Abstract
The gut microbiome plays critical roles in human health and disease. Recent studies suggest it may also be associated with chronic pain and postoperative pain outcomes. In animal models, the composition of the gut microbiome changes following general anesthesia, and affects the host response to medications, including anesthetics and opioids. In humans, the gut microbiome is associated with the development of postoperative pain and neurocognitive disorders. Additionally, the composition of the gut microbiome has been associated with pain conditions including visceral pain, nociplastic pain, complex regional pain syndrome and headaches, partly through altered concentration of circulating bacterial-derived metabolites. Furthermore, animal studies demonstrate a critical role of the gut microbiome in neuropathic pain, via immunomodulatory mechanisms. Here, we review basic concepts of the human gut microbiome and its interactions with the host, and provide a comprehensive overview of the evidence linking the gut microbiome to anesthesiology, critical care and pain medicine.
Summary Statement
Gut microbiome is increasingly recognized as a key factor implicated in the ‘gut-brain axis’. This narrative review summarizes and appraises recent advances in gut microbiome for perioperative and pain physicians and researchers.
Introduction
In recent years, new insights into the composition and function of the human microbiome are shedding new light on many fields of clinical medicine1. The human body hosts a wealth of bacterial species which play cardinal roles in its function in health and in many disease conditions. This role extends beyond mere associations, and a putative causal role of specific bacterial taxa and ensembles is constantly unraveling in fields ranging from gastroenterology to psychiatry. Until recently, little was known about the role the gut microbiome plays in anesthesiology and pain, however new studies shed interesting light on the importance of the gut microbiome in these fields, paving the way for potential clinical applications. This review aims to summarize the key findings on the bidirectional interactions between the human body and the gut microbiome pertinent to anesthesiologists, intensivists, and pain physicians. We review the interaction of the gut microbiome with four distinct clinical domains, including effects of anesthetics and analgesics, perioperative outcomes, intensive care and pain. For each domain the results of preclinical and clinical studies are summarized.
Gut microbiome – from associations to clinical applications
The human microbiome is an intricate ecosystem, typically hosting hundreds to thousands of bacterial species, as well as viruses, fungi and phages, which is constantly shaped by the host and its environment while also influencing its function, its health and diseases2. This section will review the conceptual model of microbiome research, dividing it into four distinct phases - association, causation, mechanisms and clinical applications.
While the terms microbiota and microbiome are sometimes used interchangeably, they are not synonymous. Traditionally, microbiota refers to the ecological community of microorganisms hosted by a multicellular organism, while microbiome refers to the genomes of these microorganisms. More recent definitions regard the microbiome as the microbiota within its well-defined habitat, forming its theater of activity, including microbial and external structural elements such as genes, lipids, proteins, polysaccharides and metabolites3.
The complexity of the human microbiome may be conceptualized by considering its size, diversity and dynamics: The human body hosts hundreds of bacterial species, and the number of these bacterial cells is estimated at 1.3 times the number of human cells4. The number of bacterial genes, however, is two to three orders of magnitude larger than that of human genes5. This huge bacterial community is highly variable between individuals: while the human genome is 99.5% identical between any two individuals, the composition of the of their gut microbiome differs considerably even between identical twins6,7 or individuals who share a common living environment8. The composition of the human microbiome not only varies between individuals but is also highly variable among the different body sites of a given host, with distinct communities detected in the intestinal tract, skin, urinary tract and respiratory tract, and even within different niches of these sites2. Other than being variable, the composition of the microbiome is also dynamic: the relative abundance of bacterial taxa in the human host changes over timescales of years, months, and even days, in response to a multitude of host- and environment-related factors, including age, diet, occupation, physical activity and medications2,9,10. The rate of microbiome compositional changes is also variable between individuals11. Nevertheless, emerging evidence supports certain levels of resilience in gut microbiota composition, manifesting in its ability to recover to pre-perturbation baseline composition12.
In recent years, gut microbiome research has directly benefited from the advent of next-generation sequencing technologies. Specifically, metagenomic studies fueled by 16s rRNA gene sequencing or whole genome sequencing decode the gut microbiome composition as well as community structure. Sequencing based on 16s rRNA gene is cost-effective albeit with a limited resolution, while whole genome sequencing is more expensive than 16s rRNA sequencing but provides better phylogenetic resolution13. Using these technologies, a rapidly growing body of evidence suggests an association between the composition of the human microbiome and various pathologies1. Initially directed at the gut microbiome and gastrointestinal diseases, these association studies revealed unique compositions of the gut microbiome in individuals with inflammatory bowel diseases, irritable bowel syndrome, cirrhosis and other gastroenterological disorders14. Studies in other fields of medicine established associations between the composition of the gut microbiome and a wide range of medical conditions, including metabolic, dermatologic, oncologic, cardiovascular and rheumatologic disorders to name only a few. Particularly interesting are reports on alterations of the gut microbiome in neurologic and psychiatric conditions, including Parkinson’s disease15,16, autism17, depression and anxiety18,19. The microbiome of other body sites, including the skin20, respiratory tract21,22 and genitourinary system23,24 was found to be altered in medical syndromes involving these organs.
However, it should be stressed that the association of specific microbiome compositions to a medical condition does not establish a causal relationship. Microbiome alterations could, in theory, be caused by the disease, or alternatively both microbiome alterations and the disease could be driven by a third factor (e.g. diet, genetics, physical activity etc.). With the advent of gnotobiotic animal models, the potential causal relationships between specific microbiome compositions and medical conditions are explored1. The transplantation of whole microbiome ensembles or individual bacterial taxa from human patients to germ-free (or antibiotic pre-treated) animals, while monitoring the development of disease phenotypes, allows researchers to explore the putative causal role of the gut microbiome in the disease pathogenesis. Using such methods, Prevotella copri, a bacterial species found at increased abundance in humans with arthritis, induced joint inflammation in a mouse model25. In a study on Sod-1 transgenic mice Akkermansia muciniphila ameliorated symptoms of amyotrophic lateral sclerosis, while Ruminococcus torques and Parabacteroides distasonis exacerbated symptoms26. Finally, Parabacteroides merdae was found to mediate the antiepileptic effects of the ketogenic diet in a mouse model of intractable seizures27. These, and similar studies suggest that the gut microbiome may play a role in the pathogenesis of certain pathologies. However their relevance to humans remains to be established.
After a causal relationship is demonstrated, the next step is typically exploring possible mechanisms that allow the microbiome to affect the host health. The microbiome is in a state of constant interaction with its host via a multitude of mechanisms, which are only beginning to unfold. These include bi-directional interactions with the immune system and the secretion of biologically active metabolites2. Particularly interesting is the so called ‘gut-brain axis’, a two-way biochemical communication between the gastrointestinal tract and the central nervous system18,28. The mechanisms allowing the gut microbiome to affect the function of the central nervous system remain largely unknown, however several putative mechanisms have been proposed, including the secretion of neuro-modulatory bacterial metabolic end-product to the circulation26,27, immune-mediated effects29,30 and modulation of microglia activity31 to name a few.
The available literature on the mechanisms mediating the effects of the gut microbiome on extra-intestinal pathologies remain largely in the realm of pre-clinical studies in animal models.
In this context, the gut microbiome adds a new dimension to our current understanding of the central nervous system, which is of utmost interests to anesthesiologists, intensivists and pain physicians. Importantly, many other tissues and organs have resident microbiomes, including lung, skin, oral cavity and eye, which are increasingly appreciated as modulators of health and disease. While we focus on gut microbiome in the current review, the intriguing interactions among different microbiome compartments besides gut microbiome-host interactions will likely provide a new frontier in our understandings of microbiome-host interactions32,33.
The demonstration of a causal role of the microbiome in medical disorders, and the identification of potential downstream mechanisms, may pave the way for translational studies, aiming to develop clinical applications with diagnostic and therapeutic aims. The specific gut-microbiome compositions associated with various medical conditions raise the possibility to use these biological signatures as diagnostic aids. Indeed, several preliminary reports demonstrated the feasibility of gut microbiome-based diagnosis of pathologies ranging from several types of cancer34, irritable bowel syndrome35,36 and fibromyalgia37 to name a few. Promising as these translational studies may be, to date microbiome-based diagnostic tools are still in the realm of research use, and are not yet available for clinical use. Another potential clinical utility is the manipulation of the gut microbiome to reverse dysbiosis and restore a ‘healthy’ microbiome composition. This may be achieved by the use of dietary measures, including prebiotics, probiotics, medications or by means of fecal microbiome transplantation from healthy individuals38–41. Prebiotics are nutritional compounds providing preferential advantage to selected bacterial taxa. Probiotics are live microorganisms that, when administered to patients, provide health benefits presumably through active colonization and incorporation into the host gut microbiome. Fecal microbiome transplantation is the process of enriching or replacing the host gut microbiome with that of a donor, currently indicated for refractory Clostridium difficile associated colitis42. Nevertheless, preliminary clinical studies have shown promising results in irritable bowel syndrome43, autism41, and in enhancing host response to anti-cancer treatment38,39,44. When rigorous donor screening is performed, fecal microbiome transplantation is considered safe, with most short-term adverse events being attributed to the mode of delivery (i.e. colonoscopy). Long-term risks have not been observed, and longitudinal follow-up studies are still ongoing, with no signal towards harm reported thus far. Severe adverse events have been described rarely, mostly involving the transmission of antibiotic-resistant bacteria (in cases where appropriate screening was not carried out) and even mortality in severely ill immune-compromised recipients45–47. Defining a healthy gut microbiome and screening for microorganisms that could lead to adverse infectious events will likely enhance the safety profile of fecal microbiome transplantation.
In summary, a growing body of evidence demonstrates gut-microbiome compositional changes in a variety of clinical conditions in humans. Still, evidence regarding the causal role of gut bacteria, the mechanisms by which they exert their pathophysiologic effects and the clinical applications of these observations is largely limited to preclinical studies in animal models.
When considering the role of the gut microbiome in perioperative care and in chronic pain, important first steps have been undertaken in recent years. Following the methodological steps of association, causation, mechanism and clinical application the role of the gut microbiome in these fields is constantly unravelling, and will be reviewed in the following sections.
Gut microbiome interactions with anesthetics and opioids
The gut microbiome is readily shaped by a wide spectrum of medications48, while at the same time affecting the host response to many medications. In humans, most drugs have been shown to affect the composition of the gut microbiome, and for many, the clinical efficacy is determined by its composition49,50. At this point, only preclinical literature regarding animal models is available.
General anesthesia alters the composition of the gut microbiome in rodents
Several recent animal models demonstrate that the composition of the gut microbiome is affected by exposure to anesthetics, whereby even a relatively short duration of anesthetic exposure could lead to significant changes in gut microbiota composition. For example, after 4 hours of exposure to isoflurane, mice gut microbiota composition displayed significant changes at day 1 and day 7 post exposure. These changes were characterized by a decrease in bacterial community α diversity; increase in the abundance of proteobacteria and actinobacteria, and decrease in the abundance of firmicutes and clostridiales51. By longitudinally examining the gut microbiota changes after a single episode of isoflurane exposure, Han et al. showed that changes in gut microbiota lasted for more than 14 days52. This strong perturbation was not unique to volatile anesthetics, as intravenous medication propofol was also able to induce gut microbiota changes53. Specifically, three hours of propofol infusion in rats led to a decreased abundance of Prevotella and Lactobacillus at the genus level, which showed a recovery trend on the 14th day. Besides changes in bacterial species, gut-microbiota-related metabolites were also altered by anesthetics52. Some of the compositional changes in the gut microbiome following general anesthesia were long-lasting, as neonatal (day 7) exposure to isoflurane induced gut microbiota changes that were observed at a juvenile stage (day 42). Specifically, the abundance of Firmicutes, Proteobacteria, Clostridia, Clostridiales, and Lachnospiraceae were significantly increased in exposed rats and the abundance of Bacteroidetes, Actinobacteria, Bacteroidia and Bacteroidaceae was significantly decreased compared to controls. All these studies point to a potent and lasting effect of anesthetics (isoflurane and propofol) on the composition of the gut microbiome, although the clinical significance of these perturbations is still unknown.
Gut microbiome is associated with sensitivity to anesthetics in rodents
Early evidence suggests that the gut microbiome affects sensitivity to anesthetics in animals. For example, when specific pathogen free mice and germ-free mice were exposed to pentobarbital anesthesia, germ-free mice displayed much faster recovery, as determined by righting reflex, when compared with specific pathogen-free mice17. Interestingly, conventionalization of germ-free mice did not reverse their resistance to pentobarbital. As pentobarbital undergoes xenobiotic metabolism, the differential effect of pentobarbital between germ-free and specific pathogen-free mice is likely related to liver metabolism. A microarray of 60-mer RNA from germ-free and specific pathogen-free mice livers revealed 112 genes differentially regulated in the germ-free mice vs. specific pathogen- free mice, many of which belong to the endobiotic and xenobiotic groups, which were potentially linked to pentobarbital pharmacokinetics54. It is unknown why germ-free mice conventionalization did not reverse their resistance to pentobarbital. However, from gene microarray data, it is possible that some of the liver metabolic changes induced by germ-free status were long lasting and not readily reversible 54.
Opioids and the gut microbiome in rodents
Evidence is mounting that the gut microbiome may affect the response to pain medications, and specifically opioids. The interactions between opioids, opioid receptors and the microbiome are bidirectional, whereby opioids modulate the composition of the gut microbiome, and the gut microbiome in turn affects the host response to opioids.
The administration of morphine to mice resulted in significant changes in the composition of the gut microbiome, evident as early as one day after treatment initiation55. These changes, which are regimen dependent56, were also associated with metabolomic changes, and specifically with altered serum concentrations of secondary bile acids, phosphatidylethanolamines and fatty acids. Alterations in the composition and function of the gut microbiome were partially reversible by the co-administration of the μ-receptor antagonist naltrexone. Similar results have been reported that the compromised gut barrier, associated with the administration of morphine to mice, resulted in systemic inflammation which could be reversed by fecal microbiome transplantation from placebo-treated mice 57. Moreover, following chronic treatment with oxycodone, the changes in the intestinal microbiome of mice persisted even after treatment was discontinued58.
Interestingly, the host response to morphine is dependent on the gut microbiome: in a mouse model of morphine tolerance, germ-free mice and antibiotic-treated mice were less prone to developing opioid tolerance than were naïve mice59. Opioid tolerance was associated with the depletion of two bacterial genera - Bifidobacteria and Lactobacillaeae. Microbiome reconstitution using either fecal microbiome transplantation from naïve mice or specific enrichment with Bifidobacteria and Lactobacillaeae recapitulated opioid tolerance in this model. In a similar mouse model, antibiotic-treatment prevented the development of tolerance but did not affect naloxone-induced withdrawal60. Finally, the gut microbiome, and particularly the cecal microbiome, has long been recognized to hydrolyze glucuronide conjugates, including morphine 3-glucuronide61.
It thus seems that while opioids shape the composition of the gut microbiome, the latter has an important role in the development of opioid tolerance in the host.
In conclusion, preclinical studies suggest that the composition of the gut microbiome is readily affected by host exposure to anesthetics and opioids. Moreover, gut bacteria play a role in the host sensitivity to pentobarbital and the rate of recovery after exposure to the medication, and in the development of tolerance to opioids. Notably, these preclinical findings need to be verified in clinical studies.
Gut microbiome and perioperative outcomes
Recent studies shed light on the intricate role of the microbiome in the perioperative care of the patient. Animal studies suggest that these bi-directional interactions include alterations in the composition of the gut microbiota following general anesthesia, as well as effects of gut bacteria on postoperative outcomes including postoperative pain, and postoperative cognitive dysfunction. Early human studies show an association of the gut microbiome composition and postoperative outcomes including pain and delirium.
Preclinical studies
The role of the microbiome in perioperative neurocognitive disorders in rodents
The term ‘perioperative neurocognitive disorders’ refers to cognitive impairments identified in the preoperative or postoperative period, ranging from acute transient events (postoperative delirium) to delayed neurocognitive recovery (postoperative cognitive dysfunction). The pathogenesis of perioperative neurocognitive disorders is an area of active research with significant public health relevance. In animal models, the gut microbiome has increasingly been recognized as a key player in perioperative neurocognitive disorders.
To gain mechanistic insights into the role of the gut microbiome in perioperative neurocognitive disorders, several studies have explored bacterial signatures associated with postoperative delirium and postoperative cognitive dysfunction. For postoperative delirium, when mice undergoing abdominal surgery and anesthesia were separated into non-postoperative delirium and postoperative delirium phenotypes postoperatively, major differences were observed in the composition of the gut microbiome of the two groups, at multiple taxonomic levels. For example, at the genus level, postoperative delirium mice showed significant decreases in the levels of Ruminiclostridium, Ruminococcaceae UCG 014, and Desulfovibrio compared with the non-postoperative delirium mice62. When fecal microbiome transplantation was carried out, feces from postoperative delirium mice rendered pseudo-germ-free mice (mice with largely eradicated gut microbiome using wide-spectrum antibiotics) prone to developing postoperative delirium, supporting a key role of the gut microbiome in postoperative delirium susceptibility62.
Age is a well-established risk factor for postoperative delirium, but its biological underpinnings remain poorly understood. Liufu et al. specifically examined age as a biological variable in postoperative delirium development by focusing on gut microbiome changes. They found that anesthesia and surgery induced age-dependent changes in the gut microbiome, particularly a reduction in the abundance of lactobacillus. These changes appeared to play a causal role in postoperative delirium development, as supplementation of lactobacillus could deter postoperative delirium development in aged mice after postoperatively63.
In line with its role in postoperative delirium, the gut microbiome also plays a role in postoperative cognitive dysfunction64–67. Notably, a recent animal study adopted a clinical scenario: the perioperative use of antibiotics, which are unambiguously potent microbiome modulating agents68. Cefazolin, commonly used in the perioperative setting, induced gut microbiome changes as well as cognitive dysfunction in mice not undergoing surgery. However, when cefazolin was administered once daily for five consecutive days, surgery-induced postoperative memory and learning impairment were ameliorated, supporting a potential implication of gut microbiome in postoperative cognitive dysfunction68,69 . Of note, some antibiotic agents are known to exert effects beyond mere bactericidal or bacteriostatic properties - e.g. neurotoxicity (cephalosporins), microglia inhibition (minocycline) - hence symptom reversal following antibiotics does not necessarily infer a bacterial-related mechanism of action. Similar to studies in postoperative delirium, most studies on gut microbiome and postoperative cognitive dysfunction utilized rodent models, calling for future translational studies to directly examine its role in humans.
Reported gut microbiome changes in postoperative delirium and postoperative cognitive dysfunction vary across studies62–64,66. Some of these variations are likely related to gut microbiome’s susceptibility to host genetics, dietary intake, and housing condition70.
In animal models, manipulation of the gut microbiome has the potential to alleviate postoperative delirium and postoperative cognitive dysfunction. The use of prebiotics, probiotics and fecal microbiome transplantation has been proposed. Prebiotics containing galacto-oligosaccharide were recently shown to ameliorate postoperative cognitive dysfunction and attenuate surgery-induced neuroinflammation in rats. This protective role was associated with a significant change in the β diversity of the gut microbiome and the proliferation of Bifidobacterium and other taxa with putative anti-inflammatory effect71. The probiotic Lactobacillus is a genus of gram-positive, non-spore-forming bacteria commonly found in dairy foods such as yogurt. Supplementation of lactobacillus alleviated age-dependent development of postoperative delirium in mice63. These studies support the therapeutic potential of gut microbiome-based therapeutics in perioperative neurocognitive disorders. Interestingly, the gut microbiome is also modulated by non-dietary factors, including life-styles with known health benefits. For example, in mice, caloric restriction stimulates the development of functional beige fat and extends healthy lifespan. This health benefit could be partly attributed to compositional and functional changes in the gut microbiome, which in turn dampened lipopolysaccharide absorption. Gut microbiome depleted mice derived significantly less health benefits with fasting compared with mice with intact gut microbiome72. Intermittent fasting is another life-style that may offer protection of age-associated pathologies in animals, by enhancing mitochondrial biogenesis and energy metabolism gene expression in the hippocampus, re-structuring the gut microbiome, and increasing microbial metabolites that are related to cognitive function73. It is tempting to speculate that certain health-promoting life-styles might alleviate perioperative neurocognitive disorders, through modulating the gut microbiome. Indeed, in a rat model of the metabolic syndrome, exercise reduced otherwise heightened postoperative neuroinflammation and cognitive decline, which were linked to a restored and presumably healthy gut74.
Early clinical evidence
Gut microbiome composition is associated with postoperative delirium
In a recent single center prospective observational study, twenty-one patients who underwent cardiac surgery were assessed for the development of postoperative delirium, insomnia, and pseudopsia (visual hallucination) for 1 week along with gut microbiota characterization. Staphylococcus and Pseudomonas counts were significantly higher in patients with postoperative pseudopsia75.
Gut microbiome and postoperative pain
Early evidence from a translational study suggests the microbiome may impact the incidence of chronic postoperative pain. Recently, Yao et al., explored the association between the composition of gut microbiome and chronic postoperative pain following surgery for breast cancer76. In a nested case-control study, of 132 women undergoing surgery for breast cancer, baseline gut microbiome composition was significantly different among the 66 women who developed chronic postoperative pain at 3 months, as compared to 66 patients who were pain free. Furthermore, a potential causal role of the gut microbiota was explored using fecal microbiome tranplantation from chronic postoperative pain or pain-free patients, to mice undergoing spared nerve injury. Mechanical thresholds decreased more in mice receiving fecal microbiome transplantation from chronic postoperative pain patients than from healthy controls. These changes were accompanied by significant decrease in the concentration of PPAR-γ (peroxisome proliferator- activated receptor gamma), a key metabolism modulator implicated in pain77, and a trend towards an increase in activated microglia in the spinal cord. As such, along with surgery factors, perioperative use of antibiotics etc, gut microbiome appears to be linked to postoperative pain. In a clinical study with a relatively modest number of participants (n=20), acute postoperative pain was associated with gut microbiome composition and diversity78. These effects are in line with the role of the gut microbiome in the pathogenesis of neuropathic pain, which will be discussed later.
In summary, preclinical and clinical studies demonstrate significant effects of the gut microbiome on postoperative outcomes. These include an association between the composition of the gut microbiome and the risk of postoperative delirium and pain in both animal models and humans. In animal models, manipulation of gut bacteria by antibiotics, probiotics or fecal microbiome transplantation positively affected the incidence of postoperative delirium and pain.
Gut microbiome in intensive care
Critically ill patients show significant gut microbiome alterations, which are associated with disease severity as well as their clinical outcomes. The full extent of the implications of gut microbiome composition in the intensive care setting is beyond the scope of this review, and has been thoroughly discussed elsewhere79–81. Nevertheless, several examples will be reviewed here.
Gut microbiome composition is altered in intensive-care unit (ICU) patients
Several studies explored changes in the composition of the gut microbiome in ICU patients, and their prognostic significance in the disease outcome. In a prospective cohort of 61 patients admitted to the ICU, the composition of the gut microbiome was studied and correlated with clinical indices and outcomes82. The abundance of the genus Bifidobacterium was significantly higher in patients who survived as compared to the patients who died during the hospitalization. Furthermore, when added to the clinical scores, the composition of the microbiome improved the prediction of in-hospital death as compared to Acute Physiology and Chronic Health Evaluation (APACHE) II or Sequential Organ Failure Assessment (SOFA) alone.
Sepsis is a leading cause of mortality in critically ill patients, which has been strongly associated with the gut microbiome in several studies. In a recent prospective study of 10 patients admitted to the ICU with sepsis, 10 patients admitted to the ICU without sepsis and 10 healthy controls, the composition of the gut microbiome was significantly altered in ICU patients of both groups as compared to healthy controls83. ICU patients demonstrated reduced α-diversity (indicating reduced microbiome species richness), increased abundance of Firmicutes, and decreased proportions of Bacteroidetes, Prevotella and Lachnospira. Furthermore, significant differences were observed between sepsis, and non-sepsis ICU patients, with increased abundance of Enterococcus. In a prospective study exploring the gut microbiome of 64 sepsis patients admitted to the ICU, two distinct clusters were detected84. One ff these clusters, characterized by increased abundance of Bacteroides and an unclassified genus of Enterobacteriaceae, was associated with elevated levels of serum lactate dehydrogenase and an increased risk of developing septic shock. In line with these observations, some authors have suggested a pivotal role for the gut microbiome in the development of multi-organ failure in critically ill sepsis patients80,85.
Gut microbiome and lung microbiome in critically ill patients
Gut microbiota has recently been recognized as an important factor influencing the pulmonary system, hence, the term ‘gut-lung axis’86. In a prospective study of 21 chronic obstructive pulmonary disease who were intubated and admitted to the ICU, sequential endotracheal sampling revealed that an increase in the relative abundance of the Acinetobacter genus and Acinetobacter baumannii complex species was associated with decreased chances of successful weaning 87. In another study, among patients admitted to the ICU and maintained on supplementary oxygen, exposure to hyperoxia independently predicted subsequent bacterial growth from lung specimens, with notable species-specific effects. Specifically, rates of S. aureus isolation were not affected by early exposure to hyperoxia, whereas rates of P. aeruginosa isolation decreased among patients receiving high concentrations of FiO233. Using an animal model, the same authors found experimental evidence supporting that gut bacterial communities were altered by hyperoxia and correlated with lung inflammation. More recently, Liu et al. examined human fresh lung tissue and found short chain fatty acids were present with interindividual variability32. As short-chain fatty acids are dependent on gut microbiome32,88, these findings suggest a mechanistic linkage of gut microbiome, lung microbiome, and host health.
In summary, critically ill patients admitted to the ICU demonstrate significant gut and lung dysbiosis, which is associated with various clinical outcome measures. However, the role of potential microbiome-targeted therapeutic measures has yet to beestablished in this setting.
Gut microbiome and pain
A growing body of evidence suggests that the composition of the gut microbiome is altered in humans with a variety of chronic pain conditions, including visceral pain, nociplastic pain, headache and neuropathic pain. Meanwhile, animal studies provide evidence that the gut microbiome may have a causal role in some animal pain models. The evidence for the role of the gut microbiome in experimental and clinical pain will be reviewed here.
Visceral nociplastic (functional) pain
Initial studies associating the composition of the gut microbiome with human diseases focused on the gastrointestinal tract. Nociplastic gastrointestinal diseases, including irritable bowel syndrome, are among the pathologies in which the role of the gut microbiome is best established36,89, whereby all four research milestones – association, causation, mechanisms and clinical applications – have been reached40. The association of the gut microbiome composition and irritable bowel syndrome has been demonstrated in several recent studies. While the overall composition of the gut microbiome of individuals with irritable bowel syndrome is largely similar to that of healthy controls in terms of α- and β-diversity, specific bacterial taxa are differentially abundant in these patients. These include a decreased relative abundance of the genera Bifidobacterium and Faecalibacterium and the species Faecalibabterium prausnitzii, and an increased relative abundance of Lactobacillaceae, Bacteroides and Enterobacteriaceae. Of note, the composition of the gut microbiome was associated not only with the diagnosis of irritable bowel syndrome but also with its phenotypic subtypes (diarrhea vs. constipation predominant), as well as with syndrome severity35 and with irritable bowel syndrome flares90. Specific gut microbiome alterations are not only associated with irritable bowel syndrome but have also been shown to be sufficient to cause the phenotype: fecal microbiome transplantation from humans with irritable bowel syndrome, but not from healthy controls, was shown to cause faster transit time, intestinal barrier dysfunction and anxiety in germ-free recipient mice91. Recently, in a model of chemically induced colitis in mice, the gut microbiome was implicated in visceral hypersensitivity. Fecal microbiome transplantation from viscerally hypersensitive mice, but not healthy controls, to antibiotic-treated rats resulted in long-lasting visceral hypersensitivity and alterations in luminal short-chain fatty acids concentration92. The mechanisms underlying the role of the gut microbiome in irritable bowel syndrome are still being explored, but several pathways have been suggested. Metabolomic and transcriptomic analyses revealed alterations in the gut microbiome function, including decreased levels of short-chain fatty acids (butyrate, propionate, acetate), alterations in luminal bile acids and dysregulation of the purine salvage pathway90. Finally, the utility of gut microbiome manipulation by means of dietary measures93, probiotics and fecal microbiome transplantation43,94 have been explored and some have demonstrated clinical efficacy in irritable bowel syndrome.
Chronic pelvic pain, another common syndrome of functional visceral pain, has also been associated with microbiome alterations. Several studies have demonstrated alterations in the gut- and urinary-microbiome of individuals with chronic pelvic pain, chronic prostatitis and interstitial cystitis95–98. These associations are not as well-detailed at the taxonomic level as compared to irritable bowel syndrome, and the causal and mechanistic relationships have not yet been established.
Chronic widespread pain and fibromyalgia
Fibromyalgia, a prevalent syndrome of chronic widespread pain, fatigue, sleep disorders and other somatic symptoms99, is considered a hallmark of somatic nociplastic pain. Several recent studies have demonstrated an association of the gut microbiome composition and fibromyalgia. In a study comparing the composition and function of the gut microbiome in a cohort of 77 women with fibromyalgia and 79 healthy controls, the overall composition of the gut microbiome was similar in the two groups, however higher resolution analysis revealed several bacterial species which were differentially abundant in fibromyalgia patients37. These patients appeared to have increased relative abundance of Parabacteroides merdae and decreased abundance of Prevotella copri and Akkermansia muciniphila (which was also decreased in a mouse model of chemotherapy-induced neuropathic pain100 but increased in irritable bowel syndrome). The relative abundance of several bacterial species correlated with syndrome severity, similar to the observation in irritable bowel syndrome35. Furthermore, in line with changes observed in visceral nociplastic pain, significant changes were observed in the plasma concentration of short-chain fatty acids including butyrate and propionate. The composition of the gut microbiome, when fed to a trained machine learning algorithm, accurately classified fibromyalgia patients and controls.
In a second study comparing the composition of the gut microbiome of 105 individuals with fibromyalgia and 54 matched healthy controls, the overall composition of the gut microbiome was similar in the two groups, however specific bacterial genera were differentially abundant in patients, as well as the serum concentration of metabolites, including amino acids and neurotransmitters101. The enrichment of several bacterial genera, and of certain metabolites correlated with disease severity. Of note, an earlier study investigating the gut microbiome of individuals with fibromyalgia failed to detect changes when compared to rheumatoid arthritis patients102. However this study was significantly less powered and used culture, rather than sequencing, to detect the composition of the gut microbiome.
In contrast to the changes observed in fibromyalgia, a study in patients with chronic widespread pain and two studies in patients with myalgic-encephalomyelitis/chronic fatigue syndrome, demonstrated different alterations in the gut microbiome. In a study of 113 individuals with chronic widespread pain and 1623 controls the main outcome was decreased α-diversity and a depletion of Coprococcus comes103. However participants in this study were not clinically evaluated, and their pain diagnoses were based on self-report. In two studies exploring the gut microbiome of patients with myalgic-encephalomyelitis/chronic fatigue syndrome, evidence of increased plasma lipopolysaccharide and lipopolysaccharide -binding protein was found. Taxonomic analysis showed several differentially abundant taxa, including increased abundance of Clostridium scindens (also increased in fibromyalgia), and decreased abundance of Haemophilus parainfluenza and Parabacteroides merdae, the first also depleted in fibromyalgia and the second increased37,104–106.
It would thus seem that, in nociplastic somatic pain the association of gut microbiome composition with certain syndromes seems robust, but evidence for causal and mechanistic associations is still missing, as are clinical applications.
Neuropathic pain
Several recent studies have established the important role of the gut microbiome in the development of neuropathic pain in animal models, while also shedding light on potential mechanisms of action. In an association study, the composition of the gut microbiome was significantly altered in chronic constriction injury to the sciatic nerve - treated as compared to sham-treated rats107, showing several differentially abundant taxa in the chronic constriction injury group. The relative abundance of some differentially abundant taxa correlated with the clinical phenotype, including mechanical withdrawal threshold and thermal withdrawal latency. Associated metabolomic changes were observed in both the serum and spinal cord. Specifically, decreased abundance of butyrate-producing bacteria, correlated with both phenotypic indices and with serum- and spinal cord levels of beta-hydroxy butyric acid. Alterations in butyrate-producing bacteria and their metabolic end-products were also observed in chronic pain conditions in humans37.
A causal role of the gut microbiome in the development of neuropathic pain was demonstrated in several recent studies. In an animal model of chemotherapy-induced neuropathic pain, the development of mechanical hyperalgesia following treatment with oxaliplatin was prevented in antibiotic-treated mice and in germ-free mice108, supporting an essential role for the gut microbiome in the development of chemotherapy-induced neuropathic pain. In this study, the pro-nociceptive effect of the gut-microbiome was associated with increased exposure of the dorsal-root ganglia to lipopolysaccharide , a cell wall component of gram-negative bacteria. The increased concentrations of lipopolysaccharide in the dorsal root ganglia in turn led to an increased inflammatory response, manifesting as elevated concentrations of inflammatory cytokines, macrophages and reactive oxygen species.
The important role of the microbiome in the development of chemotherapy-induced neuropathic pain was further demonstrated using fecal microbiome transplantation in germ-free mice100. Specifically, paclitaxel associated chemotherapy-induced neuropathic pain was more pronounced in C57BL/6 strain of mouse than in 129SvEv strain. The reciprocal fecal microbiome transplantation from chemotherapy-induced neuropathic pain-resistant 129SvEv strain to chemotherapy-induced neuropathic pain-sensitive C57BL/6 strain protected C57BL/6 from chemotherapy-induced neuropathic pain, while fecal microbiome transplantation from C57BL/6 strain to 129SvEv strain rendered 129SvEv susceptible to chemotherapy-induced neuropathic pain.
A recent study demonstrated that neuro-immune interactions serve as a mechanism through which gut microbiome influences pain: in a mouse chronic constriction injury model, spinal cord infiltrating T cells were found to be different between the antibiotic-treated and the sham treated groups109 whereby decreased IFNγ-producing Th1-cells and increased Foxp3+ regulatory T cells in the former, indicating an attenuated inflammatory infiltration in the antibiotic-treated mice. Antibiotics-induced T cells changes were linked to a protective effect on pain. Depletion of Foxp3+ regulatory T cells reversed the protective effect of antibiotic-treatment.
Another study confirmed a causal role of the gut microbiome in chronic constriction injury induced neuropathic pain, reproducing the protective effect of gut microbiome depletion using antibiotic-pretreatment110. Short-chain fatty acids were significantly increased in the intestines of chronic constriction injury-treated mice as compared to sham treated mice, and was almost depleted when mice were treated with antibiotics. Intestinal short-chain fatty acids concentrations correlated with microglia activation and inflammatory cytokine concentrations in both the spinal cord and the hippocampus. Finally, oral short-chain fatty acids administration partially reversed the protective effect of antibiotics on chronic constriction injury phenotype as well as on spinal cord and hippocampal microglia activation and inflammatory markers.
The role of the gut microbiome was recently demonstrated in an additional model of neuropathic pain, induced by obesity111. Mice fed on Western diet developed mechanical and thermal sensitivity, which was reversible by fecal microbiome transplantation from lean mice. Fecal microbiome transplantation resulted in alterations of gene expression in the dorsal root ganglia and the sciatic nerve, decreased RYR2-dependent calcium release in dorsal root ganglia neurons, altered macrophage polarization in the dorsal root ganglia and sciatic nerve and in a reversal of dermal small fiber loss. Serum butyrate, which increased following fecal microbiome transplantation, correlated with mechanical thresholds.
Recently, in a mouse model of lumbar disc herniation, the protective effect of Lactobacillus paracasei supplementation was demonstrated. Lumbar disc herniation mice receiving L. paracasei, but not sham, exhibited an attenuated behavioral pain response and decreased inflammatory response in the dorsal root ganglia 112.
In humans, association studies are scarce. One such study explored the composition of the gut microbiome in a small cohort of patients with complex regional pain syndrome113. The composition of the gut microbiome of 16 complex regional pain syndrome patients was altered as compared to that of 16 healthy adults, with lower diversity and decreased Firmicutes to Bacteroidetes ratio.
In summary, a growing body of evidence substantiates a causal role for the gut microbiome in murine neuropathic pain models, including chronic constriction injury, chemotherapy-induced neuropathic pain and obesity-induced, highlighting potential mechanisms involving the levels of circulating bacterial metabolites and lipopolysaccharide , immune responses and microglia activation. The clinical relevance of these observations has yet to be established.
Headache
Data on the role of the gut microbiome in headache disorders is scarce. Two studies reported on alterations of the gut- and oral microbiome in humans with migraine. One study compared the composition of the gut microbiome of 25 pairs of twins, of whom one sibling suffered from migraines114. Several differentially abundant species were observed including F. prausnitzii, B. adolescentis and M. smithii, all decreased in migraine patients. The other study found that the composition of the oral microbiome was altered in 26 migraine patients as compared to 29 healthy controls115.
Diagnoses associated with comorbid pain
The gut microbiome has also been implicated in many medical conditions, some of which are associated with comorbid pain. These include degenerative116 and inflammatory joint diseases117,118, neurological conditions including Parkinson’s disease16 and multiple sclerosis30, inflammatory bowel diseases119,120 and others. We have limited the scope of this review to include mainly primary pain conditions, however interested readers may find further reading in the references 16,94–99.
In summary, research on the role of gut microbiome in chronic pain is still taking its first steps, with accumulating evidence on correlations, some evidence on causation and as yet scant insight into mechanisms and clinical applications.
Conclusions
Evolving research on the human gut microbiome is substantially changing our understanding of pathologies in various fields of medicine. In particular, the accumulating evidence on the interactions of the gut microbiome with the central and peripheral nervous systems holds the potential to offer new mechanistic insights regarding pathologies relevant to anesthesiologists and pain physicians. Specifically, association studies demonstrate gut microbiome alterations that correlate with both chronic pain and some post-operative outcomes in humans. Animal models provide evidence that the gut microbiome may play a role in the pathogenesis of these conditions, proposing possible mechanisms mediating this effect.
The potential of this research field to transform our understanding of chronic pain and post-operative outcomes is exciting. Microbiome-related variability could, in theory, answer some long-lasting perplexing clinical dilemma: Why do certain patients develop chronic pain? Why is patients’ response to analgesic medications so variable? The variability of the human microbiome may explain some of the clinical variability we observe, in addition to established genetic and environmental factors. Nevertheless, it should be noted that research in the field is only taking its first steps, and that the available data only represent a few pieces of the puzzle. Future studies are underway aiming to explore the association of gut microbiome composition and other pain conditions, attempting to identify and separate bacterial taxa that are common to chronic pain in general from those that are specific to certain diagnoses.
Other important considerations include studies demonstrating a causal role for specific bacterial taxa or ensembles in the pathogenesis of specific pain and perioperative disorders in animals. These may also allow the characterization of mechanistic pathways mediating these roles.
Finally, a more comprehensive understanding of the role of individual bacterial taxa in the pathogenesis of pain and perioperative disorders may allow for the development of personalized clinical applications. These may include 1) objective diagnostic tools allowing for a fast and accurate diagnosis of chronic pain conditions; 2) prognostic tools, allowing to identify patients at risk for perioperative complications or those more likely to respond to certain analgesic class; 3) therapeutic tools, based on the manipulation of the gut microbiome composition and gut microbiome-derived metabolites, may allow for novel treatment modalities for patients with chronic pain and those undergoing anesthesia and surgery.
Gut microbiome studies continue to benefit from powerful computational processes and machine learning based computer algorithms, to allow for integrated multi-omic analyses (i.e. metagenomics, metabolomics, proteomics, lipidomics, transcriptomics, epitranscriptomics, etc). Fast evolving technical platforms will facilitate the identification of gut microbiome compositional changes including bacterial taxa that are functionally important for clinical phenotypes, including chronic pain, transition from acute to chronic pain, and perioperative neurocognitive disorders. Additionally, fecal biobanking might provide unprecedented opportunities to capture preoperative baseline features to identify perturbations of the gut microbiome in the perioperative setting, and to accelerate research in gut microbiome and perioperative medicine.
Promissing and exciting as this new field may be, one must bear in mind that availbale literature is mostly preclinical, and that translational clinical applications are not yet available.As perioperative and pain physicians, in the not too distant future, we may witness major breakthroughs not only in our mechanistic understanding of the pathologies we treat but also the emregence of diagnostic and therapeutic tools that may be delivered to our patients.
Figure 1:
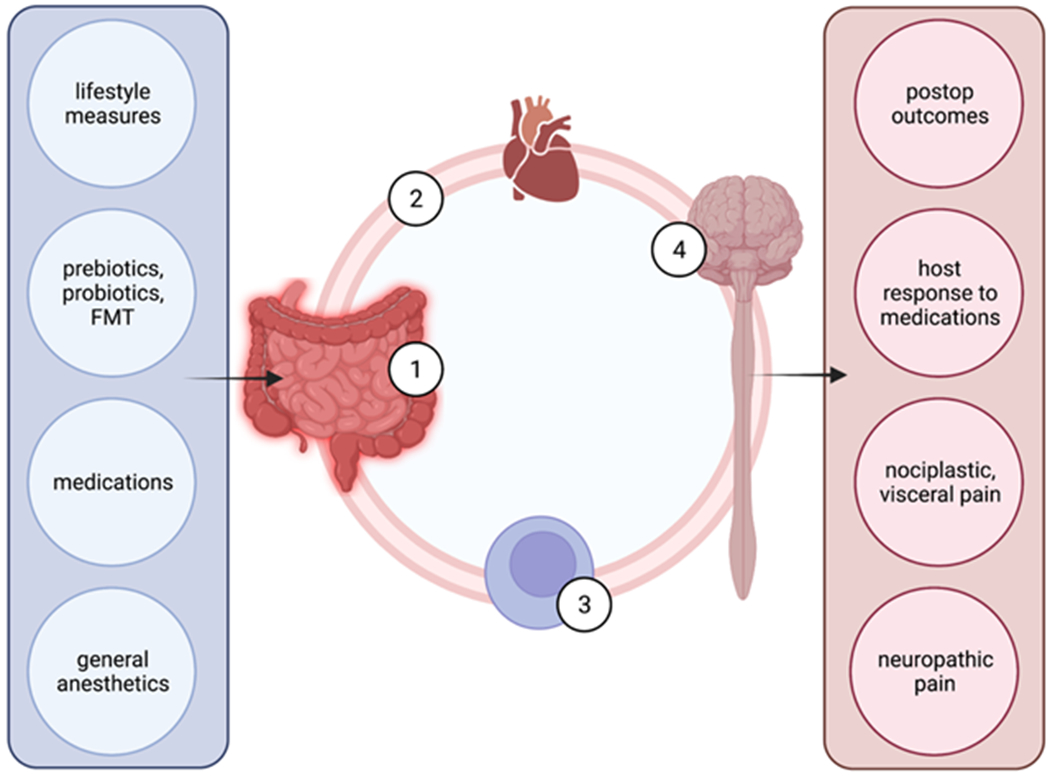
a schematic illustration of the putative mechanisms allowing the gut microbiome to affect nociception, pain modulation and postoperative outcomes. Left column lists major factors that influence gut microbiome (1). Through circulating metabolites (2) and cells, such as macrophages and T cells (3), gut microbiome influences the central nervous function (4). Right column lists functional phenotypes influenced by gut microbiome. (FMT – fecal microbiome transplantation; postop – postoperative]
Acknowledgements:
Shiqian Shen received support from NIH R35 GM128692 and RF1AG070141, in addition to NS116423, AG067947, and AG065606.
References:
- 1.Lynch SV, Pedersen O: The Human Intestinal Microbiome in Health and Disease. N Engl J Med 2016; 375:2369–79 [DOI] [PubMed] [Google Scholar]
- 2.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R: Current understanding of the human microbiome. Nature Medicine 2018; 24:392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Human Microbiome Project Consortium: Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei R, Chen X, Hu L, He Z, Ouyang X, Liang S, Dai S, Sha W, Chen C: Dysbiosis of intestinal microbiota in critically ill patients and risk of in-hospital mortality. Am J Transl Res 2021; 13:1548–57 [PMC free article] [PubMed] [Google Scholar]
- 5.Seifi N, Jafarzadeh Esfahani A, Sedaghat A, Rezvani R, Khadem-Rezaiyan M, Nematy M, Safarian M: Effect of gut microbiota modulation on feeding tolerance of enterally fed critically ill adult patients: a systematic review. Syst Rev 2021; 10:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X-J, Liu D, Ren H-Y, Zhang X-Y, Zhang J, Yang X-J: Effects of sepsis and its treatment measures on intestinal flora structure in critical care patients. World J Gastroenterol 2021; 27:2376–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah T, Baloch Z, Shah Z, Cui X, Xia X: The Intestinal Microbiota: Impacts of Antibiotics Therapy, Colonization Resistance, and Diseases. Int J Mol Sci 2021; 22:6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, Kazou M, Kinkel L, Lange L, Lima N, Loy A, Macklin JA, Maguin E, Mauchline T, McClure R, Mitter B, Ryan M, Sarand I, Smidt H, Schelkle B, Roume H, Kiran GS, Selvin J, Souza RSC de, Overbeek L van, Singh BK, Wagner M, Walsh A, Sessitsch A, Schloter M: Microbiome definition re-visited: old concepts and new challenges. Microbiome 2020; 8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sender R, Fuchs S, Milo R: Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016; 164:337–40 [DOI] [PubMed] [Google Scholar]
- 10.Tierney BT, Yang Z, Luber JM, Beaudin M, Wibowo MC, Baek C, Mehlenbacher E, Patel CJ, Kostic AD: The Landscape of Genetic Content in the Gut and Oral Human Microbiome. Cell Host & Microbe 2019; 26:283–295.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE: Human genetics shape the gut microbiome. Cell 2014; 159:789–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E: Environment dominates over host genetics in shaping human gut microbiota. Nature 2018; 555:210–5 [DOI] [PubMed] [Google Scholar]
- 13.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, Gordon JI, Fierer N, Knight R: Cohabiting family members share microbiota with one another and with their dogs. Elife 2013; 2:e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fragiadakis GK, Smits SA, Sonnenburg ED, Van Treuren W, Reid G, Knight R, Manjurano A, Changalucha J, Dominguez-Bello MG, Leach J, Sonnenburg JL: Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes 2019; 10:216–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung TC, Olson CA, Hsiao EY: Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 2017; 20:145–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, Leff JW, Vázquez-Baeza Y, Gonzalez A, Knight R, Dunn RR, Fierer N: Temporal variability is a personalized feature of the human microbiome. Genome Biol 2014; 15:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dogra SK, Doré J, Damak S: Gut Microbiota Resilience: Definition, Link to Health and Strategies for Intervention. Front Microbiol 2020; 11:572921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK-S: Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front Microbiol 2016; 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barko PC, McMichael MA, Swanson KS, Williams DA: The Gastrointestinal Microbiome: A Review. J Vet Intern Med 2018; 32:9–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Kwon S-H, Kam T-I, Panicker N, Karuppagounder SS, Lee S, Lee JH, Kim WR, Kook M, Foss CA, Shen C, Lee H, Kulkarni S, Pasricha PJ, Lee G, Pomper MG, Dawson VL, Dawson TM, Ko HS: Transneuronal Propagation of Pathologic α-Synuclein from the Gut to the Brain Models Parkinson’s Disease. Neuron 2019; 103:627–641.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet M-F, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK: Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016; 167:1469–1480.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vuong HE, Hsiao EY: Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol Psychiatry 2017; 81:411–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiuolo J, Gliozzi M, Musolino V, Carresi C, Scarano F, Nucera S, Scicchitano M, Oppedisano F, Bosco F, Ruga S, Zito MC, Macri R, Palma E, Muscoli C, Mollace V: The Contribution of Gut Microbiota-Brain Axis in the Development of Brain Disorders. Front Neurosci 2021; 15:616883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen JK, Bundgaard-Nielsen C, Hjerrild S, Nielsen RE, Leutscher P, Sørensen S: Gut microbiota variations in patients diagnosed with major depressive disorder-A systematic review. Brain Behav 2021:e02177 doi: 10.1002/brb3.2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lunjani N, Ahearn-Ford S, Dube FS, Hlela C, O’Mahony L: Mechanisms of microbe-immune system dialogue within the skin. Genes Immun 2021. doi: 10.1038/s41435-021-00133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiew PY, Jaggi TK, Chan LLY, Chotirmall SH: The airway microbiome in COPD, bronchiectasis and bronchiectasis-COPD overlap. Clin Respir J 2021; 15:123–33 [DOI] [PubMed] [Google Scholar]
- 27.Saint-Criq V, Lugo-Villarino G, Thomas M: Dysbiosis, malnutrition and enhanced gut-lung axis contribute to age-related respiratory diseases. Ageing Res Rev 2021; 66:101235. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Carrasco V, Soriano-Lerma A, Soriano M, Gutiérrez-Fernández J, Garcia-Salcedo JA: Urinary Microbiome: Yin and Yang of the Urinary Tract. Front Cell Infect Microbiol 2021; 11:617002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Punzón-Jiménez P, Labarta E: The impact of the female genital tract microbiome in women health and reproduction: a review. J Assist Reprod Genet 2021. doi: 10.1007/s10815-021-02247-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen JM: The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017; 151:363–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, Zabari M, Brik RB-Z, Kviatcovsky D, Zmora N, Cohen Y, Bar N, Levi I, Amar N, Mehlman T, Brandis A, Biton I, Kuperman Y, Tsoory M, Alfahel L, Harmelin A, Schwartz M, Israelson A, Arike L, Johansson MEV, Hansson GC, Gotkine M, Segal E, Elinav E: Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 2019; 572:474–80 [DOI] [PubMed] [Google Scholar]
- 32.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY: The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018; 173:1728–1741.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Attar N: Microbiome: Good for the gut, good for the brain. Nat Rev Microbiol 2016; 14:269 [Google Scholar]
- 34.Shahi SK, Freedman SN, Mangalam AK: Gut microbiome in multiple sclerosis: The players involved and the roles they play. Gut Microbes 2017; 8:607–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pröbstel A-K, Baranzini SE: The Role of the Gut Microbiome in Multiple Sclerosis Risk and Progression: Towards Characterization of the “MS Microbiome.” Neurotherapeutics 2018; 15:126–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erny D, Angelis ALH de, Prinz M: Communicating systems in the body: how microbiota and microglia cooperate. Immunology 2017; 150:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Tian X, Maruyama D, Arjomandi M, Prakash A: Lung immune tone via gut-lung axis: gut-derived LPS and short-chain fatty acids’ immunometabolic regulation of lung IL-1β, FFAR2, and FFAR3 expression. Am J Physiol Lung Cell Mol Physiol 2021; 321:L65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashley SL, Sjoding MW, Popova AP, Cui TX, Hoostal MJ, Schmidt TM, Branton WR, Dieterle MG, Falkowski NR, Baker JM, Hinkle KJ, Konopka KE, Erb-Downward JR, Huffnagle GB, Dickson RP: Lung and gut microbiota are altered by hyperoxia and contribute to oxygen-induced lung injury in mice. Sci Transl Med 2020; 12:eaau9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, Kanbar J, Miller-Montgomery S, Heaton R, Mckay R, Patel SP, Swafford AD, Knight R: Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020; 579:567–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré J, Störsrud S, Le Nevé B, Öhman L, Simrén M: Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology 2017; 152:111–123.e8 [DOI] [PubMed] [Google Scholar]
- 41.Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, Moayyedi P: Gut Microbiota in Patients With Irritable Bowel Syndrome-A Systematic Review. Gastroenterology 2019; 157:97–108 [DOI] [PubMed] [Google Scholar]
- 42.Minerbi A, Gonzalez E, Brereton NJB, Anjarkouchian A, Dewar K, Fitzcharles M-A, Chevalier S, Shir Y: Altered microbiome composition in individuals with fibromyalgia. Pain 2019; 160:2589–602 [DOI] [PubMed] [Google Scholar]
- 43.Bashiardes S, Tuganbaev T, Federici S, Elinav E: The microbiome in anti-cancer therapy. Semin Immunol 2017; 32:74–81 [DOI] [PubMed] [Google Scholar]
- 44.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA: The microbiome, cancer, and cancer therapy. Nat Med 2019; 25:377–88 [DOI] [PubMed] [Google Scholar]
- 45.Mars RAT, Frith M, Kashyap PC: Functional Gastrointestinal Disorders and the Microbiome—What Is the Best Strategy for Moving Microbiome-based Therapies for Functional Gastrointestinal Disorders into the Clinic? Gastroenterology 2021; 160:538–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Tian J, Yang B: Targeting gut microbiome: A novel and potential therapy for autism. Life Sci 2018; 194:111–9 [DOI] [PubMed] [Google Scholar]
- 47.Nood E van, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, Vos WM de, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, Speelman P, Dijkgraaf MGW, Keller JJ: Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407–15 [DOI] [PubMed] [Google Scholar]
- 48.El-Salhy M, Hatlebakk JG, Gilja OH, Bråthen Kristoffersen A, Hausken T: Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut 2019. doi: 10.1136/gutjnl-2019-319630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, Rotin D, Anafi L, Avivi C, Melnichenko J, Steinberg-Silman Y, Mamtani R, Harati H, Asher N, Shapira-Frommer R, Brosh-Nissimov T, Eshet Y, Ben-Simon S, Ziv O, Khan MAW, Amit M, Ajami NJ, Barshack I, Schachter J, Wargo JA, Koren O, Markel G, Boursi B.: Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021; 371:602–9 [DOI] [PubMed] [Google Scholar]
- 50.Merrick B, Allen L, Masirah M Zain N, Forbes B, Shawcross DL, Goldenberg SD: Regulation, risk and safety of Faecal Microbiota Transplant. Infection Prevention in Practice 2020; 2:100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang H, Fu L, Li X, Lu C, Su Y, Xiong K, Zhang L: Long-term efficacy and safety of monotherapy with a single fresh fecal microbiota transplant for recurrent active ulcerative colitis: a prospective randomized pilot study. Microb Cell Fact 2021; 20:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shogbesan O, Poudel DR, Victor S, Jehangir A, Fadahunsi O, Shogbesan G, Donato A: A Systematic Review of the Efficacy and Safety of Fecal Microbiota Transplant for Clostridium difficile Infection in Immunocompromised Patients. Can J Gastroenterol Hepatol 2018; 2018:1394379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Bi J-J, Guo G-J, Yang L, Zhu B, Zhan G-F, Li S, Huang N-N, Hashimoto K, Yang C, Luo A-L: Abnormal composition of gut microbiota contributes to delirium-like behaviors after abdominal surgery in mice. CNS Neurosci Ther 2019; 25:685–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liufu N, Liu L, Shen S, Jiang Z, Dong Y, Wang Y, Culley D, Crosby G, Cao M, Shen Y, Marcantonio E, Xie Z, Zhang Y: Anesthesia and surgery induce age-dependent changes in behaviors and microbiota. Aging (Albany NY) 2020; 12:1965–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang XL, Gu XY, Zhou XX, Chen XM, Zhang X, Yang YT, Qin Y, Shen L, Yu WF, Su DS: Intestinal dysbacteriosis mediates the reference memory deficit induced by anaesthesia/surgery in aged mice. Brain Behav Immun 2019; 80:605–15 [DOI] [PubMed] [Google Scholar]
- 56.Meng F, Li N, Li D, Song B, Li L: The presence of elevated circulating trimethylamine N-oxide exaggerates postoperative cognitive dysfunction in aged rats. Behav Brain Res 2019; 368:111902. [DOI] [PubMed] [Google Scholar]
- 57.Wen J, Ding Y, Wang L, Xiao Y: Gut microbiome improves postoperative cognitive function by decreasing permeability of the blood-brain barrier in aged mice. Brain Res Bull 2020; 164:249–56 [DOI] [PubMed] [Google Scholar]
- 58.Xu X, Hu Y, Yan E, Zhan G, Liu C, Yang C: Perioperative neurocognitive dysfunction: thinking from the gut? Aging (Albany NY) 2020; 12:15797–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang P, Shan W, Zuo Z: Perioperative use of cefazolin ameliorates postoperative cognitive dysfunction but induces gut inflammation in mice. J Neuroinflammation 2018; 15:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo A, Li S, Wang X, Xie Z, Li S, Hua D: Cefazolin Improves Anesthesia and Surgery-Induced Cognitive Impairments by Modulating Blood-Brain Barrier Function, Gut Bacteria and Short Chain Fatty Acids. Front Aging Neurosci 2021; 13:748637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goverman J, Woods A, Larson L, Weiner LP, Hood L, Zaller DM: Transgenic mice that express a myelin basic protein-specific T cell receptor develop spontaneous autoimmunity. Cell 1993; 72:551–60 [DOI] [PubMed] [Google Scholar]
- 62.Yang X-D, Wang L-K, Wu H-Y, Jiao L: Effects of prebiotic galacto-oligosaccharide on postoperative cognitive dysfunction and neuroinflammation through targeting of the gut-brain axis. BMC Anesthesiol 2018; 18:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabbiano S, Suárez-Zamorano N, Chevalier C, Lazarević V, Kieser S, Rigo D, Leo S, Veyrat-Durebex C, Gaïa N, Maresca M, Merkler D, Gomez de Agüero M, Macpherson A, Schrenzel J, Trajkovski M: Functional Gut Microbiota Remodeling Contributes to the Caloric Restriction-Induced Metabolic Improvements. Cell Metab 2018; 28:907–921.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Z, Dai X, Zhang H, Shi R, Hui Y, Jin X, Zhang W, Wang L, Wang Q, Wang D, Wang J, Tan X, Ren B, Liu X, Zhao T, Wang J, Pan J, Yuan T, Chu C, Lan L, Yin F, Cadenas E, Shi L, Zhao S, Liu X: Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment. Nat Commun 2020; 11:855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng X, Uchida Y, Koch L, Britton S, Hu J, Lutrin D, Maze M: Exercise Prevents Enhanced Postoperative Neuroinflammation and Cognitive Decline and Rectifies the Gut Microbiome in a Rat Model of Metabolic Syndrome. Front Immunol 2017; 8:1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maekawa M, Yoshitani K, Yahagi M, Asahara T, Shishido Y, Fukushima S, Tadokoro N, Fujita T, Ohnishi Y: Association between postoperative changes in the gut microbiota and pseudopsia after cardiac surgery: prospective observational study. BMC Surg 2020; 20:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao Z-W, Yang X, Zhao B-C, Deng F, Jiang Y-M, Pan W-Y, Chen X-D, Zhou B-W, Zhang W-J, Hu J-J, Zhu L, Liu K-X: Predictive and Preventive Potential of Preoperative Gut Microbiota in Chronic Postoperative Pain in Breast Cancer Survivors. Anesth Analg 2021. doi: 10.1213/ANE.0000000000005713 [DOI] [PubMed] [Google Scholar]
- 68.Maeda T, Kishioka S: PPAR and Pain. Int Rev Neurobiol 2009; 85:165–77 [DOI] [PubMed] [Google Scholar]
- 69.Brenner D, Cherry P, Switzer T, Butt I, Stanton C, Murphy K, McNamara B, Iohom G, O’Mahony SM, Shorten G: Pain after upper limb surgery under peripheral nerve block is associated with gut microbiome composition and diversity. Neurobiol Pain 2021; 10:100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maier L, Typas A: Systematically investigating the impact of medication on the gut microbiome. Curr Opin Microbiol 2017; 39:128–35 [DOI] [PubMed] [Google Scholar]
- 71.Weersma RK, Zhernakova A, Fu J: Interaction between drugs and the gut microbiome. Gut 2020; 69:1510–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jackson MA, Verdi S, Maxan M-E, Shin CM, Zierer J, Bowyer RCE, Martin T, Williams FMK, Menni C, Bell JT, Spector TD, Steves CJ: Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun 2018; 9:2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Serbanescu MA, Mathena RP, Xu J, Santiago-Rodriguez T, Hartsell TL, Cano RJ, Mintz CD: General Anesthesia Alters the Diversity and Composition of the Intestinal Microbiota in Mice. Anesth Analg 2019; 129:e126–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han C, Zhang Z, Guo N, Li X, Yang M, Peng Y, Ma X, Yu K, Wang C: Effects of Sevoflurane Inhalation Anesthesia on the Intestinal Microbiome in Mice. Front Cell Infect Microbiol 2021; 11:633527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo N, Zhang Z, Han C, Chen L, Zheng X, Yu K, Zhang Z, Wang C: Effects of continuous intravenous infusion of propofol on intestinal flora in rats. Biomed Pharmacother 2021; 134:111080. [DOI] [PubMed] [Google Scholar]
- 76.Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S: Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 2009; 4:e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang F, Meng J, Zhang L, Johnson T, Chen C, Roy S: Morphine induces changes in the gut microbiome and metabolome in a morphine dependence model. Sci Rep 2018; 8:3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee K, Vuong HE, Nusbaum DJ, Hsiao EY, Evans CJ, Taylor AMW: The gut microbiota mediates reward and sensory responses associated with regimen-selective morphine dependence. Neuropsychopharmacol 2018; 43:2606–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banerjee S, Sindberg G, Wang F, Meng J, Sharma U, Zhang L, Dauer P, Chen C, Dalluge J, Johnson T, Roy S: Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal Immunol 2016. doi: 10.1038/mi.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hakimian JK, Dong TS, Barahona JA, Lagishetty V, Tiwari S, Azani D, Barrera M, Lee S, Severino AL, Mittal N, Cahill CM, Jacobs JP, Walwyn WM: Dietary Supplementation with Omega-3 Polyunsaturated Fatty Acids Reduces Opioid-Seeking Behaviors and Alters the Gut Microbiome. Nutrients 2019; 11:E1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang L, Meng J, Ban Y, Jalodia R, Chupikova I, Fernandez I, Brito N, Sharma U, Abreu MT, Ramakrishnan S, Roy S: Morphine tolerance is attenuated in germfree mice and reversed by probiotics, implicating the role of gut microbiome. PNAS 2019; 116:13523–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang M, Mischel RA, Bhave S, Komla E, Cho A, Huang C, Dewey WL, Akbarali HI: The effect of gut microbiome on tolerance to morphine mediated antinociception in mice. Sci Rep 2017; 7:42658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kenyon EM, Calabrese EJ: Extent and implications of interspecies differences in the intestinal hydrolysis of certain glucuronide conjugates. Xenobiotica 1993; 23:373–81 [DOI] [PubMed] [Google Scholar]
- 84.Chen P, Billiar T: Gut Microbiota and Multiple Organ Dysfunction Syndrome (MODS). Adv Exp Med Biol 2020; 1238:195–202 [DOI] [PubMed] [Google Scholar]
- 85.Moron R, Galvez J, Colmenero M, Anderson P, Cabeza J, Rodriguez-Cabezas ME: The Importance of the Microbiome in Critically Ill Patients: Role of Nutrition. Nutrients 2019; 11:E3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu W, Cheng M, Li J, Zhang P, Fan H, Hu Q, Han M, Su L, He H, Tong Y, Ning K, Long Y: Classification of the Gut Microbiota of Patients in Intensive Care Units During Development of Sepsis and Septic Shock. Genomics, Proteomics & Bioinformatics 2020; 18:696–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klingensmith NJ, Coopersmith CM: The Gut as the Motor of Multiple Organ Dysfunction in Critical Illness. Crit Care Clin 2016; 32:203–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, Delhaes L: The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front Cell Infect Microbiol 2020; 10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang W-C, Wu M-F, Huang C-C, Liu S-Y, Chen H-C, Chen Y-Y, Hsu J-Y, Huang C-C: Dynamics of the lung microbiome in intensive care patients with chronic obstructive pulmonary disease and community-acquired pneumonia. Sci Rep 2020; 10:11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K:The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol 2019; 16:461–78 [DOI] [PubMed] [Google Scholar]
- 91.Ford AC, Lacy BE, Talley NJ: Irritable Bowel Syndrome. N Engl J Med 2017; 376:2566–78 [DOI] [PubMed] [Google Scholar]
- 92.Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, Tang X, Sun Z, Kalari KR, Korem T, Bhattarai Y, Zheng T, Bar N, Frost G, Johnson AJ, Treuren W van, Han S, Ordog T, Grover M, Sonnenburg J, D’Amato M, Camilleri M, Elinav E, Segal E, Blekhman R, Farrugia G, Swann JR, Knights D, Kashyap PC: Longitudinal Multi-omics Reveals Subset-Specific Mechanisms Underlying Irritable Bowel Syndrome. Cell 2020; 182:1460–1473.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Palma G, Lynch MDJ, Lu J, Dang VT, Deng Y, Jury J, Umeh G, Miranda PM, Pigrau Pastor M, Sidani S, Pinto-Sanchez MI, Philip V, McLean PG, Hagelsieb M-G, Surette MG, Bergonzelli GE, Verdu EF, Britz-McKibbin P, Neufeld JD, Collins SM, Bercik P: Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med 2017; 9 [DOI] [PubMed] [Google Scholar]
- 94.Lucarini E, Di Pilato V, Parisio C, Micheli L, Toti A, Pacini A, Bartolucci G, Baldi S, Niccolai E, Amedei A, Rossolini GM, Nicoletti C, Cryan JF, O’Mahony SM, Ghelardini C, Di Cesare Mannelli L: Visceral sensitivity modulation by faecal microbiota transplantation: the active role of gut bacteria in pain persistence. Pain 2021. doi: 10.1097/j.pain.0000000000002438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.El-Salhy M, Hatlebakk JG, Hausken T: Diet in Irritable Bowel Syndrome (IBS): Interaction with Gut Microbiota and Gut Hormones. Nutrients 2019; 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.El-Salhy M, Mazzawi T: Fecal microbiota transplantation for managing irritable bowel syndrome. Expert Rev Gastroenterol Hepatol 2018; 12:439–45 [DOI] [PubMed] [Google Scholar]
- 97.Arora HC, Eng C, Shoskes DA: Gut microbiome and chronic prostatitis/chronic pelvic pain syndrome. Ann Transl Med 2017; 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nickel JC, Stephens A, Landis JR, Mullins C, Bokhoven A van, Lucia MS, Ehrlich GD, MAPP Research Network: Assessment of the Lower Urinary Tract Microbiota during Symptom Flare in Women with Urologic Chronic Pelvic Pain Syndrome: A MAPP Network Study. J Urol 2016; 195:356–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shoskes DA, Wang H, Polackwich AS, Tucky B, Altemus J, Eng C: Analysis of Gut Microbiome Reveals Significant Differences between Men with Chronic Prostatitis/Chronic Pelvic Pain Syndrome and Controls. J Urol 2016; 196:435–41 [DOI] [PubMed] [Google Scholar]
- 100.Nickel JC, Stephens-Shields AJ, Landis JR, Mullins C, Bokhoven A van, Lucia MS, Henderson JP, Sen B, Krol JE, Ehrlich GD, MAPP Research Network: A Culture-Independent Analysis of the Microbiota of Female Interstitial Cystitis/Bladder Pain Syndrome Participants in the MAPP Research Network. J Clin Med 2019; 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fitzcharles M-A, Cohen SP, Clauw DJ, Littlejohn G, Usui C, Häuser W: Nociplastic pain: towards an understanding of prevalent pain conditions. Lancet 2021; 397:2098–110 [DOI] [PubMed] [Google Scholar]
- 102.Ramakrishna C, Corleto J, Ruegger PM, Logan GD, Peacock BB, Mendonca S, Yamaki S, Adamson T, Ermel R, McKemy D, Borneman J, Cantin EM: Dominant Role of the Gut Microbiota in Chemotherapy Induced Neuropathic Pain. Sci Rep 2019; 9:20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clos-Garcia M, Andrés-Marin N, Fernández-Eulate G, Abecia L, Lavín JL, Liempd S van, Cabrera D, Royo F, Valero A, Errazquin N, Vega MCG, Govillard L, Tackett MR, Tejada G, Gónzalez E, Anguita J, Bujanda, Orcasitas AMC, Aransay AM, Maíz O, López de Munain A, Falcón-Pérez JM: Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine 2019; 46:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Michalsen A, Riegert M, Lüdtke R, Bäcker M, Langhorst J, Schwickert M, Dobos GJ: Mediterranean diet or extended fasting’s influence on changing the intestinal microflora, immunoglobulin A secretion and clinical outcome in patients with rheumatoid arthritis and fibromyalgia: an observational study. BMC Complement Altern Med 2005; 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Freidin MB, Stalteri MA, Wells PM, Lachance G, Baleanu A-F, Bowyer RCE, Kurilshikov A, Zhernakova A, Steves CJ, Williams FMK: An association between chronic widespread pain and the gut microbiome. Rheumatology (Oxford) 2020. doi: 10.1093/rheumatology/keaa847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Frémont M, Coomans D, Massart S, De Meirleir K: High-throughput 16S rRNA gene sequencing reveals alterations of intestinal microbiota in myalgic encephalomyelitis/chronic fatigue syndrome patients. Anaerobe 2013; 22:50–6 [DOI] [PubMed] [Google Scholar]
- 107.Giloteaux L, Goodrich JK, Walters WA, Levine SM, Ley RE, Hanson MR: Reduced diversity and altered composition of the gut microbiome in individuals with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome 2016; 4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nagy-Szakal D, Williams BL, Mishra N, Che X, Lee B, Bateman L, Klimas NG, Komaroff AL, Levine S, Montoya JG, Peterson DL, Ramanan D, Jain K, Eddy ML, Hornig M, Lipkin WI: Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome; London 2017; 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen P, Wang C, Ren Y-N, Ye Z-J, Jiang C, Wu Z-B: Alterations in the gut microbiota and metabolite profiles in the context of neuropathic pain. Mol Brain 2021; 14:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shen S, Lim G, You Z, Ding W, Huang P, Ran C, Doheny J, Caravan P, Tate S, Hu K, Kim H, McCabe M, Huang B, Xie Z, Kwon D, Chen L, Mao J: Gut microbiota is critical for the induction of chemotherapy-induced pain. Nat Neurosci 2017; 20:1213–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ding W, You Z, Chen Q, Yang L, Doheny J, Zhou X, Li N, Wang S, Hu K, Chen L, Xia S, Wu X, Wang C, Zhang C, Chen L, Ritchie C, Huang P, Mao J, Shen S: Gut Microbiota Influences Neuropathic Pain Through Modulating Proinflammatory and Anti-inflammatory T Cells. Anesth Analg 2020. doi: 10.1213/ANE.0000000000005155 [DOI] [PubMed] [Google Scholar]
- 112.Zhou F, Wang X, Han B, Tang X, Liu R, Ji Q, Zhou Z, Zhang L: Short-chain fatty acids contribute to neuropathic pain via regulating microglia activation and polarization. Mol Pain 2021; 17:1744806921996520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bonomo RR, Cook TM, Gavini CK, White CR, Jones JR, Bovo E, Zima AV, Brown IA, Dugas LR, Zakharian E, Aubert G, Alonzo F, Calcutt NA, Mansuy-Aubert V: Fecal transplantation and butyrate improve neuropathic pain, modify immune cell profile, and gene expression in the PNS of obese mice. Proc Natl Acad Sci U S A 2020; 117:26482–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Z, Wu H, Chen Y, Chen H, Wang X, Yuan W: Lactobacillus paracasei S16 Alleviates Lumbar Disc Herniation by Modulating Inflammation Response and Gut Microbiota. Frontiers in Nutrition 2021; 8:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reichenberger ER, Alexander GM, Perreault MJ, Russell JA, Schwartzman RJ, Hershberg U, Rosen G: Establishing a relationship between bacteria in the human gut and complex regional pain syndrome. Brain Behav Immun 2013; 29:62–9 [DOI] [PubMed] [Google Scholar]
- 116.Chen J, Wang Q, Wang A, Lin Z: Structural and Functional Characterization of the Gut Microbiota in Elderly Women With Migraine. Front Cell Infect Microbiol 2019; 9:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiang W, Wang T, Liu C, Deng M, Ren X, Wang F, Zhang Y, Yu X, Yao L, Wang Y: A 16S rRNA gene sequencing based study of oral microbiota in migraine patients in China. Bioengineered 2021; 12:2523–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Favazzo LJ, Hendesi H, Villani DA, Soniwala S, Dar Q-A, Schott EM, Gill SR, Zuscik MJ: The gut microbiome-joint connection: implications in osteoarthritis. Curr Opin Rheumatol 2020; 32:92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bergot A-S, Giri R, Thomas R: The microbiome and rheumatoid arthritis. Best Pract Res Clin Rheumatol 2019; 33:101497. [DOI] [PubMed] [Google Scholar]
- 120.Xu Y-Y, Tan X, He Y-T, Zhou Y-Y, He X-H, Huang R-Y: Role of gut microbiome in ankylosing spondylitis: an analysis of studies in literature. Discov Med 2016; 22:361–70 [PubMed] [Google Scholar]
- 121.Basson A, Trotter A, Rodriguez-Palacios A, Cominelli F: Mucosal Interactions between Genetics, Diet, and Microbiome in Inflammatory Bowel Disease. Front Immunol 2016; 7:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Torun A, Hupalowska A, Trzonkowski P, Kierkus J, Pyrzynska B: Intestinal Microbiota in Common Chronic Inflammatory Disorders Affecting Children. Front Immunol 2021; 12:642166. [DOI] [PMC free article] [PubMed] [Google Scholar]


