Abstract
Species-specific 16S rRNA-targeted, Cy3 (indocarbocyanine)-labeled oligonucleotide probes were designed and validated to quantify different Eubacterium species in human fecal samples. Probes were directed at Eubacterium barkeri, E. biforme, E. contortum, E. cylindroides (two probes), E. dolichum, E. hadrum, E. lentum, E. limosum, E. moniliforme, and E. ventriosum. The specificity of the probes was tested with the type strains and a range of common intestinal bacteria. With one exception, none of the probes showed cross-hybridization under stringent conditions. The species-specific probes were applied to fecal samples obtained from 12 healthy volunteers. E. biforme, E. cylindroides, E. hadrum, E. lentum, and E. ventriosum could be determined. All other Eubacterium species for which probes had been designed were under the detection limit of 107 cells g (dry weight) of feces−1. The cell counts obtained are essentially in accordance with the literature data, which are based on colony counts. This shows that whole-cell in situ hybridization with species-specific probes is a valuable tool for the enumeration of Eubacterium species in feces.
The genus Eubacterium (41) contains anaerobic, non-spore-forming, gram-positive rods which are distinguished from other genera mainly on the basis of negative metabolic characteristics (37). In the human intestinal tract, Eubacterium is the second most common genus after the genus Bacteroides and is more common than the genus Bifidobacterium (16). The importance of members of the genus Eubacterium has been reported previously (10, 24, 38, 48). Since the identification of Eubacterium species based on phenotypic traits requires experience and is time-consuming (5, 15, 16, 36, 44), many studies involving human fecal flora composition have refrained from looking at this genus (13, 20, 34, 42).
Considerable effort has been invested in the application of molecular techniques such as PCR (19, 27, 50) and hybridization (12, 25, 52) for the identification of fecal bacteria. However, the extraction, purification, and amplification of nucleic acids by PCR from fecal samples are often selective and limited. In contrast, whole-cell hybridization with fluorescently labeled, 16S rRNA-targeted oligonucleotide probes allows the determination of the numerical abundance of bacteria, including unculturable strains (39), in various ecosystems such as water (1, 26, 32), sludge (23), and fecal samples (17, 18, 29).
In contrast to the genus Bifidobacterium, for example, which is a phylogenetically and phenotypically well-defined taxon, Eubacterium species (9, 31) are phylogenetically diverse and thus it is not possible to design a genus-specific probe for Eubacterium spp. Therefore, probes for phylogenetic clusters or species have to be considered. The development and validation of a probe for the detection of a Eubacterium species has been reported recently (45).
The purpose of this study was to develop and apply 16S rRNA-targeted oligonucleotide probes to human feces for the detection of numerically dominant fecal Eubacterium species by whole-cell hybridization. The choice of bacteria was based on the reports by Finegold et al. (16) and Moore and Holdeman (36) and included Eubacterium barkeri, E. biforme, E. cylindroides, E. contortum, E. dolichum, E. hadrum, E. lentum, E. limosum, E. moniliforme, and E. ventriosum. The specific detection and enumeration of Eubacterium species with specific oligonucleotide probes facilitates the exploration of the microbial diversity in the human gut ecosystem and is expected to contribute to resolving the ecological role of bacteria in their environment.
MATERIALS AND METHODS
Organisms and culture conditions.
All reference strains used in this study were obtained from the sources indicated in the Appendix (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ], Braunschweig, Germany; Deutsches Institut für Ernährungsforschung [DIFE], Bergholz-Rehbrücke, Germany; American Type Culture Collection [ATCC], Rockville, Md.). All DIFE strains are human fecal isolates from our laboratory and were identified by using the Vitek System (bioMérieux, Nürtingen, Germany). All strains, except for Clostridium populeti and Eubacterium lentum, which were grown on the media described in the DSMZ catalog, were cultured at 37°C under strictly anoxic conditions with N2-CO2 (80:20 [vol/vol]) as a gas phase (8, 22) in ST medium (ST), which contained (per liter): 9 g of tryptically digested peptone from meat, 1 g of proteose peptone, 3 g of meat extract, 4 g of yeast extract, 6 g of glucose, 3 g of NaCl, 2 g of Na2HPO4, 0.5 ml of Tween 80, 0.25 g of l-cystine, 0.25 g of l-cysteine · HCl, 0.1 g of MgSO4 · 7H2O, 5 mg of FeSO4 · 7H2O, and 3.4 mg of MnSO4 · 2H2O (pH 7.0).
Oligonucleotide probes.
Oligonucleotides targeting the small subunit rRNA sequences of the Eubacterium spp. were designed with the Arb software package (46). The probe sequences were then checked by using the Check-Probe function of the Ribosomal Database Project (RDP) software package (31) and the alignment function of the EMBL database. The designed probes were named according to the nomenclature suggested by the Oligonucleotide Probe Database (OPD) (2). The theoretical dissociation temperature (TD) of the designed probes was >50°C (47), and the G+C content was at least 50%. For practical use within this manuscript the probes were named in such a way that they indicate only the species name and the Escherichia coli (7) position, e.g., the probe for E. barkeri (S-S-E.bar-1237-a-A-18) was abbreviated E.bar1237.
Nucleic acid extraction.
For nucleic acid extraction the bacterial strains were grown anoxically for 12 to 36 h in 100 ml of ST. The cells were harvested by centrifugation for 15 min at 4,000 × g at 4°C. The resulting sediment was subjected to nucleic acid extraction by following protocol number five of the InViTek DNA-Isolation Kit III (InViTek GmbH, Berlin, Germany) with a slight modification. To improve lysis, cells were first incubated for 30 min in 25% sucrose (wt/vol) in H2O, centrifuged as described above, and diluted in 2 ml of Lysis-Buffer D (InViTek). The nucleic acid concentration was measured photometrically at 280 nm, and the quality was checked in ethidium bromide-stained agarose gels (1% [wt/vol]).
DNA amplification with PCR and specificity testing.
The species-specific probes were used in PCR as reverse primers; the nucleotide sequence of the forward primer was 5′-AGAGTTTGATCMTGGCTCAG-3′ (28). PCR amplification was performed with a PCR thermal cycler (Hybaid, Heidelberg, Germany) under the following conditions: denaturation at 94°C for 4 min, followed by 33 cycles of denaturation at 94°C for 1.5 min, annealing at 68°C for 1.5 min, and extension at 72°C for 1.5 min. After completion, an additional extension step was performed at 72°C for 6 min, and the samples were then chilled to 4°C. Reaction mixtures (final volume, 50 μl) contained 16 mM (NH4)2SO4, 50 mM Tris-HCl (pH 8.8), 0.1% Tween 20 (vol/vol), 5 mM MgCl2, 250 μM concentrations of each deoxynucleotide triphosphate, 50 pM concentrations of each oligonucleotide primer, 0.1% gelatin (wt/vol), and 2.5 U of Taq polymerase (InViTek). Template DNA was added to a final concentration of 1 ng μl−1. The PCR products were visualized as described for the nucleic acid extraction. Probes were tested in PCR experiments with the reference organisms given in the Appendix.
Detection limits of PCR-based identification in pure culture and feces.
To determine the detection limit of PCR-based identification in pure cultures, defined amounts of cells from the late logarithmic growth phase were serially diluted in phosphate-buffered saline (PBS) (130 mM NaCl, 10 mM NaH2PO4-Na2HPO4; pH 7.2) and nucleic acids were extracted according to the above-described protocol. To determine the detection limit of this PCR-based method in feces, cells were diluted as described above and added to 100 mg of autoclaved feces and the nucleic acids were extracted.
Cell fixation for whole-cell hybridization.
More than 90 reference strains representing the human intestinal flora were used for probe validation (see Appendix). As a positive control, the bacterium-specific probe S-D-Bact-0338-a-A-18 was used (3). Bacterial cells were grown overnight on Columbia blood agar plates (bioMérieux) incubated at 37°C in anaerobic jars under strictly anoxic conditions with N2-CO2 (80:20 [vol/vol]) as the gas phase. Bacteria that did not grow on these plates were cultured in 10 ml of ST before cell fixation and were subsequently harvested as described above. The fixation of the bacteria was dependent on their Gram stain reaction: gram-positive reference strains were diluted in PBS and adjusted to 109 cells/ml, centrifuged for 3 min at 2,000 × g, washed with 1 ml of PBS, and subsequently resuspended in 1 ml of 50% ethanol-PBS (vol/vol) (43), whereas gram-negative strains were resuspended in 1 ml of 4% paraformaldehyde (wt/vol) and stored for at least 4 h at 4°C (3). The cells were then sedimented by centrifugation and fixed by the same procedure as described for the gram-positive reference strains. The fixed cells were stored at −20°C until used.
Fecal samples.
Fresh feces were collected from healthy humans without an antibiotic therapy in the last 6 months before the start of the study. For the analysis by whole-cell hybridization a 0.1-g (wet weight) feces specimen was added to 0.9 ml of sterile PBS and then mixed by inverting and vortexing the tube for 5 to 10 min. For total cell counts, 500 μl of this dilution was fixed with paraformaldehyde and ethanol, respectively, as described above for the pure cultures.
Hybridization.
Hybridization was done on 70% ethanol rinsed and dried Teflon-coated slides (Marienfeld, Bad Mergentheim, Germany) with eight wells for independent positioning of the samples. Slides were silanized with 2% APES (3-aminopropyl-triethoxysilane) to improve cell distribution (30). Aliquots of fixed cells and fecal samples were spotted on single wells, air dried, and dehydrated by passing them through an ethanol series (60, 80, and 96% [vol/vol]) for 3 min each. Hybridizations were performed after addition of 2 μl (50 pmol/μl) of the species-specific Cy3 (indocarbocyanine; Interactiva, Ulm, Germany)-labeled probe in humid chambers overnight at the given temperature (Table 1). The hybridization buffer (10 μl) contained 0.9 M NaCl, 20 mM Tris-HCl (pH 7.2), 0.01% (wt/vol) sodium dodecyl sulfate, and no formamide if not stated otherwise.
TABLE 1.
Developed and validated 16S rRNA-targeted species-specific oligonucleotide probes for Eubacterium spp. and the corresponding sequence accession numbersa
| Probe (accession no.; TD, TH, TE [°C]) and related strains | Sequence |
|---|---|
| S-S-E.bar-1237-a-A-18 (M23927; 51, 51, 53) | 3′TTACCCAACCCTGTTTCC′ |
| 5′AAUGGGUUGGGACAAAGG′ (target) | |
| Acidaminococcus fermentans | .....UCG.CA....... |
| Ruminococcus productus | C.AU..C...U....... |
| S-S-E.bif-0462-a-A-18 (M59230; 51, 51, 56) | 3′CCCTTACTACTCACTCAC′ |
| 5′GGGAAUGAUGAGUGAGUG′ (target) | |
| Eubacterium cylindroides ATCC 27803 | .........CC...G... |
| Streptococcus pleomorphus | ..........UCG..... |
| Fusobacterium mortiferum | ...UG....CU.C..N.. |
| Fusobacterium varium | ...UG....CU.C..N.. |
| S-S-E.con-1122-a-A-20 (L34615; 61, 55, 55) | 3′GGCCTAAACCGCTGGCTACT′ |
| 5′AGUAGCCAGCGGUUUAGGCC′ (target) | |
| Eubacterium fissicatena | ..............G.C.G. |
| S-St-E.cyl-0461-a-A-18 (L34617; 53, 51, 53) | 3′TCCCTTACTAGGCACCCA′ |
| 5′AGGGAAUGAUCCGUGGGU′ (target) | |
| Eubacterium biforme | ..........GA...A.. |
| Eubacterium uniforme | G.N..G.A.NN....... |
| Eubacterium cylindroides ATCC 27528 | ...A....C.AU...... |
| Fusobacterium varium | GA..G...U....A.... |
| S-St-E.cyl-0466-a-A-18 (L34616; 53, 51, 53) | 3′TACGATACACCCACTGCC′ |
| 5′AUGCUAUGUGGGUGACGG′ (target) | |
| Clostridium innocuum | ........G.A....... |
| Eubacterium cylindroides ATCC 27803 | ..A.CC.........U.. |
| Clostridium spiroforme | ..A...CC.A........ |
| Eubacterium biforme | ..A..GA...A....U.. |
| S-S-E.dol-0183-a-A-18 (L34682; 55, 51, 53) | 3′TGTCTCCGAGATGCCTCG′ |
| 5′CGAGGCAUCUCGGAGACA′ (target) | |
| S-S-E.had-0579-a-A-20 (ARB 671C; 59, 54, 54) | 3′GACTTGCCATACCACCTACG′ |
| 5′CGUAGGUGGUAUGGCAAGUC′ (target) | |
| Eubacterium ventriosum | ......C..C.......... |
| Eubacterium uniforme | ......C.A........... |
| Bacteroides fragilis | .........AC...U..... |
| S-S-E.len-0194-a-A-18 (AB011817; 55, 51, 53) | 3′CCTTGCCGTCTGGGCTTT′ |
| 5′AAAGCCCAGACGGCAAGG′ (target) | |
| Eubacterium ventriosum | ......C..C........ |
| Eubacterium uniforme | ......C.A......... |
| Bacteroides fragilis | .........AC...U... |
| S-S-E.lim-1433-a-A-18 (M59120; 55, 51, 59) | 3′TCGGACACTCTCTTGGCG′ |
| 5′AGCCUGUGAGAGAACCGC′ (target) | |
| Eubacterium barkeri | ....A......C...... |
| Pseudoramibacter alactolyticus | ...A......C....... |
| Eubacterium cylindroides ATCC 27528 | ....G...GC.U...... |
| Eubacterium tortuosum | ....G...GC.U...... |
| S-S-E.mon-0084-a-A-18 (L34622; 53, 51, 52) | 3′CCGCTAATCCATTTCCCG′ |
| 5′CGGGAAAUGGAUUAGCGG′ (target) | |
| S-S-E.ven-0066-a-A-18 (L34421; 51, 55, 50) | 3′TCTGTCCAAGGTGCTTCG′ |
| 5′CGAAGCACCUUGGACAGA′ (target) |
Alignments of the probe sequences and their 16S rRNA target sites. Also shown are the potential 16S rRNA binding sites of nontarget organisms commonly found in human and animal feces. Accession numbers are from GenBank except for E. hadrum which only has an ARB accession number (46). Probes are named in accordance with the OPD (2). Sequences show the closest nontarget sequences available in the databases. Only nucleotides that differ from the target sequences are shown. N indicates a not yet determined nucleotide. The first TD refers to the theoretical dissociation temperature based on the contribution of GC and AT pairs to the oligonucleotide duplex stability (45). TH refers to the hybridization temperature used in the study. TE is related to the hybridization temperature as determined by the method by De Los Reyes et al. (11).
Probe specificity testing.
To test the specificity of the designed probes the bacterial reference strains listed in the Appendix were hybridized with the respective probe. Four strains per slide were tested, i.e., two wells per strain. One of the wells was used for the species-specific probe, and the other was used for the S-D-Bact-0338-a-A-18 probe (3), which detects all bacterial species. The optimal hybridization temperature (TH) was determined experimentally for each probe. TH is defined as the temperature at which a given probe hybridized exclusively with the target organism while S-D-Bact-0338-a-A-18 hybridized with all species given in the Appendix. The slides were subsequently treated with the SlowFade Antifade Kit according to the instructions of the manufacturer (Molecular Probes, Leiden, The Netherlands). In addition, the hybridization temperature (TE) of the designed probes was determined by the method of De Los Reyes et al. (11).
Analysis of fecal samples.
Prior to counting, fixed fecal samples were briefly vortexed and subsequently centrifuged at 9 × g for 3 min. The supernatant of each sample was diluted in such a way that the numbers of fluorescing cells in a microscopic field (see below) were between 10 and 30. Of each dilution, 10 μl was applied to a separate well on the slide and treated as described above for pure cultures. To improve the permeabilization of the cell envelope, samples were treated with 10 μl of lysozyme buffer (100 mM Tris-HCl [pH 8.0], 50 mM EDTA, 1 mg of lysozyme/ml [130,000 U/mg; Boehringer, Mannheim, Germany]). Slides were incubated on ice for 7 min and subsequently dehydrated and hybridized as described above.
Microscopy and documentation.
Total cell counts were determined with a Zeiss Axioplan 2 microscope (Zeiss, Jena, Germany) equipped with a 12-bit video camera (SensiCam; PCO, Kelheim, Germany) and several filters for epifluorescence microscopy. Enumeration was done by automated counting by using the KS400 3.0 software (Zeiss). The species-specific enumeration of cells was done manually with an Optiphot-2 microscope (Nikon, Düsseldorf, Germany) equipped with a G-2A filterblock for epifluorescence microscopy and a photomicrographic camera. Each well (6 mm in diameter) of the slides was subdivided into 25 different fields. Four wells were analyzed per sample (100 fields). The cell concentration (C) was calculated as follows: C (ml−1) = Nm × DFF × MF × DFS, where Nm is the mean value of the cell numbers determined in 100 microscopic fields (four wells). DFF (2,000 ml−1) is the constant dilution factor resulting from the fixation process. MF is the microscopic factor, which is defined as the total area per well divided by the area of the 25 fields inspected per well. The total area per well was 28.27 mm2, whereas the area of the 25 fields was dependent on the microscope used.
For the Zeiss Axioplan 2 microscope the area was 0.0056 mm2, and for the Optiphot-2 microscope the area was 0.0151 mm2. DFS is the dilution factor for each separate sample, which was necessary to obtain the 10 to 30 bacteria per field.
RESULTS
Probe design.
Eleven different oligonucleotide probes were designed based on comparative analysis by using the ARB program and checked with the RDP and the EMBL databases. Alignments of all chosen 16S rRNA sequences were screened for targets that enable species-specific discrimination. Table 1 shows the probe sequences; their target sites; their TD, TH, and TE values (for definitions, see Materials and Methods); and alignments of the potential 16S rRNA binding sites of nontarget organisms. Probes were named according to the OPD (2). Since the RDP contains two different 16S rRNA sequences for E. cylindroides, we designed two different species-specific probes and used an equimolar mixture of both probes for the detection of E. cylindroides. The E.lim1433 probe designed for the detection of E. limosum does not allow its discrimination from E. callanderi, a species quite closely related to E. limosum. The differentiation between the two organisms is based on minor phenotypic differences (40, 51). Hence, E.lim1433 is a dispecies-specific probe.
E. lentum was recently reclassified as Eggerthella lenta gen. nov., comb. nov. (49). However, since this organism was originally described as a numerically dominant Eubacterium species (16), we included it in our study and refer to it here as E. lentum.
Specificity of the probes in whole-cell hybridization.
Since the hybridization conditions used for fluorescence in situ hybridization (FISH) differ from those used for the determination of TE (11, 45), hybridization experiments with reference strains originating from the human and animal gastrointestinal tract were done at TH (Table 1). As a positive control, the bacterium-specific probe S-D-Bact-0338-a-A-18 (3) was used. In pure culture, all designed probes resulted in a hybridization signal of similar intensity, which worked equally well on the first try in pure cultures. The probes designed for the detection of E. barkeri (E.bar1237), E. biforme (E.bif462), E. cylindroides (E.cyl461 and E.cyl466), E. dolichum (E.dol183), E. hadrum (E.had580), E. lentum (E.len194), E. limosum (E.lim1433), E. moniliforme (E.mon84), and E. ventriosum (E.ven66) hybridized to the corresponding target organism but not to the other organisms listed in the Appendix. E.con1122, which had been designed for E. contortum, cross-hybridized with the closely related E. fissicatena. An increase in stringency by elevation of the temperature or the formamide concentration did not prevent the unspecific binding of E.con1122 to E. fissicatena. We therefore decided to use E.con1122 for the detection of both organisms. E.con1122 is therefore a dispecies-specific probe. A probe for E. rectale (S-S-E.rec-0574-a-A-18) was designed, and its specifity was tested in FISH experiments. Since this probe cross-hybridized with nontarget organisms, E.rec574 was not included in the probe panel. An increase in stringency by elevation of the temperature or the formamide concentration did not prevent unspecific binding. Examples of whole-cell hybridization with E.bar1237 in pure culture and E.bif462 in fecal samples are given in Fig. 1.
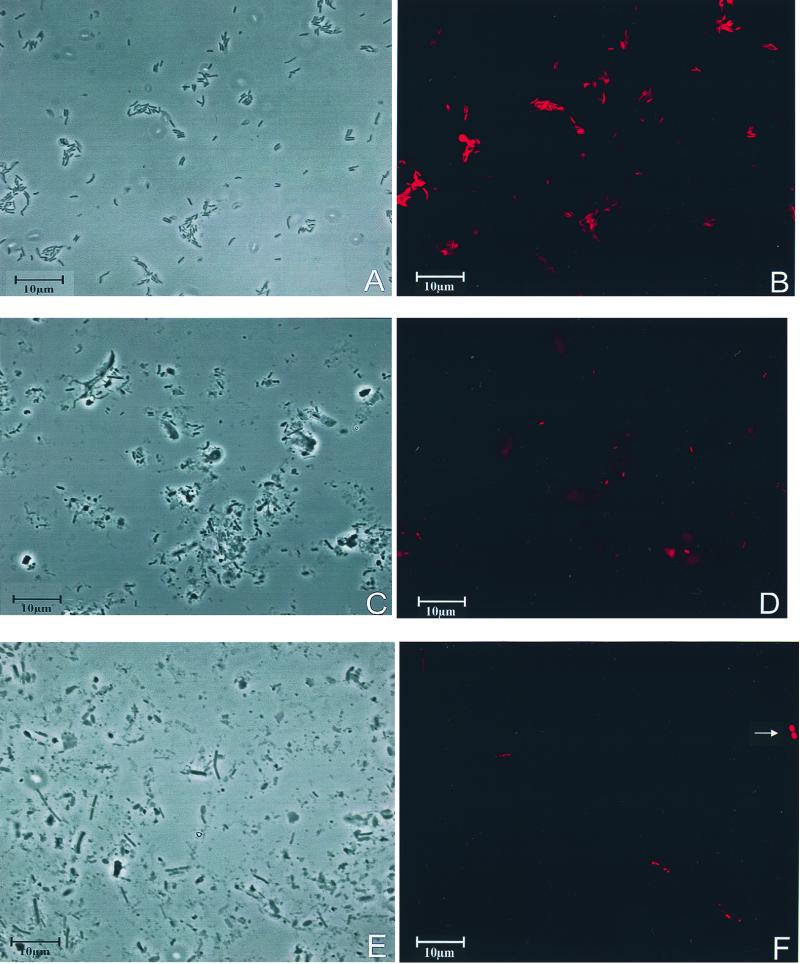
FIG. 1.
Phase-contrast and epifluorescence micrographs of pure culture and fecal samples. (A and B) Pure culture of E. barkeri hybridized with Cy3-labeled E.bar1237 (S-S-E.bar-1237-a-A-18) probe. Panels A and B show the same microscopic field viewed by phase-contrast and epifluorescence microscopy, respectively. (C and D) Detection of E. biforme in a fecal sample with E.bif462 (S-S-E.bif-0462-a-A-18). Panels C and D show the same microscopic field viewed by phase-contrast and epifluorescence microscopy, respectively. (E and F) Detection of E. hadrum in a fecal sample with E.had579 (S-S-E.had-0579-a-A-20) and cross-hybridization to an unknown coccoid-shaped bacterium. Panels E and F show the same microscopic field viewed by phase-contrast and epifluorescence microscopy, respectively. In panel E the unknown coccoid-shaped bacterium is marked with an arrow. Both visible bacteria (coccoid shaped and rod shaped) were detected with E.had579.
To determine the applicability of the validated probes to fecal samples, each probe was used to enumerate target bacteria in fecal samples from 12 healthy volunteers. Cell counts, relative proportions, and incidences of the target organisms in the 12 subjects are summarized in Table 2. The detection limit was 107 cells g (dry weight)−1.
TABLE 2.
Quantification of Eubacterium species in human feces with whole-cell hybridization compared to literature data
| Eubacterium species | Designed probe | Range in subjects testing positive (mean) | Subjects testing positive (n = 12) | Range (%) of total fecal flora in subjects testing positive | Data from Finegold et al. (15)a
|
Data from Holdeman et al. (21)
|
||
|---|---|---|---|---|---|---|---|---|
| Range in subjects testing positive (mean) | % Persons testing positive (n = 141) | Mean counta | % Total flora (n = 3) | |||||
| E. barkeri | E.bar1237 | NDb | 0 | ND | 9.1–10.5 (9.8) | 3.5 | ND | ND |
| E. biforme | E.bif462 | 7.59–9.1 (8.35) | 6 | 0.08–3.3 | 8.3–11.3 (9.7) | 8 | 9.48 | 1.2 |
| E. contortum | E.con1122 | 9.2 | 1 | 0.8 | 4.2–12.7 (9.5) | 26.2 | ND | ND |
| E. cylindroides | E.cyl461, E.cyl466 | 7.7–8.24 (7.97) | 2 | 0.03–0.07 | 3.5–11.9 (9.6) | 23 | ND | ND |
| E. dolichum | E.dol183 | ND | 0 | ND | 6.5–11.0 (8.6) | 2.8 | ND | ND |
| E. hadrum | E.had579 | 7.9–8.7 (8.3) | 11 | 0.04–0.18 | ND | ND | 8.77 | 0.23 |
| E. lentum | E.len194 | 8.1–8.5 (8.29) | 4 | 0.1–0.6 | 3.6–11.6 (9.3) | 42.6 | ND | ND |
| E. limosum | E.lim1433 | ND | 0 | ND | 5.8–10.8 (8.7) | 5.7 | 8.63 | 0.17 |
| E. moniliforme | E.mon84 | ND | 0 | ND | 8.0–11.1 (9.9) | 6.4 | ND | ND |
| E. ventriosum | E.ven66 | 7.5–9.3 (8.4) | 2 | 0.03–1.7 | 4.0–12.3 (9.2) | 12.1 | 9.1 | 0.5 |
Ranges and mean counts of bacteria are expressed as the number of organisms (log10) per gram (dry weight) of feces.
ND, not detected.
With E.had579, two types of organisms were detected in fecal samples, a rod-shaped organism and a coccoid organism. The latter was obviously not contained in the panel of reference organisms used for testing the probe specificity. Since the two types of organisms could be distinguished easily by their morphology, only the rod-shaped organisms detected with E.had579 were considered to be E. hadrum.
E. hadrum was detected in 11 of 12 subjects at 8 × 107 to 5.59 × 108 cells g (dry weight)−1. The probes E.bar1237, E.bif462, E.len194, E.cyl461, E.cyl466, E.con1123, and E.ven113 detected only bacteria with the expected morphology and were therefore considered to be species specific. The probes E.dol183, E.lim1433, and E.mon84 gave no signals in fecal samples. Results are presented in Table 2.
Use of Eubacterium probes in PCR experiments.
In order to increase the sensitivity of detection, 6 of the 11 probes were used in PCR experiments as primers. All primer sets resulted in bands of the expected size as follows: E. barkeri, 1,229 bp; E. biforme, 479 bp; E. cylindroides ATCC 27803, 480 bp; E. cylindroides ATCC 27528, 487 bp; E. limosum, 1,424 bp; and E. ventriosum, 62 bp. No amplification product could be detected with other organisms listed in the Appendix.
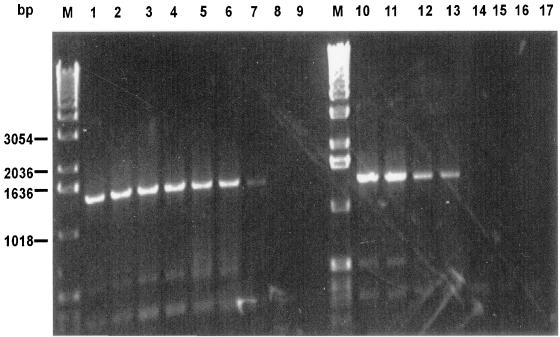
The sensitivity of the PCR methodology was first determined in pure cultures and subsequently for E. limosum in autoclaved feces (Fig. 2). In pure cultures, E. barkeri, E. biforme, and E. limosum could be detected at concentrations of 102 cells ml−1, and E. cylindroides and E. ventriosum could be detected at concentrations of 103 cells ml−1 (data not shown). The detection limit for E. limosum in autoclaved feces was by several orders of magnitude lower than in pure culture. It varied between 105 and 107 cells g (dry weight)−1.
FIG. 2.
PCR products obtained with genomic DNA of E. limosum from different cell numbers in autoclaved feces and pure culture on 1% agarose gel. Lanes M are the molecular marker (1-kb ladder). Lanes 1 to 9 show the PCR products obtained with cell numbers in the range from 108 to 100 from pure culture. Lanes 10 to 17 show PCR products obtained with cell numbers in the range from 108 to 101 which were added to autoclaved feces.
DISCUSSION
Eleven different probes were designed and applied both in whole-cell hybridization and PCR to detect various species of the genus Eubacterium in human feces. Several studies reported on the development of specific probes for various genera, including Bifidobacterium (29), Streptococcus, and Lactococcus or members of the Bacteroides fragilis, Clostridium histolyticum, Clostridium lituseburense, and Clostridium coccoides-Eubacterium rectale groups (17), respectively. Species-specific probes have been developed for a few species only (6, 14, 17, 45). The application of the newly designed probes to fecal samples allowed us to enumerate a number of Eubacterium species previously reported to be present in the human intestinal tract (15, 16, 21, 36). The cell counts obtained in our study for E. biforme, E. contortum, E. cylindroides, E. lentum, and E. ventriosum are essentially in agreement with the published data, which were obtained with classical microbiological enumeration techniques (Table 2) (15, 16, 21, 36). E. hadrum was only reported in one study (21). None of the other probes developed (E.bar1237, E.dol183, E.lim1433, and E.mon84) gave a signal in feces from any of the 12 subjects. The only exception was the probe for E. barkeri. However, with this probe the cell numbers detected in feces were too low to get reliable counts. Our experiments indicated that the detection limit for a given target organism in feces was approximately 107 cells g (dry weight)−1. Considering the high total cell counts in feces of about 1011 to 1012 cells g (dry weight)−1 (29, 45; and this study), this is not so bad as it may appear, since it is equivalent to identifying 1 cell in 100,000.
To overcome this detection limit, PCR-based amplification of 16S rDNA was used as an alternative method. The development of PCR methods for the detection of E. biforme and E. limosum with species-specific primers in fecal samples was reported earlier (50). Of the two organisms, only E. limosum was found in human fecal samples. Our species-specific primer sets for the detection of E. barkeri, E. biforme, E. cylindroides, E. limosum, and E. ventriosum identified the presence of these organisms at concentrations of 102 to 103 cells ml−1 in pure culture but not in autoclaved feces to which the target cells had been added from pure cultures. The detection limit increased to 105 to 107 cells g (dry weight)−1. The increase in the detection limit is due to fecal substances that inhibit PCR. These inhibitors have been identified as complex polysaccharides (35). None of the methods we have used for nucleic acid extraction (19, 33, 50) from feces was successful in removing these PCR inhibitors. Therefore, we decided to use the developed probes only for whole-cell hybridization.
One important difference between previous studies (16) and the present study lies in the number of subjects analyzed. Our study included 12 subjects, whereas in the Finegold et al. study (16) 141 subjects were analyzed. It is important to note that Finegold and coworkers (16) detected some Eubacterium species, e.g., E. barkeri, E. dolichum, E. limosum, and E. moniliforme, in high numbers but only in a few subjects. The inability to detect these species in our study may therefore be due to the fact that only a relatively limited number of subjects was examined.
In spite of the limitations of our study, the molecular approach used here avoids a number of problems associated with the phenotypic detection of Eubacterium species. Since most Eubacterium species are fastidious in their nutritional requirements, no special enrichment medium is available (37). Furthermore, the genus Eubacterium is phylogenetically heterogeneous and some of the Eubacterium species are pleomorphic, e.g., E. biforme, E. cylindroides, or E. dolichum, depending on the culture method. Hence, identification and determination at the species level is not a trivial task.
The advantage of whole-cell hybridization for the detection of bacteria in contrast to other molecular methods such as PCR (19, 27, 50) and dot blot hybridization (12, 25, 52), lies in the fact that the former method also allows the distinction of bacteria based on their morphology, as cells can be visualized in the microscope.
Since the probes are designed on the basis of the currently available 16S rRNA sequences (31, 46), the nonspecific detection of bacteria not included in the list of reference organisms but present in the habitat cannot be excluded. However, even then the targeted organism may still be identified based on its morphology. We were thus able to distinguish E. hadrum from a coccoid bacterium, which was obviously not among the 90 reference strains but was present in human feces.
The application of whole-cell hybridization to the fecal samples also has its limitations, the major one being the lack of sensitivity. This may be partly due to the fact that the number of rRNA target molecules in cells occurring in natural habitats is lower than in pure cultures because nutritional limitation and other competitive factors influence the cellular ribosome content (4, 29). It is therefore not surprising that the fluorescence signal of cells in fecal samples is lower than in pure culture (29). In addition, nonspecific fluorescence may hamper the visualization of bacteria. To increase the sensitivity of whole-cell hybridization, we used probes labeled with Cy3, whose signal strength is superior to that of other fluorescent molecules (1). In addition, fecal samples were treated with lysozyme to improve cell permeabilization, and the SlowFade Antifade Kit was used to reduce fading during microscopy.
In conclusion, whole-cell hybridization with species-specific oligonucleotide probes can assist in fast and accurate analysis of the Eubacterium spp. in fecal samples, provided the concentration of the target organisms is above the detection limit of 107 cells g (dry weight)−1. The oligonucleotide probes developed in this study will help to refine our present knowledge of this important group of bacteria in the intestinal tract.
ACKNOWLEDGMENT
This study was supported by the European Commission (FAIR-CT97-3035).
Appendix
The strains considered in this study included Acidaminococcus fermentans (DSMZ 2073), Actinobaculum suis (DSMZ 20639), Bacteroides distasonis (DSMZ 20701), Bacteroides ovatus (DSMZ 1896), Bacteroides sp. (B. fragilis, DIFE), Bacteroides sp. (B. vulgatus, DIFE),* Bacteroides thetaiotaomicron (DSMZ 2079), Bifidobacterium adolescentis (ATCC 15703), Bifidobacterium angulatum (ATCC 27535), Bifidobacterium animalis (ATCC 25527), Bifidobacterium bifidum (ATCC 29521), Bifidobacterium breve (ATCC 15700),* Bifidobacterium catenulatum (ATCC 27539), Bifidobacterium dentium (ATCC 27534), Bifidobacterium infantis (ATCC 15697), Bifidobacterium infantis (ATCC 15702),* Bifidobacterium infantis (ATCC 25962), Bifidobacterium longum (ATCC 15707), Bifidobacterium longum (ATCC 15708), Butyvibrio fibrisolvens (DSMZ 3071),* Clostridium acetobutylicum (ATCC 824), Clostridium barati (DSMZ 601), Clostridium bifermentans (DSMZ 46282), Clostridium butyricum (DSMZ 10702), Clostridium cellobioparum (DSMZ 1351), Clostridium clostridiforme (DSMZ 933), Clostridium coccoides (DSMZ 935),* Clostridium innocuum (DSMZ 1286), Clostridium pasteurianum (DSMZ 525), Clostridium perfringens (DSMZ 756), Clostridium populeti (DSMZ 5832), Clostridium propionicum (DSMZ 1682), Clostridium saccharolyticum (DSMZ 2544), Clostridium sartagoformum (DSMZ 1292), Clostridium sordellii (DSMZ 2141), Clostridium sp. (DSMZ 523), Clostridium sp. (C. butyricum, DIFE), Clostridium sp. (C. perfringens, DIFE), Clostridium spiroforme (DSMZ 1552),* Clostridium sporosphaeroides (DSMZ 1294), Clostridium tyrobutyricum (DSMZ 663), Clostridium xylanolyticum (DSMZ 6555), Coprococcus catus (ATCC 27761), Enterococcus hirae (DSMZ 20160),* Enterococcus sp. (E. cassiliflavus, DIFE), Enterococcus sp. (E. durans, DIFE), Enterococcus sp. (E. faecalis, DIFE), Enterococcus sp. (E. faecium, DIFE), Escherichia hermanii (ATCC 33650), Escherichia sp. (E. coli, DIFE), Eubacterium aerofaciens (DSMZ 3979), Eubacterium barkeri (ATCC 25849), Eubacterium biforme (DSMZ 3989), Eubacterium contortum (DSMZ 3982), Eubacterium cylindroides (ATCC 27528), Eubacterium cylindroides (ATCC 27803), Eubacterium dolichum (DSMZ 3991), Eubacterium eligens (DSMZ 3376), Eubacterium fissicatena (DSMZ 3598), Eubacterium hadrum (DSMZ 3319),* Eubacterium hallii DSMZ 3353),* Eubacterium lentum (DSMZ 2243), Eubacterium limosum (DSMZ 20543), Eubacterium moniliforme (DSMZ 3984), Eubacterium multiforme (DSMZ 20694), Eubacterium ramulus (ATCC 29099), Eubacterium rectale (ATCC 33656), Eubacterium saburreum (DSMZ 3986), Eubacterium siraeum (DSMZ 3996), Eubacterium sp. (E. hadrum, DIFE), Eubacterium sp. (E. ramulus, DIFE), Eubacterium sp. nov. (DIFE),* Eubacterium tenue (DSMZ 20695), Eubacterium tortuosum (DSMZ 3987), Eubacterium uniforme (ATCC 35992), Eubacterium ventriosum (ATCC 27560), Fusobacterium mortiferum (ATCC 25557), Fusobacterium naviforme (DSMZ 20699), Fusobacterium necrogenes (ATCC 25556), Fusobacterium nucleatum subsp. polymorphum (DSMZ 20482), Fusobacterium varium (ATCC 8501), Klebsiella sp. (K. pneumoniae, DIFE), Lactobacillus fermentum (DSMZ 20052), Lactobacillus gasseri (DSMZ 20243), Lactobacillus murinus (DSMZ 20452), Lactobacillus plantarum (DSMZ 20174), Lactobacillus reuteri (DSMZ 20016), Lactobacillus sp. (L. acidophilus, DIFE), Megasphaera sp. (DIFE), Peptostreptococcus anaerobius (DSMZ 2949),* Peptostreptococcus asaccharolyticus (DSMZ 20463), Peptostreptococcus prevotii (DSMZ 20548),* Pseudoramibacter alactolyticus (DSMZ 3980), Ruminococcus hansenii (DSMZ 20583),* Ruminococcus productus (DSMZ 2950), Streptococcus intermedius (DSMZ 20573), Streptococcus pleomorphus (DSMZ 20574), Veillonella parvula (DSMZ 2008), and Veillonella sp. (DIFE). Strains marked with an asterisk were not used in PCR validation experiments.
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:299–306. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attebery H R, Sutter V L, Finegold S M. Normal human intestinal flora. In: Balows A, DeHaan R M, Dowell V R, Guze L B, editors. Anaerobic bacteria-role in disease. Springfield, Ill: Charles C Thomas; 1974. pp. 81–97. [Google Scholar]
- 6.Boye M, Jensen T K, Moller K, Leser T D, Jorsal S E. Specific detection of the genus Serpulina, S. hyodysenteriae and S. pilosicoli in porcine intestines by fluorescent rRNA in situ hybridization. Mol Cell Probes. 1998;12:323–330. doi: 10.1006/mcpr.1998.0193. [DOI] [PubMed] [Google Scholar]
- 7.Brosius J, Dull T L, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 8.Bryant M P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 9.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combination. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 10.Cresci A, Orpianesi C, Silvi S, Mastrandrea V, Dolara P. The effect of sucrose or starch-based diet on short-chain fatty acids and faecal microflora in rats. J Appl Microbiol. 1999;86:245–250. doi: 10.1046/j.1365-2672.1999.00668.x. [DOI] [PubMed] [Google Scholar]
- 11.De Los Reyes F L, Ritter W, Raskin L. Group-specific small-subunit rRNA hybridization probes to characterize filamentous foaming in activated sludge systems. Appl Environ Microbiol. 1997;63:1107–1117. doi: 10.1128/aem.63.3.1107-1117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doré J, Sghir A, Hannequart-Garmet G, Corthier G, Pochart P. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantification of human faecal Bacteroides populations. System Appl Microbiol. 1998;21:65–71. doi: 10.1016/S0723-2020(98)80009-X. [DOI] [PubMed] [Google Scholar]
- 13.Drasar B S, Shiner M, McLeod G M. Studies on the intestinal flora. I. The bacterial flora of the gastrointestinal tract in healthy and achlorhydric persons. Gastroenterology. 1969;56:71–79. [PubMed] [Google Scholar]
- 14.DuTeau N M, Rogers J D, Bartholomany C T, Reardon K F. Species-specific oligonucleotides for enumeration of Pseudomonas putida F1, Burkholderia sp. strain JS150, and Bacillus subtilis ATCC 7003 in biodegradation experiments. Appl Environ Microbiol. 1998;64:4994–4999. doi: 10.1128/aem.64.12.4994-4999.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finegold S M, Attebery H R, Sutter V L. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456–1469. doi: 10.1093/ajcn/27.12.1456. [DOI] [PubMed] [Google Scholar]
- 16.Finegold S M, Sutter V L, Mathisen G E. Normal indigenous intestinal flora. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press, Inc.; 1983. pp. 3–31. [Google Scholar]
- 17.Franks A H, Harmsen H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA, targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmsen H J M, Prieur D, Jeanthon C. Group-specific 16S rRNA-targeted oligonucleotide probes to identify thermophilic bacteria in marine hydrothermal vents. Appl Environ Microbiol. 1997;18:67–73. doi: 10.1128/aem.63.10.4061-4068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengstler M, Kronberg U, Fahr A M. A reliable method for the removal of PCR inhibitors during the preparation of bacterial DNA from stool specimens. Clin Lab. 1998;44:447–450. [Google Scholar]
- 20.Hentges D J, Maier B R, Burton G C, Elynn M A, Tsutakawa R K. Effect of a high-beef diet on fecal bacterial flora of humans. Cancer Res. 1977;37:568–571. [PubMed] [Google Scholar]
- 21.Holdeman L V, Good I J, Moore W E C. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl Environ Microbiol. 1976;31:359–375. doi: 10.1128/aem.31.3.359-375.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hungate R E. A roll tube method for cultivation of strict anaerobes. In: Norris J R, Ribbons D W, editors. Methods in microbiology. 3B. New York, N.Y: Academic Press, Inc.; 1969. pp. 117–132. [Google Scholar]
- 23.Juretschko S, Timmermann G, Schmid M, Schleifer K H, Pomme-renning-Röser A, Koops H P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanauchi O, Fujiyama Y, Mitsuyama K, Araki Y, Ishii T, Nakamura T, Hotomi Y, Agata K, Saiki T, Andoh A, Toyonaga A, Bamba T. Increased growth of Bifidobacterium and Eubacterium by germinated barely foodstuff, accompanied by enhanced butyrate production in healthy volunteers. Int J Mol Med. 1999;3:175–179. doi: 10.3892/ijmm.3.2.175. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko T, Kurihara H. Digoxigenin-labeled deoxyribonucleic acid probes for the enumeration of bifidobacteria in fecal samples. J Dairy Sci. 1997;80:1254–1259. doi: 10.3168/jds.S0022-0302(97)76054-5. [DOI] [PubMed] [Google Scholar]
- 26.Kenzaka T, Yamaguchi N, Tani K, Nasu M. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology. 1998;144:2085–2093. doi: 10.1099/00221287-144-8-2085. [DOI] [PubMed] [Google Scholar]
- 27.Kreader C A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol. 1995;61:1171–1179. doi: 10.1128/aem.61.4.1171-1179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 29.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H F, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;6:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddox P H, Jenkins D. 3-Aminopropyltriethoxysilane (APES): a new advance in section adhesion. J Clin Pathol. 1987;40:1256–1257. doi: 10.1136/jcp.40.10.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz W, Szewzyk U, Ericsson P, Amann R, Schleifer K H, Stenström T A. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl Environ Microbiol. 1993;59:2293–2298. doi: 10.1128/aem.59.7.2293-2298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marmur J. Procedure for the isolation of desoxyribonucleic acids from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 34.Mitsuoka T, Ohno K, Benno Y, Suzuki K, Namba K. Die Faecalflora bei Menschen. IV. Mitteilung: Vergleich des neu entwickelten Verfahrens mit dem bisherigen üblichen Verfahren zur Darmfloraanalyse. Zentbl Bakteriol Hyg Abt 1 Orig A. 1976;234:219–233. [PubMed] [Google Scholar]
- 35.Monteiro L, Bonnemaison D, Vekris A, Petry K G, Bonnet J, Vidal R, Cabrita J, Mégraud F. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol. 1997;35:995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore W E C, Holdeman L V. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore W E C, Holdeman Moore L V. Genus Eubacterium Prévot 1938. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1353–1373. [Google Scholar]
- 38.Mori A, Ishiyama I, Akita H, Suzuki K, Mitsuoka T, Oishi T. Formation of amphetamine from its nitro analogue by anaerobic intestinal bacteria. Xenobiotica. 1990;20:629–634. doi: 10.3109/00498259009046878. [DOI] [PubMed] [Google Scholar]
- 39.Moter A, Leist G, Rudolph R, Schrank K, Choi B K, Wagner M, Göbel U. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology. 1998;144:2459–2467. doi: 10.1099/00221287-144-9-2459. [DOI] [PubMed] [Google Scholar]
- 40.Mountfort D O, Grant W D, Clarke R, Asher R A. Eubacterium callanderi sp. nov. that demethoxylates o-methoxylated aromatic acids to volatile fatty acids. Int J Syst Bacteriol. 1988;38:254–258. [Google Scholar]
- 41.Prevot A R. Études de systématique bactérienne. IV. Critique de la conception actuelle du genre Clostridium. Ann Inst Pasteur. 1938;61:72–91. [Google Scholar]
- 42.Reddy B S, Weisburger J H, Wynder E L. Effect of high risk and low risk diets for colon carcinogenesis on fecal microflora and steroids in man. J Nutr. 1975;105:878–884. doi: 10.1093/jn/105.7.878. [DOI] [PubMed] [Google Scholar]
- 43.Roller C, Wagner M, Amann R I, Ludwig W, Schleifer K H. In situ probing of gram-positive bacteria with high G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 44.Schneider H, Schwiertz A, Collins M D, Blaut M. Anaerobic transformation of quercetin-3-glucoside by bacteria from the human intestinal tract. Arch Microbiol. 1999;171:81–91. doi: 10.1007/s002030050682. [DOI] [PubMed] [Google Scholar]
- 45.Simmering R, Kleessen B, Blaut M. Quantification of the flavonoid-degrading bacterium Eubacterium ramulus in human fecal samples with a species-specific oligonucleotide hybridization probe. Appl Environ Microbiol. 1999;65:3705–3709. doi: 10.1128/aem.65.8.3705-3709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strunk O, Ludwig W. A software environment for sequence data. Munich, Germany: Technische Universität München; 1996. [Google Scholar]
- 47.Suggs S V, Hirose T, Miyake T, Kawashima E H, Johnson M J, Itakura K, Wallace R B. Use of synthetic oligonucleotides for the isolation of specific cloned DNA sequences. In: Brown D, Fox C F, editors. Developmental biology using purified genes. New York, N.Y: Academic Press, Inc.; 1981. pp. 683–693. [Google Scholar]
- 48.Van Eldere J, Robben J, De Pauw G, Merckx R, Eyssen H. Isolation and identification of intestinal steroid-desulfating bacteria from rats and humans. Appl Environ Microbiol. 1988;54:2112–2117. doi: 10.1128/aem.54.8.2112-2117.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wade W G, Downes J, Dymock D, Hiom S J, Weightman A J, Dewhirst F E, Paster B J, Tzellas N, Coleman B. The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov., and Slackia heliotrinireducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol. 1999;49:595–600. doi: 10.1099/00207713-49-2-595. [DOI] [PubMed] [Google Scholar]
- 50.Wang R F, Cao W W, Cerniglia C E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willems A, Collins M D. Phylogenetic relationships of the genera Acetobacterium and Eubacterium sensu stricto and reclassification of Eubacterium alactolyticum as Pseudoramibacter alactolyticus gen. nov., comb. nov. Int J Syst Bacteriol. 1996;46:1083–1087. doi: 10.1099/00207713-46-4-1083. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto T, Morotomi M, Tanaka R. Species-specific oligonucleotide probes for five Bifidobacterium species detected in human intestinal microflora. Appl Environ Microbiol. 1992;58:4076–4079. doi: 10.1128/aem.58.12.4076-4079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]