.1; AtMYB106, NP_186763.2; AtMYB17, OAP04938.1; NtMYB39, XP_016468968.1; AmMIXTA1, CAA55725.1; AmMYBML1, CAB43399.1; GhMYB25, AAK19616.1.
Funding Statement
This work was supported by funding from Fine Fettle LLC (G.A.B.), and award 2022-67014-37049 from the Foundational Knowledge of Plant Products program at the USDA National Institute of Food and Agriculture (Y.M. and G.A.B), and Hatch project CONS01027 (G.A.B.).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huchelmann A., Boutry M., Hachez C. Plant Glandular Trichomes: Natural Cell Factories of High Biotechnological Interest. Plant Physiol. 2017;175:6–22. doi: 10.1104/pp.17.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuurink R., Tissier A. Glandular Trichomes: Micro-organs with Model Status? New Phytol. 2020;225:2251–2266. doi: 10.1111/nph.16283. [DOI] [PubMed] [Google Scholar]
- 3.Raman V., Lata H., Chandra S., Khan I.A., ElSohly M.A. Morpho-Anatomy of Marijuana (Cannabis sativa L.) In: Chandra S., Lata H., ElSohly M.A., editors. Cannabis sativa L.—Botany and Biotechnology. Springer International Publishing; Cham, Switzerland: 2017. pp. 123–136. [Google Scholar]
- 4.Dayanandan P., Kaufman P.B. Trichomes of Cannabis sativa L. (Cannabaceae) Am. J. Bot. 1976;63:578–591. doi: 10.1002/j.1537-2197.1976.tb11846.x. [DOI] [Google Scholar]
- 5.Hammond C.T., Mahlberg P.G. Morphology of glandular hairs of Cannabis sativa from scanning electron microscopy. Am. J. Bot. 1973;60:524–528. doi: 10.1002/j.1537-2197.1973.tb05953.x. [DOI] [Google Scholar]
- 6.Hammond C.T., Mahlberg P.G. Morphogenesis of capitate glandular hairs of Cannabis sativa (Cannabaceae) Am. J. Bot. 1977;64:1023–1031. doi: 10.1002/j.1537-2197.1977.tb11948.x. [DOI] [Google Scholar]
- 7.Gülck T., Møller B.L. Phytocannabinoids: Origins and Biosynthesis. Trends Plant Sci. 2020;25:985–1004. doi: 10.1016/j.tplants.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Kim E.S., Mahlberg P.G. Secretory Vesicle Formation in the Secretory Cavity of Glandular Trichomes of Cannabis sativa L. (Cannabaceae) Mol. Cells. 2003;15:387–395. [PubMed] [Google Scholar]
- 9.Matías-Hernández L., Aguilar-Jaramillo A.E., Cigliano R.A., Sanseverino W., Pelaz S. Flowering and Trichome Development Share Hormonal and Transcription Factor Regulation. J. Exp. Bot. 2016;67:1209–1219. doi: 10.1093/jxb/erv534. [DOI] [PubMed] [Google Scholar]
- 10.Livingston S.J., Quilichini T.D., Booth J.K., Wong D.C.J., Rensing K.H., Laflamme-Yonkman J., Castellarin S.D., Bohlmann J., Page J.E., Samuels A.L. Cannabis Glandular Trichomes Alter Morphology and Metabolite Content during Flower Maturation. Plant J. 2020;101:37–56. doi: 10.1111/tpj.14516. [DOI] [PubMed] [Google Scholar]
- 11.Chalvin C., Drevensek S., Dron M., Bendahmane A., Boualem A. Genetic Control of Glandular Trichome Development. Trends Plant Sci. 2020;25:477–487. doi: 10.1016/j.tplants.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Shi P., Fu X., Shen Q., Liu M., Pan Q., Tang Y., Jiang W., Lv Z., Yan T., Ma Y., et al. The Roles of AaMIXTA1 in Regulating the Initiation of Glandular Trichomes and Cuticle Biosynthesis in Artemisia annua. New Phytol. 2018;217:261–276. doi: 10.1111/nph.14789. [DOI] [PubMed] [Google Scholar]
- 13.Yan T., Li L., Xie L., Chen M., Shen Q., Pan Q., Fu X., Shi P., Tang Y., Huang H., et al. A Novel HD-ZIP IV/MIXTA Complex Promotes Glandular Trichome Initiation and Cuticle Development in Artemisia annua. New Phytol. 2018;218:567–578. doi: 10.1111/nph.15005. [DOI] [PubMed] [Google Scholar]
- 14.Glover B.J., Perez-Rodriguez M., Martin C. Development of Several Epidermal Cell Types Can Be Specified by the Same MYB-Related Plant Transcription Factor. Development. 1998;125:3497–3508. doi: 10.1242/dev.125.17.3497. [DOI] [PubMed] [Google Scholar]
- 15.Apicella P.V., Sands L.B., Ma Y., Berkowitz G.A. Delineating the Genetic Regulation of Cannabinoid Biosynthesis during Female Flower Development in Cannabis sativa. Plant Direct. 2022. accepted . [DOI] [PMC free article] [PubMed]
- 16.Deguchi M., Bogush D., Weeden H., Spuhler Z., Potlakayala S., Kondo T., Zhang Z.J., Rudrabhatla S. Establishment and Optimization of a Hemp (Cannabis sativa L.) Agroinfiltration System for Gene Expression and Silencing Studies. Sci. Rep. 2020;10:3504. doi: 10.1038/s41598-020-60323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galán-Ávila A., Gramazio P., Ron M., Prohens J., Herraiz F.J. A Novel and Rapid Method for Agrobacterium-Mediated Production of Stably Transformed Cannabis sativa L. Plants. Ind. Crops Prod. 2021;170:113691. doi: 10.1016/j.indcrop.2021.113691. [DOI] [Google Scholar]
- 18.Zhang X., Xu G., Cheng C., Lei L., Sun J., Xu Y., Deng C., Dai Z., Yang Z., Chen X., et al. Establishment of an Agrobacterium -mediated Genetic Transformation and CRISPR/Cas9-mediated Targeted Mutagenesis in Hemp (Cannabis Sativa L.) Plant Biotechnol. J. 2021;19:1979–1987. doi: 10.1111/pbi.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uzelac B., Stojičić D., Budimir S. Glandular Trichomes on the Leaves of Nicotiana tabacum: Morphology, Developmental Ultrastructure, and Secondary Metabolites. In: Ramawat K.G., Ekiert H.M., Goyal S., editors. Plant Cell and Tissue Differentiation and Secondary Metabolites. Springer International Publishing; Cham, Switzerland: 2021. pp. 25–61. (Reference Series in Phytochemistry). [Google Scholar]
- 20.Lashbrooke J.G., Adato A., Lotan O., Alkan N., Tsimbalist T., Rechav K., Fernandez Moreno J.-P., Widemann E., Grausem B., Pinot F., et al. The Tomato MIXTA-like Transcription Factor Coordinates Fruit Epidermis Conical Cell Development and Cuticular Lipid Biosynthesis and Assembly. Plant Physiol. 2015;169:2553–2571. doi: 10.1104/pp.15.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oshima Y., Shikata M., Koyama T., Ohtsubo N., Mitsuda N., Ohme-Takagi M. MIXTA-Like Transcription Factors and WAX INDUCER1/SHINE1 Coordinately Regulate Cuticle Development in Arabidopsis and Torenia Fournieri. Plant Cell. 2013;25:1609–1624. doi: 10.1105/tpc.113.110783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu B., Taylor L., Pucker B., Feng T., Glover B.J., Brockington S.F. The Land Plant-specific MIXTA-MYB Lineage Is Implicated in the Early Evolution of the Plant Cuticle and the Colonization of Land. New Phytol. 2021;229:2324–2338. doi: 10.1111/nph.16997. [DOI] [PubMed] [Google Scholar]
- 23.Payne T., Clement J., Arnold D., Lloyd A. Heterologous Myb Genes Distinct from GL1 Enhance Trichome Production When Overexpressed in Nicotiana Tabacum. Development. 1999;126:671–682. doi: 10.1242/dev.126.4.671. [DOI] [PubMed] [Google Scholar]
- 24.Guo R., Guo H., Zhang Q., Guo M., Xu Y., Zeng M., Lv P., Chen X., Yang M. Evaluation of Reference Genes for RT-QPCR Analysis in Wild and Cultivated Cannabis. Biosci. Biotechnol. Biochem. 2018;82:1902–1910. doi: 10.1080/09168451.2018.1506253. [DOI] [PubMed] [Google Scholar]
- 25.Voinnet O., Rivas S., Mestre P., Baulcombe D. Retracted: An Enhanced Transient Expression System in Plants Based on Suppression of Gene Silencing by the P19 Protein of Tomato Bushy Stunt Virus: An Enhanced Transient Expression System in N. Benthamiana. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.