Abstract
One of the most common bacterially mediated diarrheal infections is caused by enterotoxigenic Escherichia coli (ETEC) strains. ETEC-derived plasmids are responsible for the distribution of the genes encoding the main toxins, namely, the heat-labile and heat-stable enterotoxins. The origins and transfer modes (intra- or interplasmid) of the toxin-encoding genes have not been characterized in detail. In this study, we investigated the DNA regions located near the heat-labile enterotoxin-encoding genes (eltAB) of several clinical isolates. It was found that the eltAB region is flanked by conserved 236- and 280-bp regions, followed by highly variable DNA sequences which consist mainly of partial insertion sequence (IS) elements. Furthermore, we demonstrated that rearrangements of the eltAB region of one particular isolate, which harbors an IS91R sequence next to eltAB, could be produced by a recA-independent but IS91 sequence-dependent mechanism. Possible mechanisms of dissemination of IS element-associated enterotoxin-encoding genes are discussed.
Enterotoxigenic Escherichia coli (ETEC) infections are the major cause of bacterium-associated diarrheal diseases in developing countries (5, 15) and among travelers (4, 37). They are the subject of development of novel vaccines (19, 45). The main virulence determinants of ETEC strains are heat-labile enterotoxin (LT) and heat-stable enterotoxin (ST) (2, 40) and specific colonization factors (CFs) (13). ETEC strains colonize the small intestine and encode more than 20 different CFs (13). Indeed, combinations of CFs together with either ST or ST and LT in ETEC strains are the main risk factors for acquisition of acute ETEC-associated diarrheal diseases (14, 26).
About one-third of the clinically relevant ETEC strains express both LT and ST, whereas the remaining two-thirds express either LT or ST (13). LT and ST can be encoded together or separately on large, variable plasmids called Ent plasmids (42), along with CFs, antibiotic resistance markers, and conjugation systems (11, 41).
Genes encoding LT (8) and cholera toxin (27) presumably have a common ancestor, since considerable amino acid and DNA sequence homologies are apparent (8). It has also been proposed that the LT genes are foreign genes which were acquired by horizontal gene transfer to form an enteropathogen (33, 50). The activities and structures of LT and cholera toxin are nearly identical. Both toxins consist of two subunits, the catalytically active subunit A and the receptor domain subunit B. The toxic activity of LT is caused by the catalytic activity of subunit A, which is able to catalyze the ADP-ribosylation of protein Gsα in eukaryotic cells. This in turn constitutively induces adenylate cyclase to produce elevated intracellular cyclic AMP concentrations (12).
LT-encoding ETEC strains can be isolated from humans (LTh or LT-I) and animals (porcine LT [LTp] or LT-II) (2, 40). Although the overall similarity is high, some distinct differences at the DNA sequence level were observed (21, 47). The authors concluded that there might be little or no plasmid transfer between the two host systems. Instead, the coencoded colonization factors are probably responsible for this tropism, since they are host specific, allowing the colonization of either the porcine or the human small intestine (13).
Regarding the mechanism of dissemination of LT-encoding genes, a particular Ent plasmid carrying two copies of elt genes was characterized (33). This plasmid contains elt genes of different origins which are flanked by partial IS600 elements and a nearby complete IS3411 sequence. In subsequent analyses it was shown that transposition activity did not result in a movement of the elt genes. Instead, the authors suggested the participation of a putative former IS600-based composite transposon to be responsible for LT transmission. They further showed that the isolated Ent plasmid contained two origins of replication, suggesting that this particular plasmid represents a cointegrate form of two former Ent plasmids.
The recent discovery of cholera toxin genes encoded by the filamentous phage CTX⊘ (48) and the work of Murphy and Dallas (33) prompted us to review the mobility of ETEC-derived Ent plasmids, with particular focus on the identification of dissemination routes. The flanking DNA regions of eight different Ent plasmids of clinical ETEC isolates were analyzed. The results show that the encoding elt genes are embedded within two highly conserved regions of 236 and 280 bp. These conserved regions are then followed by partial insertion sequence (IS) types of about five different IS elements. In particular, we investigated the dissemination processes of a Ent plasmid carrying a partial IS91R-associated elt gene cluster.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown on Luria broth medium at 37°C under aerobic conditions. Plasmids pACYC184 (36) and pCVD442 (9) were used as control and recipient plasmids, respectively, for the construction of pJBS620. In the growth medium the following antibiotics were used: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 12.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant characteristics | Toxin status | Source or reference |
|---|---|---|---|
| Human ETEC isolates | |||
| S1-008C | O6:H− | LT+ ST+ | P. Echeverria |
| S2-018B | O78:H− | LT+ ST+ | P. Echeverria |
| S6-044A | O25:H− | LT+ ST− | P. Echeverria |
| 102-34344F | NDa | LT+ ST+ | J. Hacker |
| K1 164/82 | O148:H28 | LT+ ST− | H. Karch |
| K2 297/87 | O25:H42 | LT+ ST− | H. Karch |
| K3 G1253 | O147:H19:K88 | LT+ ST− | H. Karch |
| K4 117/86 | O6:H− | LT+ ST+ | H. Karch |
| Porcine ETEC isolate K5 284/97 | ND | LT+ ST− | H. Karch |
| E. coli K-12 laboratory strains | |||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR | 6 | |
| MC4100 recA | F−araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR recA::kan | J. J. Mekalanos | |
| MC4100 λpir | F−araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR λpir | 10 | |
| MC4100 λpir recA | MC4100 λpir recA::kan | This work | |
| XL1-Blue | F′::Tn10 proA+B+ lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17(rK− mK+) supE44 relA1 lac | New England Biolabs | |
| Plasmids | |||
| pACYC184 | Rep(p15A) Cmr Tetr | 7 | |
| pSU2600 | Rep(p15A) IS91 Cmr | 28 | |
| pCVD442 | Rep(R6K) mobRP4 sacB Apr | 9 | |
| pJBS620 | pCVD442 IS91R eltAB Apr | This work |
ND, not determined.
Genetic methods.
Plasmid DNA preparations were carried out according to the Qiagen kit protocol. Cloning and restriction analysis were done by procedures described by Maniatis et al. (24).
PCR amplification of the IS91R eltAB- and eltAB-carrying DNA fragment was performed with the Elongase kit protocol (Gibco BRL-Life Technologies) and thermal DNA cycler protocol (MWG-Biotech GmbH, Ebersberg, Germany), based on the method of Mullis and Faloona (32). The specific oligomers (MWG-Biotech GmbH) used for PCR are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′→3′) | Purpose |
|---|---|---|
| ETAseq | TCAGCACGGTATAATTTGTCGCCA | Sequencing of flanking regions of eltAB |
| ETA2seq | AAGCGATAAAGGAAGAGTGC | |
| ETBseq | GCCATTGAAAGGATGAAGGACACA | |
| IS91L | TGTCATTGTCTTTCAGGTAGTT | PCR amplification of the IS91 element |
| IS91R | TTTTTCTCACCGTCTCCGTTAT | |
| IS91R2 | TTGTCGACCATATGAGCGGAAA | |
| ETA1 | CCGGATTGTCTTCTTGTATGATA | PCR amplification of the enterotoxin genes eltAB |
| ETB2 | GGTCTCGGTCAGATATGTGATTC | |
| S6LSalI | TGAAAAACCAGTCGACACCATTGTCTATCG | PCR amplification of the IS91R eltAB fragment from isolate S6 |
| S6RSphI | TTCAAATGCAGCATGCGGCTCATTA | |
| Cat5′ | AACTGCAGTACGTAGCACCTCAAAAACACCATCATACAC | Detection of cointegrate formation together with ETB2 by PCR |
| Cat3′ | AATACGTACTGCAGCAGGCGTTTAAGGGCACCAATAACT | Detection of cointegrate formation together with ETB2 by PCR |
Southern blot analysis was performed as described by the manufacturer (Amersham Life Science) and according to the method of Southern (44). DNA was cut with appropriate restriction enzymes and separated on an 0.7% agarose gel. DNA was then transferred onto a nylon membrane (Amersham Life Science). By using specifically labeled LT or IS91 probe DNA, detection of hybridizing fragments was done by the ECL protocol (Amersham Life Science).
DNA sequencing.
DNA sequences were determined by the dideoxy nucleotide chain termination method of Sanger et al. (38). The sequence reactions were performed with the PCR cycling reaction (Amersham Life Science). The sequencing and detection were done with an infrared dye-labeled primer (IRD41) and monitored by the automatic sequencing method of the LiCor system (MWG-Biotech GmbH). The sequencing primers used are listed in Table 2.
Construction of pJBS620.
The suicide plasmid pCVD442 (9) served as a recipient plasmid for cloning of the IS91R eltAB fragment from an Ent plasmid of human ETEC isolate S6. The primers S6LSalI and S6RSphI (Table 2) were used to amplify by PCR a 1.9-kb fragment comprising the 391 bp of the IS91 right end together with the complete eltAB sequences. This IS91R eltAB fragment with engineered SalI and SphI restriction sites at the fragment ends was digested with SalI and SphI and then ligated into the SalI/SphI-opened pCVD442 plasmid, resulting in pJBS620.
Generation of E. coli MC4100 λpir recA::Kan.
E. coli MC4100 λpir recA::Kan was constructed via P1 transduction (30). P1 infection of strain MC4100 recA::Kan resulted in a P1 phage lysate, which was used for transduction of E. coli MC4100 λpir. Transductants were selected for kanamycin resistance and tested for UV sensitivity, as described by Maniatis et al. (24), by using the Stratalinker UV-crosslinker 1800 (Stratagene, La Jolla, Calif.). The obtained transductants (MC4100 λpir recA::Kan) were then used as the recipient strain for recombination experiments with pSU2600 and pJBS620.
Recombination assays and identification of joint plasmids.
Strains MC4100 λpir recA::Kan and MC4100 λpir were first transformed with pJBS620, isolated, purified, and subsequently transformed with pSU2600. Double transformants were then plated onto Luria broth agar supplemented with antibiotics chloramphenicol and ampicillin, and 100 colonies each were purified under the same selecting conditions. After growth these cells were pooled and plasmid DNA was prepared. Subsequently, the plasmid DNA was retransformed into MC4100 recA::Kan and transformants were selected for Cmr and Apr. Fifty isolates each, originally derived from a recA+ strain and a recA mutant strain, were further subjected to PCR analysis. The plasmid-specific oligonucleotides ETB2 (pJBS620) and Cat5′ or Cat3′ (pSU2600) (Table 2) were used to screen for cointegrate formation.
Nucleotide sequence accession numbers.
DNA sequences have been deposited in GenBank: the accession numbers for the A region are AF190920 to AF190927, and those for the B region are AF190912 to AF190919, respectively.
RESULTS
Plasmid isolation and Ent plasmid identification.
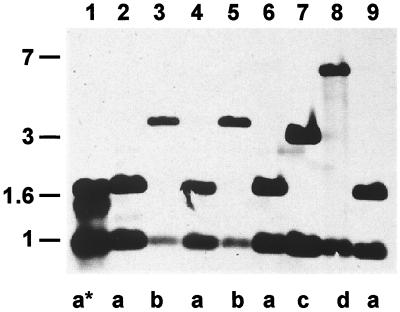
Plasmids derived from different clinical ETEC strains were investigated for plasmid parameters such as antibiotic profile, restriction fragment pattern, and conjugational behavior (data not shown). Most importantly, all isolated plasmids were analyzed for the presence of the enterotoxin-encoding genes eltAB (LT genes). The genes were identified by PCR, utilizing specific oligonucleotides ETA1 and ETA2, according to the established PCR protocol of O'Meara et al. (34). All isolated plasmids produced the specific 1.1-kb eltAB fragment of LT (data not shown). In addition, numerous strains contained more than one plasmid with different antibiotic resistance markers, such as those for ampicillin, kanamycin, chloramphenicol, tetracycline, and streptomycin. The locations of the eltAB genes on the isolated Ent plasmids were identified by Southern blot analysis (Fig. 1), and plasmids were digested with HindIII and hybridized with a specific 1.1-kb eltAB fragment as labeled probe DNA. Since a HindIII restriction site is contained in eltA at bp 569, it was expected that two hybridizing fragments would appear. Indeed, eight of the isolated plasmid pools showed two hybridizing fragments, whereas isolate K1 showed an additional one, indicating two copies of the elt genes (Fig. 1). The 800-bp HindIII fragments appeared in all isolates, whereas the larger HindIII fragments showed variation and were found to be represented at least in four distinct classes (a, b, c, and d).
FIG. 1.
Southern blot analysis of isolated Ent plasmids from clinical ETEC strains. Lanes contained HindIII-digested plasmid DNAs from the following isolates hybridized with LT probe DNA: 1, K1; 2, K2; 3, K3; 4, K4; 5, K5; 6, S1; 7, S2; 8, S6; 9, 102. Differentiation into types a, a*, b, c, and d is indicated for restriction site polymorphism of the upstream eltAB region (a* indicates a plasmid harboring two eltAB copies). Numbers on the left indicate fragment size (in kilobase pairs).
Characterization of DNA adjacent to the elt gene.
Eight isolates were further subjected to DNA analysis. In determining the DNA regions adjacent to eltAB, two conserved regions were identified; these were termed the A box, of 236 bp and located in the upstream region of eltA, and the B box, of 280 bp and located downstream of eltB. The two sequences were aligned as shown in Fig. 2. The GC contents for the A-box, B-box, and LT genes were determined to be 36, 41, and 37%, respectively. A closer analysis revealed a significant formation of two stem-loop structures in the A and B boxes; the latter was also recognized by Dallas and Falkow (8) and was suggested to act as the presumed transcriptional termination site (Fig. 2). Recently, Trachman and Maas (46) have described a temperature-regulated and H-NS-dependent synthesis of the eltA gene, along with indications of the location of the start site of mRNA synthesis. Interestingly, we found an adequately matching E. coli promoter, based on an algorithm of Mulligan et al. (31), of about 70%, which is thought to be a very strong E. coli promoter, located right before the mRNA start site. Interestingly, exactly around the predicted −35 region a significant stem-loop structure is observed, which may indicate some regulatory function (Fig. 2).
FIG. 2.
Sequence characteristics of boxes A and B. Shown are the aligned sequences of the eight isolates; sequence variation among the isolates is indicated in parentheses. Stem-loop structures are indicated by arrows, and the putative promoter region of eltA is marked by underlining. The A box ends with bp 236 immediately before initiator codon ATG of subunit eltA. The B box starts with bp 1 immediately downstream of stop codon TAG.
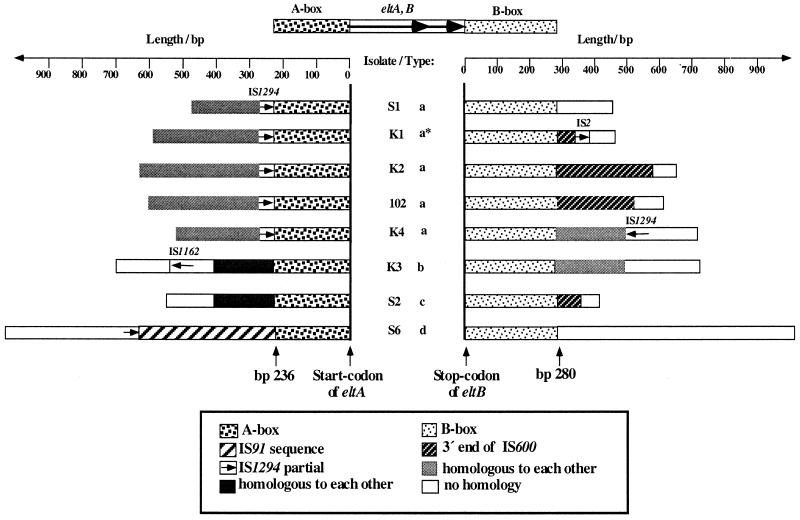
Further sequencing revealed that the extended distal and proximal sequences were different for all eight isolates. They consisted of either unknown DNA or partial or incomplete IS elements. In Fig. 3, a scheme indicating the positions, lengths, and identifications of the DNA regions is shown. As also described by Murphy and Dallas (33), we found different partial sequences of IS600 (Shigella sonneii) (25) in four isolates and also cryptic IS2 (35), IS1162 (Pseudomonas fluorescens) (43), IS1294 (E. coli) (N. Tavakoli, et al., direct submission to GenBank), and 391 bp of the complete right end of IS91 (E. coli) (29).
FIG. 3.
Schematic survey of sequences flanking the conserved eltAB operon of different Ent plasmids. DNA sequencing revealed an upstream-located A box (236 bp) and a downstream-located B box (280 bp), which are conserved in all analyzed plasmids. Mainly partial copies of different IS elements are closely associated with the eltAB operon structure. Isolate S6 carries a plasmid with the right end of IS91 fused to the A box.
Rearrangements of IS91-based eltAB derivatives in a recA-dependent assay.
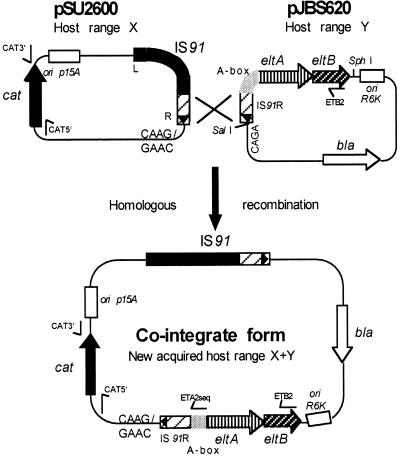
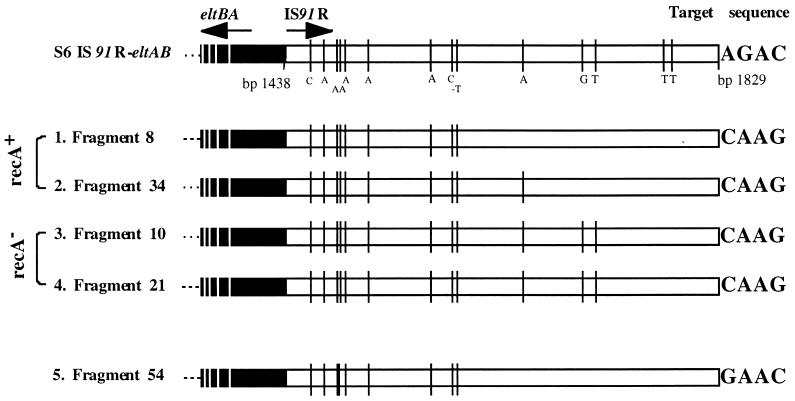
The complete 391 bp of IS91R, identified next to the A box in isolate S6, prompted us to investigate the ability of this nearby element to be mobilized along with the elt genes. Defined activities of the IS91 terminus were reported previously (28), indicating that IS91R sequences can be activated by trans-active intact IS91 elements. To test the possibility of IS91-activated transposition, the S6 isolate harboring the IS91R eltAB-containing fragment was subcloned into the suicide plasmid pCVD442 (9), resulting in pJBS620 (see Materials and Methods) (Fig. 4). By use of the recombination assay (Materials and Methods), cointegrated plasmids were subsequently isolated. Some cointegrate isolates were further analyzed by PCR and DNA sequencing. This procedure was directed to obtain specific joint fragments of cointegrate isolates by using pSU2600 and eltB-specific oligonucleotides (Fig. 4). Two joint fragments of 3.2 kb (data not shown) generated from cointegrate plasmids from MC4100 λpir recA::Kan (fragments 10 and 21 [Fig. 5]) and two from MC4100 λpir (fragments 8 and 34 [Fig. 5]) were then subjected to DNA sequencing. For sequencing, the oligonucleotide ETA2seq (Table 2; Fig. 4) was used. The four sequences obtained showed that recombination between the intact IS91 and the IS91R sequences has occurred within the homologous region of the 391 bp of IS91R, as indicated in Fig. 4. Due to the cointegrate formation, all four IS91R eltAB isolates have received a reconstituted IS91R region attached to the intact target site of CAAG. By analyzing the cointegrate formation due to transposition activity, we searched for joint fragments outside the original IS91-carrying region of pSU2600. By using the oligonucleotides Cat5′ and ETB2 and cointegrate plasmids, derived from a recA+ strain selected as Cmr Ampr cells, a joint fragment of about 2 kb was generated. After sequence analysis, using oligonucleotide ETA2seq, it was found that the IS91R eltAB was inserted at bp 4443 of pSU2600 at the target sequence GAAC (fragment 54 [Fig. 5]).
FIG. 4.
Cointegrate formation of IS91-based plasmids. Plasmid pSU2600 (host range X) with a complete IS91 can form a cointegrate plasmid with a new host range (X+Y) after recombination with IS91R of the suicide plasmid pJBS620 (host range Y). Oligonucleotides Cat3′, Cat5′, and ETB2 for PCR amplification and sequencing primer ETA2seq are shown as small arrows. The target sequences at the ends of IS91 and IS91R and relevant restriction sites are indicated.
FIG. 5.
Recombinational events of plasmid crossing in IS91 coding regions. PCR-generated joint fragments of cointegrated plasmids pSU2600 and pJBS620 are shown (for details, see the text). The distribution of point mutations within the IS91R sequence of ETEC isolate S6 and cointegrate plasmid-derived fragments 1 to 5 is shown. Single point mutations are indicated by the mutated base and depicted as vertical lines. Target sequences are also shown.
Characterization of IS91 insertions among ETEC-derived plasmids.
The 391 bp of IS91R identified next to the A box prompted us to look for further associations of IS91 sequences within ETEC-derived plasmids. Accordingly, whole IS91 element sequences and IS91 sequences truncated at the right end were used as labeled probe DNAs. Southern blot analysis, utilizing ETEC-derived plasmid DNAs of human and porcine isolates, indicated the presence of numerous copies of IS91 sequences in some isolates (data not shown). In addition, by searching the GenBank database for IS91-associated sequences, several sequence entries for E. coli virulence factors which were associated with yet-unrecognized partial sequences of the IS91 element on plasmids could be located. For example, IS91 sequences were located near cfaD (39), CS6 (49), CS3 (18), and faeA (17) and also on an enterohemorrhagic E. coli-specific virulence plasmid, pO157 (23).
DISCUSSION
Mobile genetic and associated elements, like plasmids, bacteriophages, or IS elements, are ubiquitous in nature and are responsible for generating pathogenic novel variants by horizontal gene transfer and genetic rearrangements (1, 3, 16, 20, 22). Although cholera is one of the most severe forms of bacterial diarrhea, ETEC-caused diarrhea seems to be the most frequent one (13). ETEC-derived enterotoxin- and cholera toxin-encoding genes are closely related, and it was recently demonstrated that cholera toxin-encoding genes are harbored on the genome of a filamentous phage and are therefore horizontally transmissible to form toxigenic Vibrio cholerae strains (48). Earlier, Yamamoto et al. (50) attempted to describe the evolutionary origins of LT and cholera toxin. Based on conserved regions of the DNA sequences, they proposed that the corresponding genes may have been separated in the late Jurassic period or the beginning of the Cretaceous period, about 120 × 106 years ago. It is tempting to speculate that specific transmission or dissemination modes have served to facilitate the species-specific pathogenic development and evolution of V. cholerae and ETEC. Dallas and coworkers also described the origin of the eltAB genes (8). They indicated some discrimination between different animal sources (21, 47) and further described gene duplication or a merodiploid state of eltAB-harboring plasmids (33).
Our initial aim was to investigate, whether filamentous replicative-form plasmids are associated with eltAB genes, similar to the case for cholera toxin and phage CTX⊘ (48). Although some of the ETEC isolates investigated in this study harbored temperate bacteriophages, none of the isolates had the capacity to transduce the elt genes via phage routes (unpublished results).
Next, the heterogeneities of the proximal and distal DNA regions of the elt genes were analyzed. It was found that the eltAB genes are flanked by highly conserved short DNA sequences of 236 and 280 bp, termed the A and B boxes, respectively. The GC contents of these boxes approximately matched that obtained for the eltAB region. The average GC content of the A box-eltAB-B box region (38%) was significantly different from that of E. coli (51%), indicating that the A and B boxes along with the coding regions were not acquired from E. coli and remained stably attached with the eltAB genes. The A and B boxes are followed by highly variable sequences, mostly containing partial IS elements. No particular features other than inverted repeats were identified within the A and B boxes, suggesting that they may contain the proper promoter (46) and termination (8) structures, as indicated earlier.
In an earlier analysis, partial sequences of element IS600 were located right next to the elt genes, carried on an Ent plasmid which contained two copies of the eltAB genes (33). In our studies we found some more partial IS elements next to the A and B boxes. Besides IS600 we found sequences belonging to IS91, IS1162, IS2, and IS1294. The arguments for the putative role of such IS elements in dissemination were essentially described by Murphy and Dallas (33). Our goal was to extend these results by attempting to exemplify the dissemination processes by using isolate S6, which contains an IS91R element of 391 bp directly linked with the A box. As reported earlier (28), the right end of IS91 represents the initiator of transposition via a suggested replicative sequential transposition mechanism of the so-called one-ended transposition. IS91R sequences are then mobilized by a trans-acting IS91 transposase (28). A prerequisite of transposition activation of IS91 is the target site specificity of IS91 at the insertion site (CAAG/GAAC), which was found to be necessary for tnp activity (28). We analyzed the features of mobility of the putative genetic element IS91R eltAB of isolate S6. Sequence analysis of the IS91R target site on the S6 isolate revealed no CAAG specificity but an intact terminal repeat sequence. Considering the lack of the intact target site, we assumed that this element is probably not a substrate for trans activation of transposition. However, we found that indeed rearrangements were produced in the upstream region of the eltAB genes in a recA-independent recombination via IS91 sequences. As a result, the intact target sequence of the eltAB-associated IS91R element was restored. Additionally, we could identify a transposed IS91R eltAB insertion on pSU2600 at a location different from that of the original IS91 insertion. This indicates a combination of (i) recombination activity to generate an intact target site via homologous recombination between the wild-type IS91 and IS91R eltAB and (ii) a subsequent transposition event which had moved that IS91R eltAB to a new destination with target site specificity of GAAC.
Based on the results of the IS91 analysis, we propose that recA-dependent as well as -independent recombination and subsequent transposition events might be the genetic driving force for selecting new variants of Ent plasmids associated with antibiotic resistance markers and CFs. Our data indicate that IS elements, such as IS91, are involved in the rearrangement processes of the eltAB genes, warranting the variation of cointegrate formation of new plasmids with new characteristics of host range, conjugation, and antibiotic resistance systems. Interestingly, we found that IS91 elements are widely distributed among the investigated ETEC strains, as we were also able to subclone intact and active IS91 elements from such Ent plasmids (data not shown). We propose that the formation of a particular IS91R eltAB isolate is evolutionarily ancient, since 14-bp exchanges have accumulated in the 391-bp region of the IS91R sequence. How the particular IS91R sequence had been moved so close to the A box remains unknown, but it might indicate a former composite transposon consisting of IS91 elements and eltAB genes, similar to the case for hly operon on pHly152 (51).
ACKNOWLEDGMENTS
We like to thank U. Hentschel and K. Erb for critical reading of the manuscript. For the clinical E. coli strains used in this study, we thank P. Echeverria, J. Hacker, H. Karch, and J. J. Mekalanos. For plasmid pSU2600, we will thank V. Mendiola and F. de la Cruz.
This work was funded by BMBF grant 01KI8906.
Stefan Schlör and Sabine Riedl contributed equally to this work.
REFERENCES
- 1.Arber W. Evolution of prokaryotic genomes. Gene. 1993;135:49–56. doi: 10.1016/0378-1119(93)90048-8. [DOI] [PubMed] [Google Scholar]
- 2.Betley M J, Miller V L, Mekalanos J J. Genetics of bacterial enterotoxins. Annu Rev Microbiol. 1986;40:577–605. doi: 10.1146/annurev.mi.40.100186.003045. [DOI] [PubMed] [Google Scholar]
- 3.Bishai W R, Murphy J R. Bacteriophage gene products that cause human disease. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum; 1988. pp. 683–724. [Google Scholar]
- 4.Black R E. Epidemiology of travelers' diarrhea and relative importance of various pathogens. Rev Infect Dis. 1990;12:73–79. doi: 10.1093/clinids/12.supplement_1.s73. [DOI] [PubMed] [Google Scholar]
- 5.Black R E. Persistent diarrhea in children of developing countries. Pediatr Infect Dis J. 1993;12:751–761. doi: 10.1097/00006454-199309000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban M J. Transposition and fusion of the lac genes to select promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 7.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallas W S, Falkow S. Amino acid sequence homology between cholera toxin and Escherichia coli heat-labile toxin. Nature. 1980;288:499–501. doi: 10.1038/288499a0. [DOI] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dziejman M, Mekalanos J J. Analysis of membrane protein interaction: ToxR can dimerize the amino terminus of phage lambda repressor. Mol Microbiol. 1994;13:485–494. doi: 10.1111/j.1365-2958.1994.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 11.Echeverria P, Verneart L, Ulyangco C V, Komalarini S, Ho M T. Antimicrobial resistance and enterotoxin production among isolates of Escherichia coli in the Far East. Lancet. 1978;8090:589–592. doi: 10.1016/s0140-6736(78)92820-9. [DOI] [PubMed] [Google Scholar]
- 12.Fishmann P H. Mechanism of action of cholera toxin. In: Moss J, Vaughan M, editors. ADP-ribosylating toxins and G proteins. Washington, D.C.: American Society for Microbiology; 1990. pp. 127–137. [Google Scholar]
- 13.Gaastra W, Svennerholm A M. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4:444–452. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 14.Giron J A, Xu J G, Gonzalez C R, Hone D, Kaper J B, Levine M M. Simultaneous expression of CFA/I and CS3 colonization factor antigens of enterotoxigenic Escherichia coli by delta aroC, delta aroD Salmonella typhi vaccine strain CVD 908. Vaccine. 1995;13:939–946. doi: 10.1016/0264-410x(95)00003-j. [DOI] [PubMed] [Google Scholar]
- 15.Gorbach S L, Banwell J G, Chatterjee B D, Jacobs B, Sack R B. Acute undifferentiated human diarrhea in the tropics. J Clin Invest. 1971;50:881–889. doi: 10.1172/JCI106560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 17.Huisman T T, Baker D, Klaasen P, de Graf F K. Leucine-responsive regulatory protein, IS1 insertions, and the negative regulator FeaA control the expression of the fae (K88) operon in Escherichia coli. Mol Microbiol. 1994;11:525–536. doi: 10.1111/j.1365-2958.1994.tb00333.x. [DOI] [PubMed] [Google Scholar]
- 18.Jalajakumari M B, Thomas C J, Halter R, Manning P A. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol Microbiol. 1989;3:1685–1695. doi: 10.1111/j.1365-2958.1989.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 19.Jertborn M, Ahren C, Holmgren J, Svennerholm A M. Safety and immunogenicity of an oral inactivated enterotoxigenic Escherichia coli vaccine. Vaccine. 1998;16:255–260. doi: 10.1016/s0264-410x(97)00169-2. [DOI] [PubMed] [Google Scholar]
- 20.Lee C A. Pathogenicity islands and the evolution of bacterial pathogens. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 21.Leong J, Vinal A C, Dallas W S. Nucleotide sequence comparison between heat-labile toxin B-subunits from Escherichia coli of human and porcine origin. Infect Immun. 1985;48:73–77. doi: 10.1128/iai.48.1.73-77.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahillon J, Chandler M. Insertions sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yatsudo H C, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han C, Ohtsubo A, Kasamatsu M, Kuhara T H, Shinagawa H. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 1998;5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 25.Matsutani S, Ohtsubo H, Meada Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 26.McConnell M M, Thomas L V, Day N P, Rowe B. Enzyme-linked immunosorbent assays for the detection of adhesion factor antigens of enterotoxigenic Escherichia coli. J Infect Dis. 1985;152:1120–1127. doi: 10.1093/infdis/152.6.1120. [DOI] [PubMed] [Google Scholar]
- 27.Mekalanos J J, Swartz D J, Pearson G D, Harford N, Groyne F, deWilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 28.Mendiola V, Bernales I, de la Cruz F. Differential roles of the transposon termini in IS91 transposition. Proc Natl Acad Sci USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendiola V, Jubete Y, de la Cruz F. DNA sequence of IS91 and identification of the transposase gene. J Bacteriol. 1992;174:1345–1351. doi: 10.1128/jb.174.4.1345-1351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Mulligan M E, Hawley D K, Entriken R, McClure W R. E. coli promoter sequences predict in vitro RNA-polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullis K B, Faloona F. Specific synthesis of DNA in vitro via a polymerase chain reaction. Methods Enzymol. 1987;155:335–340. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 33.Murphy G L, Dallas W. Analysis of two genes encoding heat-labile toxins and located on a single plasmid from Escherichia coli. Gene. 1991;103:37–43. doi: 10.1016/0378-1119(91)90388-r. [DOI] [PubMed] [Google Scholar]
- 34.O'Meara D, O'Shaughnessy E, Cryan B, Fanning S. Colorimetric detection of toxin-encoding gene of enterotoxigenic Escherichia coli by PCR. J Clin Microbiol. 1995;33:1957–1960. doi: 10.1128/jcm.33.7.1957-1960.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronecker H J, Rak B. Genetic organization of insertion element IS2 based on a revised nucleotide sequence. Gene. 1987;59:291–296. doi: 10.1016/0378-1119(87)90337-4. [DOI] [PubMed] [Google Scholar]
- 36.Rose R E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe B, Taylor J, Bettelheim K A. An investigation of travellers diarrhoea. Lancet. 1970;i:1–5. doi: 10.1016/s0140-6736(70)90520-9. [DOI] [PubMed] [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savelkoul P H, Willshaw G A, McConnell M M, Smith H R, Hamers A M, van der Zeijst B A, Gaastra W. Expression of CFA/I fimbriae is positively regulated. Microb Pathog. 1990;8:91–99. doi: 10.1016/0882-4010(90)90073-y. [DOI] [PubMed] [Google Scholar]
- 40.Sears C L, Kaper J B. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol Rev. 1996;60:167–215. doi: 10.1128/mr.60.1.167-215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith H R. Genetics of enterotoxin production in Escherichia coli. Biochem Soc Trans. 1984;12:187–189. doi: 10.1042/bst0120187. [DOI] [PubMed] [Google Scholar]
- 42.Smith W H, Halls S. The transmissible nature of the genetic factor in Escherichia coli that controls enterotoxin production. J Gen Microbiol. 1968;52:319–334. doi: 10.1099/00221287-47-1-153. [DOI] [PubMed] [Google Scholar]
- 43.Solinas F, Maraconi A M, Ruzzi M, Zemaro E. Characterization and sequence of a novel insertion sequence, IS1162, from Pseudomonas fluorescens. Gene. 1995;155:77–82. doi: 10.1016/0378-1119(94)00922-f. [DOI] [PubMed] [Google Scholar]
- 44.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;51:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 45.Svennerholm A M, Holmgren J, Sack D A. Development of oral vaccines against enterotoxinogenic Escherichia coli diarrhoea. Vaccine. 1989;7:196–198. doi: 10.1016/0264-410x(89)90228-4. [DOI] [PubMed] [Google Scholar]
- 46.Trachmann J D, Maas W K. Temperature regulation of the heat-labile enterotoxin (LT) synthesis in Escherichia coli is mediated by an interaction of H-NS protein with the LT A-subunit. J Bacteriol. 1998;180:3715–3718. doi: 10.1128/jb.180.14.3715-3718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vinal A C, Dallas W S. Partition of heat-labile enterotoxin genes between human and animal Escherichia coli isolates. Infect Immun. 1987;55:1329–1331. doi: 10.1128/iai.55.5.1329-1331.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldor K W, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 49.Wolf M K, de Haan L A, Cassels F J, Willshaw G A, Boedecker W R, Gaastra W. The CS6 colonisation factor of human enterotoxigenic Escherichia coli contains two heterologous major subunits. FEBS Lett. 1997;148:35–42. doi: 10.1111/j.1574-6968.1997.tb10263.x. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto T, Gojobori T, Yokota T. Evolutionary origin of pathogenic determinants in enterotoxigenic Escherichia coli and Vibrio cholerae O1. J Bacteriol. 1987;169:1352–1357. doi: 10.1128/jb.169.3.1352-1357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zabala J C, Garcia-Lobo J M, Diaz-Aroca E, de la Cruz F, Ortiz J M. Escherichia coli alpha-hemolysin synthesis and export genes are flanked by a direct repetition or IS91-like elements. Mol Gen Genet. 1984;197:90–97. doi: 10.1007/BF00327927. [DOI] [PubMed] [Google Scholar]