Abstract
Coronaviruses are a class of single-stranded, positive-sense RNA viruses that have caused three major outbreaks over the past two decades: Middle East respiratory syndrome–related coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). All outbreaks have been associated with significant morbidity and mortality. In this study, we have identified and explored conserved binding sites in the key coronavirus proteins for the development of broad-spectrum direct acting anti-coronaviral compounds and validated the significance of this conservation for drug discovery with existing experimental data. We have identified four coronaviral proteins with highly conserved binding site sequence and 3D structure similarity: PLpro, Mpro, nsp10-nsp16 complex(methyltransferase), and nsp15 endoribonuclease. We have compiled all available experimental data for known antiviral medications inhibiting these targets and identified compounds active against multiple coronaviruses. The identified compounds representing potential broad-spectrum antivirals include: GC376, which is active against six viral Mpro (out of six tested, as described in research literature); mycophenolic acid, which is active against four viral PLpro (out of four); and emetine, which is active against four viral RdRp (out of four). The approach described in this study for coronaviruses, which combines the assessment of sequence and structure conservation across a viral family with the analysis of accessible chemical structure – antiviral activity data, can be explored for the development of broad-spectrum drugs for multiple viral families.
Keywords: Coronaviruses, SARS-CoV-2, Broad-spectrum antivirals, Protein conservation
1. Introduction
Coronaviruses are a class of single stranded, positive-sense RNA viruses that have caused significant morbidity and mortality in recent years. Coronaviruses of the genus Alphacoronaviridae, which contains some of the common cold coronaviruses, and Betacoronaviridae, which contains the three well-known coronaviruses MERS-CoV, SARS-CoV, and SARS-CoV-2, have been isolated from wildlife such as bats, palm civets, and camels (Corman et al., 2018; Monchatre-Leroy et al., 2017). The alphacoronaviruses HCoV-NL63 and HCoV-229E and the betacoronaviruses HCoV-OC43 and HCoV-HKU1 are the endemic human coronaviruses that typically result in the common cold (Corman et al., 2018). These viruses usually cause only mild respiratory illness but can result in more severe disease in immunocompromised individuals, the elderly, and infants (Corman et al., 2018). A third genus, including porcine delta-coronavirus holds zoonotic potential, as strains have recently been identified in plasma samples of three Haitian children (Lednicky et al., 2021).
The noted potential for zoonotic transmission and the emergence of novel coronaviruses creates an urgent need to rapidly develop new broad-spectrum antiviral therapies in addition to those that currently circulate, e.g., SARS-CoV-2 (Graham and Baric, 2020; Menachery et al., 2015). In the previous MERS-CoV and SARS-CoV outbreaks, the fatality rates were 34.4% and 10%, respectively, but the number of infected individuals was relatively low with 2,585 and 8,422 cases, respectively (WHO, 2022a; 2022b). A striking deviation from this pattern has been observed for SARS-CoV-2, which, since December 2019 and as of March 2022, has infected over 450 million people and killed over 6 million people globally (“COVID-19 by CSSE at Johns Hopkins,” 2022, “WHO Coronavirus (COVID-19) Dashboard,” 2022; Dong et al., 2020). As the pandemic continues ravaging the world, its effects on society, poverty, the environment, and the economy grow (Chakraborty and Maity, 2020; Saladino et al., 2020). Despite the desperate need for therapies, there are currently no approved drugs to treat any of these coronaviruses, though several including tocilizumab, sotrovimab, bamlanivimab, etesevimab, casirivimab, imdevimab, and baricitinib have been approved through Emergency Use Authorization (EUA) by the FDA during the COVID-19 pandemic (“Coronavirus Disease 2019 (COVID-19) EUA Information,” 2021).
Through extensive research into the mechanisms of coronavirus replication and function, scientists have begun to discern important nuances concerning coronaviruses. One such variation is that the host receptors for coronaviruses can differ even between genera. For instance, in the genus Betacoronavirus, MERS-CoV uses dipeptidyl peptidase-4 (DPP4) for host cell entry whereas SARS-CoV and SARS-CoV-2 use the angiotensin-converting enzyme 2 (ACE2) as their host receptor (Fehr and Perlman, 2015). After binding, the spike (S) protein on the outside of the coronavirus virion must be primed by host cell proteases to catalyze a conformational change that will enable its fusogenic activity, thus permitting the virus to enter the host cell cytosol. This priming is typically accomplished by transmembrane serine protease 2 (TMPRSS2), cathepsin L, or some other cellular protease (Fehr and Perlman, 2015; Hoffmann et al., 2018). Next, the two polyproteins, pp1a and pp1ab, must be translated from the virion genomic RNA. These two polyproteins are cleaved by viral proteases to produce the 16 non-structural proteins (nsps) essential for the replication of the coronavirus (Astuti and Ysrafil, 2020). This complex shares a conserved S-adenosyl methionine (SAM)-binding pocket between SARS-CoV, MERS-CoV, and SARS-CoV-2 (Lin et al., 2020). The replicase-transcriptase complex (RTC) is composed of many of these nsps. Mature virions can be formed after the viral genomic RNA buds into membranes of the endoplasmic reticulum–Golgi intermediate compartment containing the viral structural proteins, S, E, and M (Astuti and Ysrafil, 2020; V'kovski et al., 2021).
The nsps that constitute the RTC are tractable drug targets for coronaviruses, e.g., nsp12, which encodes the RNA-dependent RNA polymerase (RdRp) domain responsible for replicating viral RNA. Remdesivir, a nucleoside analog that has been approved by the FDA through an EUA, has a mechanism of action that works through the inhibition of RdRp (Shannon et al., 2020). Encoded by nsp15, the RNA endonuclease (NendoU) is conserved among Nidovirales, the virus order containing coronaviruses. The nsp10-nsp16 complex (ribose-2′-O-methyltransferase) has been implicated in modulating the actions of NendoU, though much is still unknown about how coronaviruses regulate the RNA cleavage activity of this protein (Ricagno et al., 2006). Due to its unique conservation among coronaviruses, NendoU is a unique target for broad-spectrum, coronavirus-specific antiviral drug development. The drug Tipiracil, which is used to treat colorectal cancer, was recently shown to bind within the NendoU active site and modestly inhibit SARS-CoV-2 replication in whole cell assays (Kim et al., 2021). Tipiracil was not tested against other viruses, so it remains unknown whether it has broad-spectrum activity. The nsp10-nsp16 complex principally functions to cap viral mRNAs, thereby protecting them from host innate immune responses. Though it has been noted that, most likely, the interface could not be targeted by small-molecule drugs due to its large area and complex network of molecular interactions, in 2020 the SARS-CoV-2 nsp10-nsp16 complex bound to a pan-methyltransferase inhibitor sinefungin was crystallized (Krafcikova et al., 2020). Other viral nsps that show promise as antiviral drug targets are the two proteases of coronaviruses, the papain-like protease (PLpro) and the main protease (Mpro), which work to cleave transcribed polyproteins into 16 nsps with distinct functions (Astuti and Ysrafil, 2020; Zumla et al., 2016).
Given the high potential of recurrent coronavirus outbreaks, the development of broad-spectrum antivirals is crucial to control both the present and future coronavirus epidemics (Totura and Bavari, 2019). A database containing all known broad-spectrum antiviral compounds has been compiled at https://drugvirus.info/(Andersen et al., 2020). Several compounds have demonstrated broad-spectrum antiviral activity against Human Immunodeficiency Virus (HIV), Hepatitis C virus (HCV), and influenza viruses (Vigant et al., 2015). Compounds such as umifenovir, a viral fusion inhibitor, and nitazoxanide, a pyruvate ferredoxin oxidoreductase enzyme inhibitor, have been and continue to be used against influenza viruses as well as other viral respiratory illness (Boriskin et al., 2008; Rossignol, 2014). Other compounds are in development and trials, such as GS-5734 (also known as remdesivir), a non-toxic and potent broad-spectrum antiviral against endemic and zoonotic coronaviruses. This compound was found effective against SARS-CoV and MERS-CoV in vitro as well as against bat CoVs, pre-pandemic bat CoVs, and circulating contemporary human CoVs in primary human lung cells (Sheahan et al., 2017).
Amino acid residues constituting active sites of enzymes, especially crucial catalytic residues, have a tendency to be highly conserved over evolutionary time (Jack et al., 2016). A radical change in those sites would likely confer a change in functionality, reducing the fitness of the virus. Thus, the analysis of specific binding sites with a more focused consideration of individual proteins conserved in (beta)coronaviruses may help guiding broad-spectrum antiviral discovery (Li, 2016; Tilocca et al., 2020).
In this study, we have investigated whether homologous coronavirus proteins could be exploited as targets for the development of broad-spectrum anti-coronaviral compounds. To this end, we have analyzed the sequence similarity for all available coronavirus proteins. In addition, we also analyzed binding site similarities for four homologous coronavirus proteins with known 3D structures deposited in the Protein Data Bank (PDB) including Papain-Like Protease (PLpro), Main Protease (Mpro), Methyltransferase (nsp10-nsp16), and Endoribonuclease (NendoU). Below we review the current data about both the sequence and structure conservation of these proteins across coronaviruses and molecules that have been tested for the activity against these proteins. We provide a perspective on how the conservation analysis of viral proteins’ sequence and structure could support the discovery of broad-spectrum antivirals in response to future coronavirus epidemics.
2. Material and methods
2.1. Comparison of homologous coronavirus protein ligand binding sites
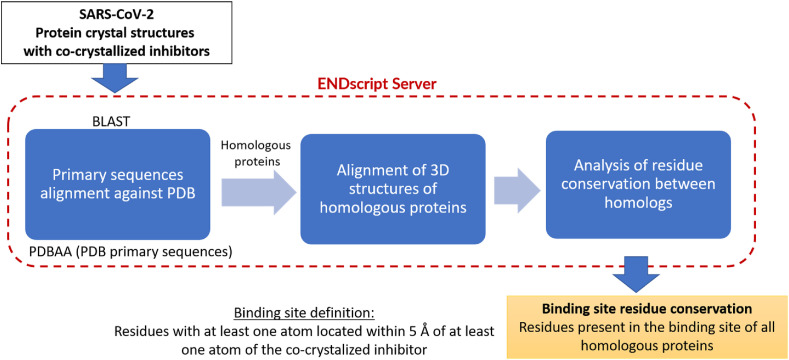
We analyzed the similarity between SARS-CoV-2 proteins and their related counterparts from other coronaviruses, focusing specially on the comparison of ligand binding sites. SARS-CoV-2 proteins were chosen as the reference and query sequences for each search. The general binding site comparison workflow is presented in Fig. 1 . The details of the analysis are described in the following sections.
Fig. 1.
General workflow of the protein binding sites comparison. The ENDscript server (https://endscript.ibcp.fr/ESPript/ENDscript/) was employed; it is publicly accessible tool for multiple sequence alignment of proteins homologous to the query, alignment of their corresponding crystal structures, and coloring the query structure according to residue conservation (Robert and Gouet, 2014).
2.1.1. Protein selection and collection
Coronavirus proteins were selected based on the availability of their crystal structures in Protein Data Bank (PDB, http://www.rcsb.org/) (Berman, 2000). The primary sequence of each protein was obtained from UniProt (Bateman, 2019). Furthermore, since we focused on binding site comparisons, proteins with co-crystallized ligands were prioritized, namely, papain-like protease (PLpro), main protease (Mpro), nsp10-nsp16 (methyltransferase), endoribonuclease (NendoU), and RNA-dependent RNA polymerase (RdRP). The list of all selected proteins including their UniProt IDs and PDB codes can be found in Supplemental Tables S2–S3.
2.1.2. Homolog search and structural alignment
To speed up the analysis and visualization of the primary sequence alignments, homology searches, and 3D binding site alignments we used the ENDscript server (Robert and Gouet, 2014). This publicly accessible server (https://endscript.ibcp.fr/ESPript/ENDscript/) was used to execute the following steps. 1) Primary sequence alignment, using the Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990) of a given SARS-CoV-2 protein against the PDBAA database (NCBI, 2020) containing all primary sequences corresponding to all entries in PDB. It is important to highlight that all homologous proteins identified at this stage would not necessarily cover all possible existing homologs because the search was limited to the set of primary sequences with available structures in PDB (PDBAA). 2) The structures of all homologs identified in the previous step were then extracted from PDB and subsequently aligned both to each other and with the respective query SARS-CoV-2 protein. To avoid overestimation of residue conservation due to replicate entries of the same protein from the same viral species, only one representative crystal structure was considered for each protein. We prioritized the structure with a co-crystallized inhibitor for comparison of the protein's ligand binding sites in an inactive conformation. In the absence of a co-crystallized ligand, we chose the one with the resolution. 3) A 3D structure of the query SARS-CoV-2 protein with a heat-map color-coded representation of residue conservation across homologous proteins was generated. In this study, the measure of conservation was based on the frequency of co-occurrence of residues across homologous proteins.
2.1.3. Binding site similarity
We focused on the conservation of experimentally defined orthosteric ligand binding sites as having potential for antiviral drug development, although conservation of potential allosteric binding sites could also be analyzed in a similar manner in the future. For consistency, a binding site was defined as a collection of residues with at least one atom within 5 Å distance from any atom of the co-crystallized inhibitor.
2.2. Primary sequence comparison of remaining proteins
Primary sequences of all 26 SARS-CoV-2 proteins, including 21 proteins with no co-crystallized ligands, i.e., without experimentally defined binding sites and not included in the previous analysis, were used as queries for primary sequence comparisons against the “UniProtKB reference proteomes plus Swiss-Prot” using BLAST (Altschul et al., 1990) service available at Uniprot (https://www.uniprot.org/blast/). After the search, only homologous proteins flagged as “Swiss-Prot reviewed”, i.e., those that passed through a quality inspection in Swiss-Prot, were selected. The resulting list of homologous proteins for each target was aligned in Clustal Omega 1.2.4 (Sievers et al., 2011) available at (https://www.uniprot.org/align/).
3. Results and discussion
3.1. Binding site similarity
Except for RdRp, all proteins with crystal structures containing co-crystallized inhibitors returned results after submission to the ENDscript server. Despite the detection of homologs of RdRp at the primary sequence level, the server did not achieve any acceptable (by internal metrics that are not visible to the user) 3D alignment. Thus, two limitations are prevalent: (i) the number of existing homologous proteins and their level of binding site similarity presented in this section may not entirely reflect the real number of coronavirus homologous proteins because there are limited PDB structures available and (ii) many mutant proteins exist, and because one representative structure was chosen, this work does not reflect conservation amongst each possible mutant protein for these coronaviruses. The results of this analysis are discussed below for each query protein separately.
3.1.1. Papain-like protease (PLpro)
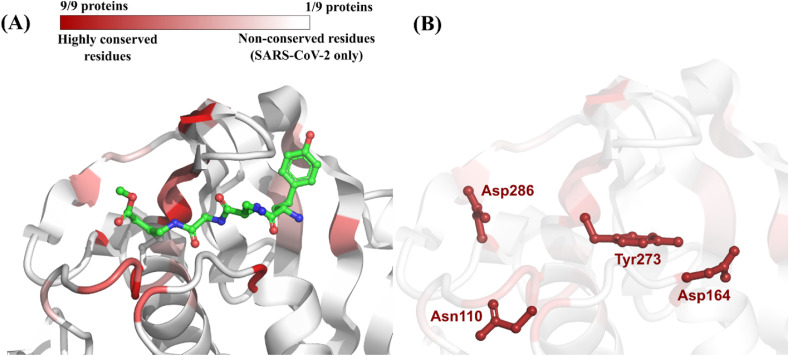
Eight homologous proteins of SARS-CoV-2 papain-like protease (PLpro) were identified (Table 1 ). These proteins were found in coronaviruses from three different genera (Betacoronavirus, Gammacoronavirus, and Alphacoronavirus) associated with diverse hosts (human, mice, pigs, and birds). Based on primary sequence alignment, two homologous PLpro with the greatest sequence identity to the SARS-CoV-2 counterpart were derived from different strains of SARS-CoV and presented identities of 82.54% and 82.22% (Table 1; Fig. S1, Supplementary Material). After alignment of their 3D structures, regions of high conservation (i.e., the same residue is present, in the same position, in all nine homologs) were identified in the binding site defined around the co-crystallized peptide-like inhibitor VIR251(Fig. 2 A) (Rut et al., 2020). In total, four residues in the binding site of PLpro from SARS-CoV-2, representing 19% of all residues in the binding site, were conserved in all homologous proteins (Table 1; Fig. 2B).
Table 1.
SARS-CoV-2 PLpro primary sequence identity and binding site residues conservation against all nine corresponding homologous proteins identified through ENDscript (Robert and Gouet, 2014).
| PDB ID | Virus | Genus | Host | Global primary sequence identity to SARS-CoV-2 PLpro (%) | SARS-CoV-2 PLpro binding site residues conserved in all species |
|---|---|---|---|---|---|
| 6WX4 | SARS-CoV-2 | Betacoronavirus | Human | Used as query | Asn110 Asp164 Tyr273 Asp286 (19% of all binding site residues) |
| 3E9S | SARS-CoV | Betacoronavirus | Human | 82.54 | |
| 4OVZ | SARS-CoV (Urbani) | Betacoronavirus | Human | 82.22 | |
| 4P16 | MERS-CoV (2c EMC/2012) | Betacoronavirus | Human | 29.91 | |
| 4R3D | MERS-CoV (England 1) | Betacoronavirus | Human | 29.57 | |
| 4REZ | MERS-CoV (2c Jordan-N3/2012) | Betacoronavirus | Human | 30.03 | |
| 4X2Z | Avian Infectious Bronchitis Virus (Strain Beaudette) | Gammacoronavirus | Chicken | 21.47 | |
| 4YPT | Murine Hepatitis Virus (strain A59) | Betacoronavirus | Mouse | 30.00 | |
| 6L5T | Swine Acute Diarrhea Syndrome Coronavirus | Alphacoronavirus | Pig | 20.77 |
Fig. 2.
(A) Color-coded depiction of residue conservation at the binding site of all identified SARS-CoV-2 PLpro homologous proteins. Regions in dark red represent residues with high co-occurrence among homologous proteins (i.e., nine out of nine proteins share the same residue). Regions in light red represent residues with moderate co-occurrence (i.e., between 2 and 8 out of nine proteins share the same residue). Regions in white represent residues with no co-occurrence (i.e., the residue is present only in SARS-CoV-2). The protein structure used as template is the PLpro from SARS-CoV-2 (PDB ID: 6WX4) co-crystallized with the peptide-like inhibitor VIR251 (in green) (Rut et al., 2020); (B) Binding site residues of SARS-CoV-2 PLpro conserved in all homologous proteins listed in Table 1.
3.1.2. Main protease (Mpro)
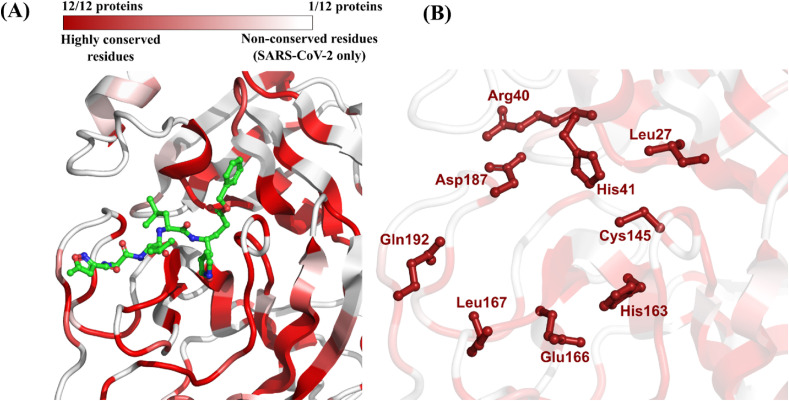
We identified eleven homologous proteins of SARS-CoV-2 main protease (Mpro) (Table 2 ). These proteins were derived from the same genera of coronavirus previously identified in the PLpro analysis, namely, Betacoronavirus, Gammacoronavirus, and Alphacoronavirus. The list of coronavirus species and associated hosts was also similar except for the Tylonycteris Bat Coronavirus HKU4, which is related to MERS-CoV (Lau et al., 2013). Regarding primary sequence comparison, the SARS-CoV Mpro presented the highest identity to the SARS-CoV-2 counterpart (96.1%) followed by MERS-CoV (50.7%) (Table 2; Fig. S2, Supplementary Material). Subsequently, the 3D structural alignment of all homologs revealed regions of high conservation in the binding site defined around the co-crystallized peptide-like inhibitor N3 (Fig. 3 A) (Jin et al., 2020). In total, eight residues in the binding site of Mpro from SARS-CoV-2, which correspond to 37.5% of all residues forming the binding site, were conserved in all homologous proteins (Table 2; Fig. 3B).
Table 2.
SARS-CoV-2 Mpro primary sequence identity and binding site residues conservation against all twelve corresponding homologous proteins identified through ENDscript(Robert and Gouet, 2014).
| PDB ID | Virus | Genus | Host | Global primary sequence identity to SARS-CoV-2 Mpro (%) | SARS-CoV-2 Mpro binding site residues conserved in all species |
|---|---|---|---|---|---|
| 6LU7 | SARS-CoV-2 | Betacoronavirus | Human | Used as query | His41 Arg40 Leu27 Asp187 Cys145 Gln192 Leu167 Glu166 His163 (37.5% of all binding site residues) |
| 1WOF | SARS-CoV | Betacoronavirus | Human | 96.08 | |
| 4RSP | MERS-CoV | Betacoronavirus | Human | 50.65 | |
| 3D23 | HKU1 (isolate N1) | Betacoronavirus | Human | 49.17 | |
| 1P9S | 229E | Alphacoronavirus | Human | 39.47 | |
| 3TLO | NL63 | Alphacoronavirus | Human | 44.30 | |
| 2AMP | Transmissible Gastroenteritis Virus | Alphacoronavirus | Pig | 44.44 | |
| 4XFQ | Porcine Epidemic diarrhea virus | Alphacoronavirus | Pig | 44.77 | |
| 2YNA | Tylonycteris Bat Coronavirus HKU4 | Betacoronavirus | Bat | 49.68 | |
| 2Q6D | Avian Infectious Bronchitis Virus | Gammacoronavirus | Chicken | 40.82 | |
| 4ZRO | Feline Infectious Peritonitis Virus (strain 79-1146) | Alphacoronavirus | Cat | 44.22 | |
| 6JIJ | Murine Hepatitis Virus (strain A59) | Betacoronavirus | Mouse | 50.33 |
Fig. 3.
(A) Color-coded depiction of residue conservation at the binding site of all identified SARS-CoV-2 Mpro homologous proteins. Regions in dark red represent residues with high co-occurrence among homologous proteins (i.e., 12 out of 12 proteins share the same residue). Regions in light red represent residues with moderate co-occurrence (i.e., between 2 and 11 out of 12 proteins share the same residue). Regions in white represent residues with no co-occurrence (i.e., the residue is present only in SARS-CoV-2). The protein structure used as template is the Mpro from SARS-CoV-2 (PDB ID: 6LU7) co-crystallized with the peptide-like inhibitor N3 (in green) (Jin et al., 2020); (B) Binding site residues of SARS-CoV-2 Mpro conserved in all homologous proteins.
3.1.3. nsp10-nsp16 (methyltransferase)
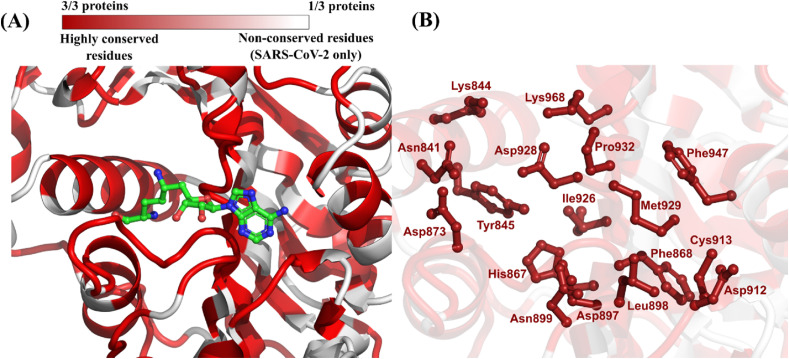
Only two homologous proteins of SARS-CoV-2 methyltransferase were identified in this study. These proteins correspond to the closely related SARS-CoV and MERS-CoV (strain 2c EMC/2012), both members of the Betacoronavirus genus that are known to infect humans and cause severe respiratory disease. The primary sequence alignment showed that SARS-CoV methyltransferase shares a high identity with its SARS-CoV-2 homolog (93.8%). The MERS-CoV homolog also shares a notable primary sequence identity with SARS-CoV-2 (66.3%) (Table 3 ; Fig. S3, Supplementary Material). Afterwards, the 3D structural alignment revealed a remarkable conservation in the binding site defined around the co-crystallized inhibitor sinefungin (Fig. 4 A) (Rosas-Lemus et al., 2020). Although the high conservation could possibly be attributed to the reduced number of proteins compared in this case, it is notable how those three betacoronaviruses have important matches in binding site compositions. In total, 17 residues in the binding site of methyltransferase from SARS-CoV-2, which correspond to 77.3% of binding site composition, were conserved in all three homologous proteins (Table 3; Fig. 4B). Lin et al., also compared their crystal structure of the SARS-CoV-2 nsp10/nsp16 2′-O-methylase structure (PDB: 7C2I, 7C2J) to both SARS-CoV (PDB: 3R24) and MERS-CoV (PDB: 5YNM). While their analysis showed highly similar structures for SARS-CoV-2 and MERS-CoV, there were some differences observed in the RNA-binding groove of SARS-CoV which the authors attribute to a possible artifact in the structure for this region. Although the crystal structure provided by Lin et al. was not used in this study, this comparison highlights the observed conservation of both the primary sequence and secondary structure between the coronaviruses nsp10-nsp16 proteins (Lin et al., 2020).
Table 3.
SARS-CoV-2 methyltransferase primary sequence identity and binding site residues conservation against all corresponding homologous proteins identified through ENDscript (Robert and Gouet, 2014).
| PDB ID | Virus | Genus | Host | Global primary sequence identity to SARS-CoV-2 methyltransferase (%) | SARS-CoV-2 Methyltransferase binding site residues conserved in all species |
|---|---|---|---|---|---|
| 6WKQ | SARS-CoV-2 | Betacoronavirus | Human | Used as query | Lys844, Asn841, Asp873, Lys968, Asp928, Tyr845, His867, Asn899, Asp897, Ile926, Pro932, Leu898, Phe868, Met929, Phe947, Cys913, Asp912 (77.3% of all binding site residues) |
| 2XYR | SARS-CoV | Betacoronavirus | Human | 93.81 | |
| 5YN5 | MERS-CoV (2c EMC/2012) | Betacoronavirus | Human | 66.33 |
Fig. 4.
(A) Color-coded depiction of residue conservation at the binding site of all identified SARS-CoV-2 methyltransferase homologous proteins. Regions in dark red represent residues with high co-occurrence among homologous proteins (i.e., three out of three proteins share the same residue). Regions in light red represent residues with moderate co-occurrence (i.e., two out of three proteins share the same residue). Regions in white represent residues with no co-occurrence (i.e., the residue is present only in SARS-CoV-2). The protein structure used as template is the nsp10-nsp16 methyltransferase from SARS-CoV-2 (PDB ID: 6WKQ) co-crystallized with the inhibitor sinefungin (in green) (Rosas-Lemus et al., 2020); (B) Binding site residues of SARS-CoV-2 methyltransferase conserved in all homologous proteins.
3.1.4. Endoribonuclease (NendoU)
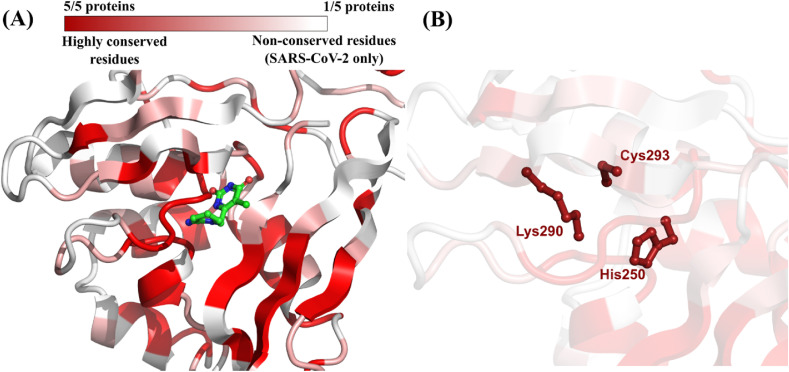
Four homologs of the SARS-CoV-2 endoribonuclease (NendoU) were identified: three betacoronaviruses (SARS-CoV, MERS-CoV, and Murine Hepatitis Virus) and one human alphacoronavirus (hCoV-229E). The results of primary sequence alignment showed that SARS-CoV NendoU shares a high identity (87.9%) with its SARS-CoV-2 homolog (Table 4 ; Fig. S4, Supplementary Material). After 3D structural alignment, a moderate conservation in the binding site, defined around the co-crystallized inhibitor tipiracil, was observed (Fig. 5 A) (Kim et al., 2021). In total, three residues in the binding site of NendoU from SARS-CoV-2, namely His250, Lys290, and Cys293, representing 37.5% of all residues in the binding site, were conserved in all five homologous proteins (Table 4; Fig. 5B).
Table 4.
SARS-CoV-2 Endoribonuclease primary sequence identity and binding site residues conservation against all corresponding homologous proteins identified through ENDscript (Robert and Gouet, 2014).
| PDB ID | Virus | Genus | Host | Global primary sequence identity to SARS-CoV-2 NendoU (%) | SARS-CoV-2 NendoU binding site residues conserved in all species |
|---|---|---|---|---|---|
| 6WXC | SARS-CoV-2 | Betacoronavirus | Human | Used as query | His250 Lys290 Cys293 (37.5% of all binding site residues) |
| 2H85 | SARS-CoV | Betacoronavirus | Human | 87.86 | |
| 5YVD | MERS-CoV | Betacoronavirus | Human | 50.72 | |
| 4S1T | 229E | Alphacoronavirus | Human | 42.30 | |
| 2GTH | Murine Hepatitis Virus (strain A59) | Betacoronavirus | Mouse | 43.88 |
Fig. 5.
(A) Color-coded depiction of residue conservation at the binding site of all identified SARS-CoV-2 NendoU homologous proteins. Regions in dark red represent residues with high co-occurrence among homologous proteins (i.e., five out of five proteins share the same residue). Regions in light red represent residues with moderate co-occurrence (i.e., between 2 and 4 out of five proteins share the same residue). Regions in white represent residues with no co-occurrence (i.e., the residue is present only in SARS-CoV-2). The protein structure used as template is the NendoU from SARS-CoV-2 (PDB ID: 6WXC) co-crystallized with the inhibitor tipiracil (in green) (Kim et al., 2021); (B) Binding site residues of SARS-CoV-2 NendoU conserved in all homologous proteins.
3.2. Primary sequence comparison of remaining targets
A total of 21 additional SARS-CoV-2 proteins, not included in the three-dimensional binding site comparisons, due to the lack of co-crystallized inhibitors, were used to search for homologs based only on their primary sequences. The results of primary sequence analysis, for 24 SARS-CoV-2 proteins, are summarized in Fig. 6 and Table S1 (Supplementary Material). Two proteins did not return any results after BLAST(Altschul et al., 1990) search (nsp11 and orf10). Protein orf10 has not yet been confirmed at the experimental level and has the lowest annotation score in the Swiss-Prot database (Apweiler et al., 2004).
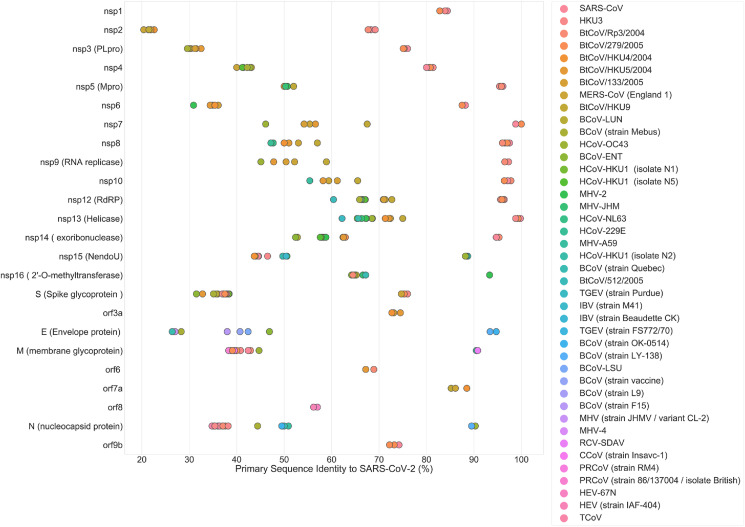
Fig. 6.
Primary sequence identity between homologs, from various coronaviral species, and their counterparts in SARS-CoV-2 identified by BLAST (Altschul et al., 1990). HKU3: Bat coronavirus HKU3; BtCoV: Bat coronavirus; HCoV: Human coronavirus; MHV: Murine Hepatitis Virus; BCoV: Bovine coronavirus; TGEV: Porcine transmissible gastroenteritis coronavirus; IBV: Avian infectious bronchitis virus; RCV: Rat coronavirus; CCoV: Canine coronavirus; PRCoV: Porcine respiratory coronavirus; HEV: Porcine hemagglutinating encephalomyelitis virus; TCoV: Turkey enteric coronavirus.
Although sequence similarity is not analogous to homology, it does provide valuable insight into the possible functions of specific sequences in under-researched coronaviruses in animals such as bats, rats, cows, pigs, turkeys, and others. Higher percent sequence identities are more likely to result in shared Gene Ontology (GO) annotations such as Molecular Function, which may indicate homology between proteins (Joshi and Xu, 2007). The high sequence identity demonstrated between some of the under-researched coronavirus sequences and that of specific proteins in SARS-CoV-2 indicates that these might also be tractable protein targets for antiviral therapies (Fig. 6).
The SARS-CoV-2 pandemic has served as a reminder of the threat posed by highly contagious, emerging viruses. Lack of consistent investment and research into the development of antiviral agents is disappointingly common, often leaving the scientific community struggling to discover therapies and create vaccines in time to treat patients and protect others once an outbreak occurs (e.g., Ebola virus, Zika virus) (Bobrowski et al., 2020). Despite this, the antiviral research prior to the COVID-19 pandemic enabled the scientific community to develop highly effective vaccines in record time as well as quickly place remdesivir into clinical trials and receive emergency use authorization. In many ways the scientific community's response to the pandemic is a success story. Establishing a similar basis for successful treatments of previous and potentially similar newly emerging viruses is crucial to rapidly develop both specific and broad-spectrum antivirals. In this study, we demonstrate an approach to identifying conserved binding site residues across homologous viral proteins as potential target sites for the discovery of broad-spectrum coronavirus antiviral drugs. The rationale for this approach is illustrated by Merck's RDRP inhibitor molnupiravir that successfully passed Phase 3 of clinical trials and recently gained positive FDA advisory committee vote for treatment of mild to moderate COVID-19 in high-risk adults. Molnupiravir case follows the same approach as discussed in this study, i.e., identify conserved target (RDRP in this case), test drugs, find the one that works, ensure it works across multiple strains, subject to in vivo experiments and clinical trials (Fischer et al., 2021; Kabinger et al., 2021; Willyard, 2021).
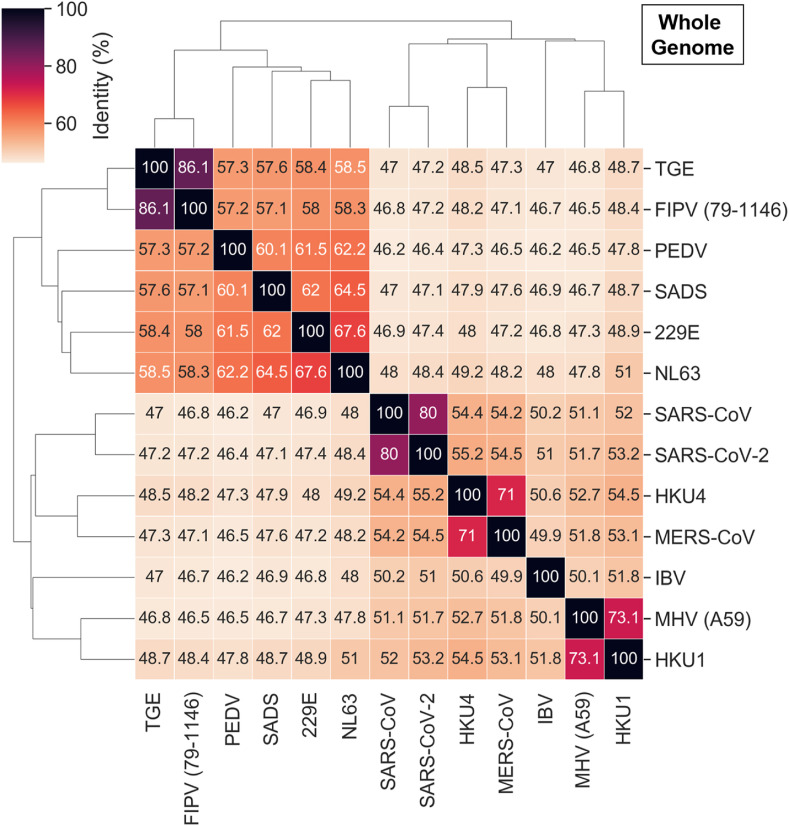
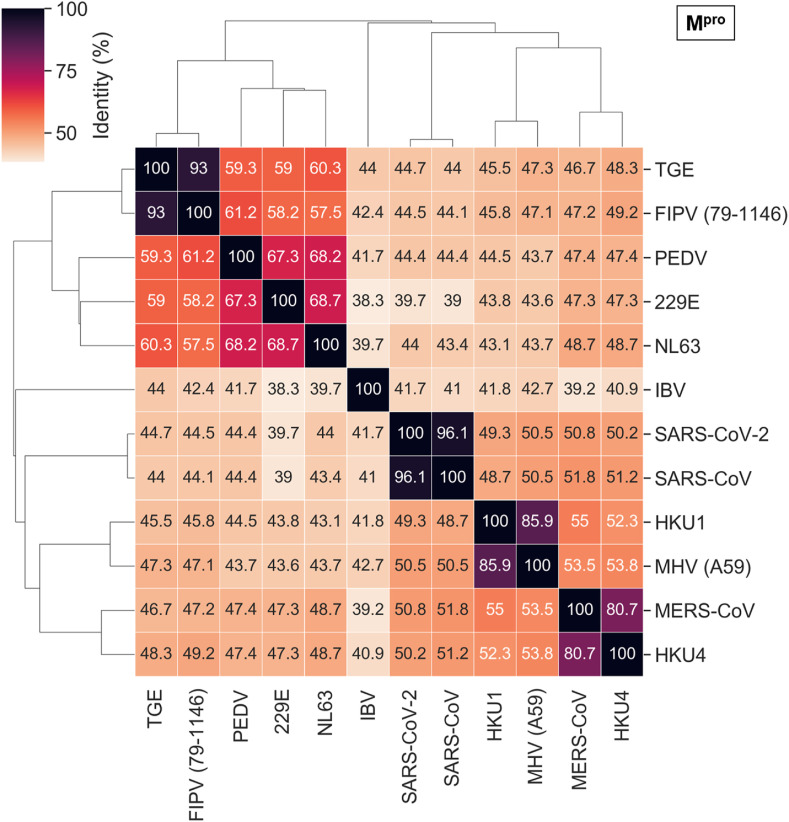
As detailed above, the conservation at the levels of sequence, structure, and binding sites among betacoronavirus proteins was especially high for SARS-CoV-2, SARS-CoV, and MERS-CoV. While perhaps not as strong, the additional binding site similarities for other coronaviruses should not be disregarded. Exploring the homologs with the highest sequence and, especially, binding site similarity could provide crucial insight for the development of broad-spectrum antivirals, including viral outbreaks yet to come. The pairwise whole genome and Mpro sequence similarity between the coronaviruses with available crystal structures, that we used for binding site comparison, are represented in Fig. 7 and Fig. 8 . Similar analysis for PLpro, nsp10-nsp16, and NendoU proteins is depicted in Figs. S5–S7.
Fig. 7.
Identity between all coronaviruses considered in our study based on whole genome sequence comparison.
Fig. 8.
Primary sequence comparison of Mpro from different coronaviruses.
Identifying conserved viral proteins can help both find similar proteins that would respond to the same (or similar) treatments as well as shed light on the key differences that might affect treatment efficacy. However, because our data compared the residue homology against the binding sites of proteins in SARS-CoV-2, our set is limited in that there could be homology with other coronaviruses that do not have proteins with existing crystal structures. This potential pitfall is represented in our study by comparing percent sequence identity of understudied coronaviruses in animals, revealing possible homologous proteins in these coronaviruses and pointing to the need for the elucidation of additional viral protein crystal structures. This elucidation could be assisted by the prediction of respective protein structures using recently developed computational tools such as AlphaFold 2 (Jumper et al., 2021) that showed high accuracy in the most recent CASP competition (CASP14, 2020).
Analogous efforts have been made with respect to specific proteases or proteins. Prior to the SARS-CoV-2 outbreak, Kim et al. concluded that the homologous Mpro orthosteric residues of various positive-sense RNA viruses were viable candidate drug target sites for potential wide spectrum treatments (Kim et al., 2012a). In 2004, Hillisch et al., attempted homology modeling of the relatively novel SARS-CoV Mpro but at this point they were unsuccessful and deemed the modeling insufficient (Hillisch et al., 2004). Yet, in 2003, Anand et al., were able to identify considerable conservation of the SARS-CoV Mpro binding site with that of the transmissible gastroenteritis virus, a porcine coronavirus (Anand, 2003). Interestingly, Yang et al. reviewed drugs that were developed for SARS-CoV and referenced the potential for Mpro inhibitors as wide-spectrum antivirals in 2006 (Yang et al., 2006).
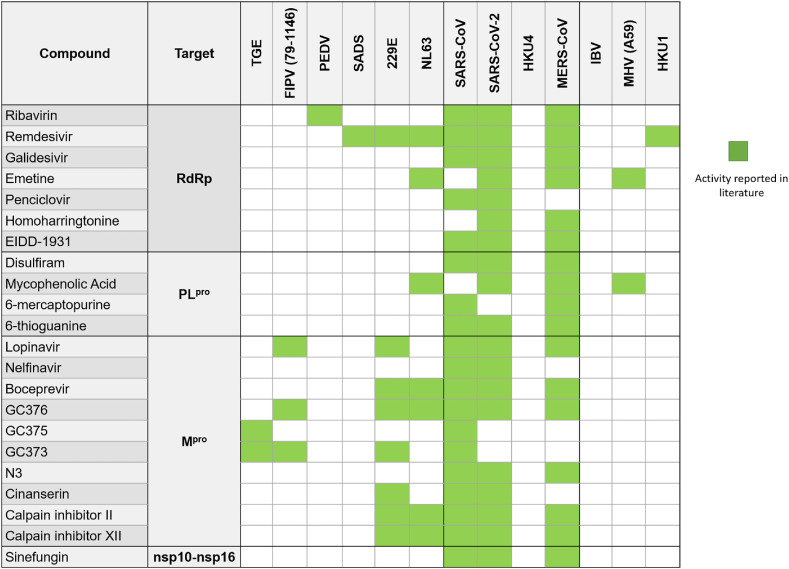
The response of scientists to the SARS-CoV-2 outbreak stands as a testament to the exponential advances in scientific knowledge in short periods of time. A great example of this is Pfizer's development of a (relatively) selective Mpro inhibitor that is active against multiple coronaviruses. Their compound, identified as PF-07304814, is metabolized in the body into a potent Mpro inhibitor that has gone into Phase 1b clinical trials (Boras et al., 2020) (albeit recently Pfizer stopped developing this drug for the lack of efficacy in patients) (Taylor, 2022). To this point, many such examples of Mpro inhibitors targeting multiple coronaviruses from our study exist (Fig. 9 ) including protease inhibitors of hepatitis C (boceprevir), and feline infectious peritonitis virus (GC376). Similarly, inhibitors of the RNA-dependent RNA polymerase (RdRp) have shown activity against multiple coronaviruses (Fig. 9) including the approved broad-spectrum antiviral remdesivir, a nucleotide analog prodrug that incorporates into the growing RNA and induces a translocation barrier to stall RdRp (Beigel et al., 2020; Kokic et al., 2021). Other nucleoside analogs including molnupiravir and galidesivir have also shown to be effective inhibitors of RdRp. Molnupiravir has recently successfully passed Phase 3 of clinical trials and recently gained positive FDA advisory committee vote for treatment of mild to moderate COVID-19 in high-risk adults (Fischer et al., 2021; Kabinger et al., 2021; Willyard, 2021). Thus, further exploring nucleoside analogs that exploit remdesivir's mechanism of action for multiple coronaviruses could provide additional treatment options, lowering the high demand for remdesivir, therefore making treatment more affordable for patients.
Fig. 9.
Examples of inhibitors of homologous proteins targeting multiple coronaviruses. RdRP (Bassendine et al., 2020; Brown et al., 2019; Chen et al., 2004; Choy et al., 2020; de Wit et al., 2020; Edwards et al., 2020; Imran et al., 2021; Jeon et al., 2020; Kim and Lee, 2013; Saijo et al., 2005; Sharif-Yakan and Kanj, 2014; Sheahan et al., 2020; Wang et al., 2020; Xu et al., 2021; Zhao et al., 2021); PLpro (Armstrong et al., 2021; Chan et al., 2013; Cheng et al., 2015; Chou et al., 2008; F et al., 2020; Jan et al., 2021; Lin et al., 2018; Swaim et al., 2020; Xu et al., 2021); Mpro (Choy et al., 2020; de Wilde et al., 2014; Fu et al., 2020; Hu et al., 2020; Jan et al., 2021; Jeon et al., 2020; Jin et al., 2020; Kim et al., 2012b, 2016; Ma et al., 2020; Theerawatanasirikul et al., 2020; Xu et al., 2020, 2021; Yamamoto et al., 2004; Chen et al., 2005); nsp10-nsp16 (Aouadi et al., 2017; Benoni et al., 2020; Bouvet et al., 2010; Decroly et al., 2011; Perveen et al., 2021). The table contains all available experimental data for drugs and compounds known to target the respective proteins and tested against different coronaviruses. TGE: Porcine transmissible gastroenteritis coronavirus; FIPV: Feline Infectious Peritonitis Virus; PEDV: Porcine Epidemic Diarrhea Virus; SADS: Swine Acute Diarrhea Syndrome Coronavirus; HKU4: Bat Coronavirus HKU4; IBV: Avian Infectious Bronchitis Virus; MHV: Murine Hepatitis Virus; HKU1: Human HKU1 Coronavirus.
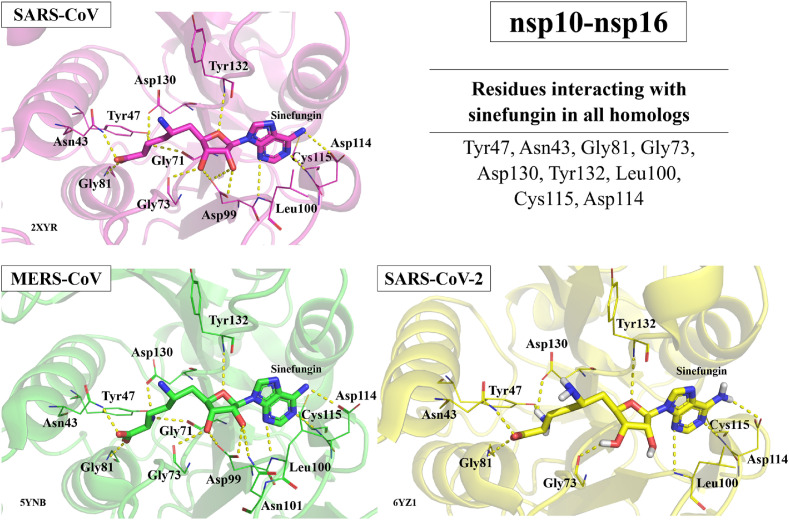
As mentioned above, PLpro or nsp10-16 displayed homology at levels equal to or above that of Mpro. This bodes well for the possibility of finding other drugs that will act similarly against PLpro or nsp10-16. While less explored than Mpro and RdRp, a few compounds targeting the PLpro, or the nsp10-16 complex of multiple coronaviruses have been identified (Fig. 9). One interesting example of a PLpro inhibitor effective against SARS-CoV, MERS-CoV, and SARS-CoV-2 is alcohol-aversive medication disulfiram, which is currently in Phase 2 clinical trials for the treatment of SARS-CoV-2. Thiopurine analogs, like 6-mercaptopurine and 6-thioguanine also inhibit PLpro in SARS-CoV and MERS-CoV, and 6-thioguanine also inhibits PLpro in SARS-CoV-2. As for nsp10-16, the nucleoside sinefungin is effective against SARS-CoV, MERS-CoV, and SARS-CoV-2. Sinefungin acts as a methyltransferase inhibitor and is effective against coronaviruses due to its relation to S-adenosylmethionine (SAM), the methyl-donor required for the RNA capping process which is essential for viral replication and allows coronaviruses to evade the human immune system (Lin et al., 2020). Thus, although heretofore understudied as broad-spectrum coronavirus inhibitor targets, the high level of homology of both PLpro and nsp10-16 as well as the proof of concept displayed by the few known inhibitors to date suggest PLpro and nsp10-16 could be promising targets for future development of broad-spectrum coronavirus inhibitors. Sinefungin, for example, is a well-known inhibitor of nsp10-nsp16 protein in SARS-CoV, SARS-CoV-2, and MERS-CoV. As observed in Fig. 10 , based on available crystal structures deposited in PDB, sinefungin interacts with the same nine binding site residues in all three coronaviruses.
Fig. 10.
Interactions between sinefungin and binding site residues of nsp10-nsp16 preserved in all three homologous proteins from SARS-CoV, SARS-CoV-2, and MERS-CoV. PDB IDs: 2XYR, 5YNB, 6YZ1.
Unfortunately, there are no compounds tested against all or even the majority of the viruses of interest. As represented in Fig. 9, each compound was tested on average against 3–4 coronaviruses out of 13, ranging from 2 to 7 viruses per compound. SARS-CoV-2, SARS-CoV, and MERS-CoV were the most frequently tested viruses. On the other hand, no information was available on the binding of any of the 13 compounds to the viral targets for 2 of the 13 viruses. The compilation of all the available data results in a matrix with the sparsity degree of 72% which drives forth the question as to how many coronaviruses these compounds could actually work against. There were other drugs that have either been tested only in SARS-CoV-2 or negative results have not been published. These drugs were not reported in this study. Table 5 contains data on compounds (including molnupiravir) that inhibited at least one of the coronavirus’ homologous proteins discussed above. These compounds could potentially be tested against homologs from different coronavirus species. Moreover, combinations of the compounds from Table 5 that bind to protein targets responsible for different stages of viral lifecycle may be helpful in creating synergistic drug combinations preserving their broad-spectrum activity (Muratov and Zakharov, 2020).
Table 5.
Examples of a SAR-CoV-2 EUA candidate, molnupiravir, and in vitro inhibitors, reported in ChEMBL database, targeting proteins analyzed in our study.
| Compound | Structure | Target | Virus | References |
|---|---|---|---|---|
| Molnupiravir | 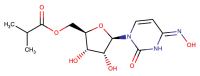 |
RdRP | SARS-CoV-2 | (Fischer et al., 2021; Kabinger et al., 2021; Willyard, 2021) |
| CHEMBL4522602 | 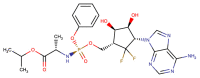 |
RdRP | MERS-CoV | Yoon et al. (2019) |
| CHEMBL4544781 | 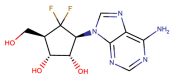 |
RdRP | MERS-CoV | Yoon et al. (2019) |
| CHEMBL2115462 | 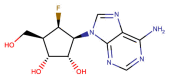 |
RdRP | MERS-CoV | Yoon et al. (2019) |
| CHEMBL421 (Sulfasalazine) | 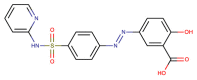 |
PLpro | MERS-CoV | Lee et al. (2019) |
| CHEMBL1368663 | 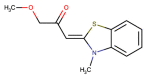 |
PLpro | MERS-CoV | Lee et al. (2019) |
| CHEMBL1595473 | 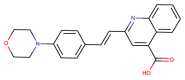 |
PLpro | MERS-CoV | Lee et al. (2019) |
| CHEMBL480 (Lansoprazole) | 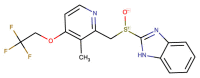 |
Mpro | SARS-CoV-2 | Kuzikov et al. (2021) |
| CHEMBL297453 | 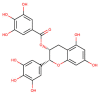 |
Mpro | SARS-CoV-2 | Kuzikov et al. (2021) |
| CHEMBL1271993 | 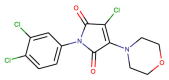 |
Mpro | SARS-CoV-2 | Kuzikov et al. (2021) |
Recently, several studies similar to ours have been published. Also exploring the conservation of coronaviruses, Schapira et al. aimed to identify drug binding sites within the SARS-CoV-2 proteome (Yazdani et al., 2021). Druggable binding pockets were mapped onto experimental structures of SARS-CoV-2 proteins and analyzed for their conservation across all available PDB structures of α- and β-coronaviruses, as well as samples from patients with SARS-CoV-2. The present study complements that of Schapira et al., whereby it further explores the idea that similarities between homologous coronavirus proteins could be exploited for target prioritization and the development of broad-spectrum anti-coronaviral compounds, while also putting this into the context of potential broad-spectrum inhibitors of conserved targets from literature. Despite the findings of both Schapira et al., and this work, a recent molecular dynamics simulation study was published comparing ligand-binding sites available for SARS-CoV2, SARS-CoV, and MERS-CoV Mpro (Cho et al., 2021). From their simulations, the authors concluded that developing a pan-inhibitor of Mpro based on protein conservation could be extremely challenging due to differences in the dynamics of the binding sites. While this study depicts an interesting consideration in the design of future antiviral medications, it is not supported by any experimental results. In contrast, we identified drugs inhibiting the targets discussed in this study, carefully collected, and analyzed all known experimental data on their antiviral activity and estimated their potential as broad-spectrum drugs.
Unfortunately, targeting highly conserved targets does not always translate into broad-spectrum antiviral activity. While conservation does give a good idea of the breadth of the potential spectrum of activity, there are numerous factors that create anomalies and discrepancies. As described (Prichard, 2007), there are nuances within selected molecules that could alter their activity in a broad-spectrum application. Some of these include but are not limited to spectrum specificity, ligand activity, binding regions of similar viruses, and post-translational modifications of proteins (e.g., different phosphorylation patterns). These differences are generally enough to explain the exceptions and compounds that do not work as expected (Prichard, 2007). While understanding the impact of each of these nuances will be crucial to interpreting any further results derived from conservation studies, it should not discourage the creation of a base of knowledge so that researchers do not have to start from nothing with every new viral epidemic (Muratov et al., 2021).
Collecting data as we encounter new viral threats can help in future efforts. The recent emergence and spread of SARS-CoV-2 reminded the public of the momentous threat held by zoonotic viruses (Bobrowski et al., 2020). However, SARS-CoV-2 is just one of over 250 viruses to have jumped from animal to humans and caused disease (Grange et al., 2021). Once a zoonotic virus jumps to humans, the threat to global public health is immense. To understand these spillover events, research has been performed worldwide to understand the risk of each known zoonotic virus and to predict how likely these viruses are to jump to humans. One such tool called SpillOver (Grange et al., 2021; “Spillover,” n.d.) was developed to identify host, viral, and environmental risk factors contributing to zoonoses. SpillOver uses these risk factors to provide a risk assessment score to 887 known viruses for their potential to jump to humans; the first 12 of which were known to have already made the jump. Knowledge collections like these, in combination with our work, are invaluable in our preparation for the next, inevitable virus to jump to humans.
4. Conclusions and perspectives
Exploring the conservation between homologous coronavirus proteins is a valuable strategy for drug target selection that could assist the development of broad-spectrum anti(corona)viral compounds. We analyzed the primary sequence similarity between all known SARS-CoV-2 proteins and their homologs from several human and animal coronaviruses. Furthermore, we investigated 3D binding site similarities, using the ENDscript server, between four SARS-CoV-2 proteins and their several homologs with three-dimensional structures available in the PDB: Papain-Like Protease (PLpro), Main Protease (Mpro), Methyltransferase (nsp10-nsp16), and Endoribonuclease (NendoU). All the aforementioned proteins presented important binding site conservation between SARS-CoV-2 and different human and animal coronaviruses. It is important to highlight that all results of the binding site conservation analysis are limited by the availability of the corresponding homologous protein structures in PDB. To demonstrate the potential of exploring conserved homologous proteins for the development of broad-spectrum antivirals, we found several examples of bioactive compounds and approved drugs, known to inhibit those proteins, with reported activity against different animal and human coronaviruses. Some examples include the RdRp inhibitor remdesivir, the PLpro inhibitor disulfiram, and the nsp10-nsp16 inhibitor sinefungin.
Examining the homology of ligand binding sites in coronavirus proteins could provide an immense support in searching for broad spectrum direct antiviral agents as novel viruses continue to emerge. With this goal in mind, initiatives such as NIH's Antiviral Program for Pandemics (“NIH Antiviral Program for Pandemics,” 2021) and READDI (“The Rapidly Emerging Antiviral Drug Development Initiative (READDI),” 2021) at UNC Chapel Hill are working to develop broad-spectrum antivirals and bring them to phase I/II clinical trials so they are readily available for future viral outbreaks. This way, the scientific community does not have to start ex nihilo in regard to antiviral drug development and may already have a head start on managing outbreaks before they reach pandemic levels (“Open science drug discovery partnership, READDI, aims to invest $125 million to prevent future pandemics,” 2020).
One major advantage of surveying conservation is the ability to consider individual protein targets. In doing so, the common proteins responsible for different viral functions, such as replication, can be targeted and applied across a greater number of viruses. As opposed to targeting viral structural proteins (which may be more important targets for vaccine development), targeting replication proteins for small molecule therapies in homologous binding sites should be evaluated in a more nuanced study to determine if they may be pertinent in the search for both selective and broad-spectrum inhibitors.
Moving forward, the next step would be to attempt to compare more homologs within the Coronaviridae as well as potentially moving outside this family. While we did this on a small scale, more expansive research should be done. Targeting common host proteins and pathways involved in viral entry and replication is another potential strategy for broad-spectrum antivirals design (including the combination therapy), e.g., exploring the link between both T-cell immunity in SARS-CoV and SARS-CoV-2 patients as well as the shared binding to the ACE2 receptor that could provide a potential therapeutic overlap (Bobrowski et al., 2021; da Costa et al., 2020; Dutta, 2022).
In summary, we note that finding chemicals active against highly conserved targets in laboratory tests does not always translate into new broad-spectrum antivirals. However, our studies suggest that this strategy could result in new treatments both for current and future viral epidemics and therefore the protein targets that contain conserved sequence and at least partially conserved binding sites should continue to be explored for the discovery of broad-spectrum direct antivirals.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Acknowledgements
This study was supported in part by NIH grants R01GM140154, U19AI171292, and RO1 AI108197.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105360.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anand K. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Andersen P.I., Ianevski A., Lysvand H., Vitkauskiene A., Oksenych V., Bjørås M., Telling K., Lutsar I., Dumpis U., Irie Y., Tenson T., Kantele A., Kainov D.E. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int. J. Infect. Dis. 2020;93:268–276. doi: 10.1016/j.ijid.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi W., Blanjoie A., Vasseur J., Debart F., Canard B., Decroly E. Binding of the methyl donor S -adenosyl- l -methionine to Middle East respiratory syndrome coronavirus 2′- O -methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J. Virol. 2017;91:1–18. doi: 10.1128/JVI.02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M.J., Natale D. a, O'Donovan C., Redaschi N., Yeh L.-S.L. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L.A., Lange S.M., Dee Cesare V., Matthews S.P., Nirujogi R.S., Cole I., Hope A., Cunningham F., Toth R., Mukherjee R., Bojkova D., Gruber F., Gray D., Wyatt P.G., Cinatl J., Dikic I., Davies P., Kulathu Y. Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its inhibition by nanobodies. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti I., Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metabol. Syndr.: Clin. Res. Rev. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- F K., S M., M K., T H., H K., M T. Antiviral activities of mycophenolic acid and IMD-0354 against SARS-CoV-2. Microbiol. Immunol. 2020;64:635–639. doi: 10.1111/1348-0421.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassendine M.F., Bridge S.H., McCaughan G.W., Gorrell M.D. COVID‐19 and comorbidities: a role for dipeptidyl peptidase 4 (DPP4) in disease severity? J. Diabetes. 2020;12:649–658. doi: 10.1111/1753-0407.13052. [DOI] [PubMed] [Google Scholar]
- Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19 — final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoni R., Krafcikova P., Baranowski M.R., Kowalska J., Boura E., Cahova H. Substrate specificity of SARS-CoV-2 nsp10-nsp16 methyltransferase Roberto. bioRxiv : the preprint server for biology. 2020;21:1–9. doi: 10.1101/2020.07.30.228478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski T., Melo-Filho C.C., Korn D., Alves V.M., Popov K.I., Auerbach S., Schmitt C., Moorman N.J., Muratov E.N., Tropsha A. Learning from history: do not flatten the curve of antiviral research. Drug Discov. Today. 2020:1–10. doi: 10.1016/j.drudis.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski T., Chen L., Eastman R.T., Itkin Z., Shinn P., Chen C.Z., Guo H., Zheng W., Michael S., Simeonov A., Hall M.D., Zakharov A.V., Muratov E.N. Synergistic and antagonistic drug combinations against SARS-CoV-2. Mol. Ther. 2021;29:873–885. doi: 10.1016/j.ymthe.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., Hammond J., Hoffman R., Kadar E.P., Kania R., Kimoto E., Kirkpatrick M.G., Lanyon L., Lendy E.K., Lillis J.R., Luthra S.A., Ma C., Noell S., Obach R.S., O'Brien M.N., O'Connor R., Ogilvie K., Owen D., Pettersson M., Reese M.R., Rogers T., Rossulek M.I., Sathish J.G., Steppan C., Ticehurst M., Updyke L.W., Zhu Y., Wang J., Chatterjee A.K., Mesecar A.D., Anderson A.S., Allerton C. 2020. Discovery of a Novel Inhibitor of Coronavirus 3CL Protease as a Clinical Candidate for the Potential Treatment of COVID-19. bioRxiv : the Preprint Server for Biology. [DOI] [Google Scholar]
- Boriskin Y., Leneva I., Pecheur E.-I., Polyak S. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr. Med. Chem. 2008;15:997–1005. doi: 10.2174/092986708784049658. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J., Won J.J., Graham R.L., Dinnon K.H., Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASP14 . 2020. 14th community wide experiment on the critical assessment of techniques for protein structure prediction [WWW Document]https://predictioncenter.org/casp14/index.cgi 1.17.22. URL. [Google Scholar]
- Chakraborty I., Maity P. COVID-19 outbreak: migration, effects on society, global environment and prevention. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Chan K.-H., Kao R.Y.T., To K.K.W., Zheng B.-J., Li C.P.Y., Li P.T.W., Dai J., Mok F.K.Y., Chen H., Hayden F.G., Yuen K.-Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Chan K.H., Jiang Y., Kao R.Y.T., Lu H.T., Fan K.W., Cheng V.C.C., Tsui W.H.W., Hung I.F.N., Lee T.S.W. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E., Drosten C., Bai D., He X., Ludewig B., Chen J., Luo H., Yang Yiming, Yang Yifu, Zou J., Thiel V., Chen K., Shen J., Shen X., Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K.-W., Cheng S.-C., Chen W.-Y., Lin M.-H., Chuang S.-J., Cheng I.-H., Sun C.-Y., Chou C.-Y. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antivir. Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E., Rosa M., Anjum R., Mehmood S., Soban M., Mujtaba M., Bux K., Moin S.T., Tanweer M., Dantu S., Pandini A., Yin J., Ma H., Ramanathan A., Islam B., Mey A.S.J.S., Bhowmik D., Haider S. Dynamic profiling of β-coronavirus 3CL M pro protease ligand-binding sites. J. Chem. Inf. Model. 2021;61:3058–3073. doi: 10.1021/acs.jcim.1c00449. [DOI] [PubMed] [Google Scholar]
- Chou C.-Y., Chien C.-H., Han Y.-S., Prebanda M.T., Hsieh H.-P., Turk B., Chang G.-G., Chen X. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75:1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.-T., Wong A.Y.-L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P.-H., Huang X., Peiris M., Yen H.-L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Advances in Virus Research. 2018. Hosts and sources of endemic human coronaviruses; pp. 163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus Disease 2019 COVID-19 EUA information [WWW Document] 2021. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization#covid19euas 10.5.21, URL.
- COVID-19 dashboard by the Center for systems science and engineering (CSSE) at Johns Hopkins [WWW Document] 2022. https://coronavirus.jhu.edu/map.html URL, 3.25.22. [DOI] [PMC free article] [PubMed]
- da Costa V.G., Moreli M.L., Saivish M.V. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020;165:1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta K. Allosteric site of ACE-2 as a drug target for COVID-19. ACS Pharm. Translat. Sci. acsptsci. 2022:2c00003. doi: 10.1021/acsptsci.2c00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C.E., Yount B.L., Graham R.L., Leist S.R., Hou Y.J., Dinnon K.H., Sims A.C., Swanstrom J., Gully K., Scobey T.D., Cooley M.R., Currie C.G., Randell S.H., Baric R.S. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:26915–26925. doi: 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. In: Coronaviruses: Methods and Protocols, Methods in Molecular Biology. Maier H.J., Bickerton E., Britton P., editors. Springer New York; New York, NY: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Eron J.J., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R., Duke E.R., Azizad M.M., Borroto-Esoda K., Wohl D.A., Loftis A.J., Alabanza P., Lipansky F., Painter W.P. 2021. Molnupiravir, an Oral Antiviral Treatment for COVID-19. medRxiv : the Preprint Server for Health Sciences. [DOI] [Google Scholar]
- Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. SARS-CoV-2: combating coronavirus emergence. Immunity. 2020;52:734–736. doi: 10.1016/j.immuni.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange Z.L., Goldstein T., Johnson C.K., Anthony S., Gilardi K., Daszak P., Olival K.J., O'Rourke T., Murray S., Olson S.H., Togami E., Vidal G., Mazet J.A.K. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2002324118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillisch A., Pineda L.F., Hilgenfeld R. Utility of homology models in the drug discovery process. Drug Discov. Today. 2004;9:659–669. doi: 10.1016/S1359-6446(04)03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Pöhlmann S. Priming time: how cellular proteases arm coronavirus spike proteins. Activat. Viruses Host Proteases. 2018;71 doi: 10.1007/978-3-319-75474-1_4. [DOI] [Google Scholar]
- Hu Y., Ma C., Szeto T., Hurst B., Tarbet B., Wang J. Boceprevir, calpain inhibitors II and XII, and GC-376 have broad-spectrum antiviral activity against coronaviruses in cell culture. bioRxiv. 2020 doi: 10.1101/2020.10.30.362335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran M., Alshrari A.S., Asdaq S.M.B., Abida Trends in the development of remdesivir based inventions against COVID-19 and other disorders: a patent review. J. Infection Public Health. 2021;14:1075–1086. doi: 10.1016/j.jiph.2021.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack B.R., Meyer A.G., Echave J., Wilke C.O. Functional sites induce long-range evolutionary Constraints in enzymes. PLoS Biol. 2016;14:1–23. doi: 10.1371/journal.pbio.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan J.-T., Cheng T.-J.R., Juang Y.-P., Ma H.-H., Wu Y.-T., Yang W.-B., Cheng C.-W., Chen X., Chou T.-H., Shie J.-J., Cheng W.-C., Chein R.-J., Mao S.-S., Liang P.-H., Ma C., Hung S.-C., Wong C.-H. Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2021579118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang Xinglou, You T., Liu Xiaoce, Yang Xiuna, Bai F., Liu H., Liu Xiang, Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Joshi T., Xu D. Quantitative assessment of relationship between sequence similarity and function similarity. BMC Genom. 2007;8:222. doi: 10.1186/1471-2164-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabinger F., Stiller C., Schmitzová J., Dienemann C., Kokic G., Hillen H.S., Höbartner C., Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lee C. Ribavirin efficiently suppresses porcine nidovirus replication. Virus Res. 2013;171:44–53. doi: 10.1016/j.virusres.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.-C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.-O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/jvi.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.-C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.-O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Liu H., Galasiti Kankanamalage A.C., Weerasekara S., Hua D.H., Groutas W.C., Chang K.-O., Pedersen N.C. Reversal of the progression of fatal coronavirus infection in Cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Wower J., Maltseva N., Chang C., Jedrzejczak R., Wilamowski M., Kang S., Nicolaescu V., Randall G., Michalska K., Joachimiak A. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Communications Biology. 2021;4:193. doi: 10.1038/s42003-021-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Höbartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzikov M., Costanzi E., Reinshagen J., Esposito F., Vangeel L., Wolf M., Ellinger B., Claussen C., Geisslinger G., Corona A., Iaconis D., Talarico C., Manelfi C., Cannalire R., Rossetti G., Gossen J., Albani S., Musiani F., Herzog K., Ye Y., Giabbai B., Demitri N., Jochmans D., de Jonghe S., Rymenants J., Summa V., Tramontano E., Beccari A.R., Leyssen P., Storici P., Neyts J., Gribbon P., Zaliani A. Identification of inhibitors of SARS-Cov2 M-Pro enzymatic activity using a small molecule repurposing screen. 2021. [DOI] [PMC free article] [PubMed]
- Lau S.K.P., Li K.S.M., Tsang A.K.L., Lam C.S.F., Ahmed S., Chen H., Chan K., Woo P.C.Y., Yuen K. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence Divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese pipistrelle : implications for the origin of the novel Middle East. Respiratory. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J.A., Tagliamonte M.S., White S.K., Elbadry M.A., Alam M.M., Stephenson C.J., Bonny T.S., Loeb J.C., Telisma T., Chavannes S., Ostrov D.A., Mavian C., De Rochars V.M.B., Salemi M., Morris J.G. 2021. Emergence of Porcine Delta-Coronavirus Pathogenic Infections Among Children in Haiti through Independent Zoonoses and Convergent Evolution. medRxiv : the Preprint Server for Health Sciences. [DOI] [Google Scholar]
- Lee H., Ren J., Pesavento R.P., Ojeda I., Rice A.J., Lv H., Kwon Y., Johnson M.E. Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via high-throughput screening and molecular modeling. Bioorg. Med. Chem. 2019;27:1981–1989. doi: 10.1016/j.bmc.2019.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-H., Moses D.C., Hsieh C.-H., Cheng S.-C., Chen Y.-H., Sun C.-Y., Chou C.-Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antivir. Res. 2018;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Chen H., Ye F., Chen Z., Yang F., Zheng Y., Cao Y., Qiao J., Yang S., Lu G. Crystal structure of SARS-CoV-2 nsp10/nsp16 2′-O-methylase and its implication on antiviral drug design. Signal Transduct. Targeted Ther. 2020;5:131. doi: 10.1038/s41392-020-00241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchatre-Leroy E., Boué F., Boucher J.-M., Renault C., Moutou F., Ar Gouilh M., Umhang G. Identification of alpha and beta coronavirus in wildlife species in France: bats, rodents, rabbits, and hedgehogs. Viruses. 2017;9:364. doi: 10.3390/v9120364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratov E., Zakharov A. 2020. Viribus Unitis: Drug Combinations as a Treatment against COVID-19. ChemRxiv : the Preprint Server for Chemistry. [Google Scholar]
- Muratov E.N., Amaro R., Andrade C.H., Brown N., Ekins S., Fourches D., Isayev O., Kozakov D., Medina-Franco J.L., Merz K.M., Oprea T.I., Poroikov V., Schneider G., Todd M.H., Varnek A., Winkler D.A., Zakharov A.V., Cherkasov A., Tropsha A. A critical overview of computational approaches employed for COVID-19 drug discovery. Chem. Soc. Rev. 2021;50:9121–9151. doi: 10.1039/d0cs01065k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI The BLAST databases [WWW Document] 2020. https://ftp.ncbi.nlm.nih.gov/blast/documents/blastdb.html 4.1.21, URL.
- NIH Antiviral Program for pandemics [WWW Document] 2021. https://www.niaid.nih.gov/research/antivirals 9.6.21, URL.
- Open science drug discovery partnership . 2020. READDI, Aims to Invest $125 Million to Prevent Future Pandemics [WWW Document]https://pharmacy.unc.edu/2020/04/open-science-drug-discovery-partnership-readdi-aims-to-invest-125-million-to-prevent-future-pandemics/ 2.2.21. URL. [Google Scholar]
- Perveen S., Khalili Yazdi A., Devkota K., Li F., Ghiabi P., Hajian T., Loppnau P., Bolotokova A., Vedadi M. A high-throughput RNA Displacement assay for screening SARS-CoV-2 nsp10-nsp16 complex toward developing therapeutics for COVID-19. SLAS DISCOVERY: Adv. Sci. Drug Discov. 2021 doi: 10.1177/2472555220985040. 247255522098504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard M.N. In: Human Herpesviruses Biology, Therapy, and Immunoprophylaxis. Arvin A., Campadelli-Fiume G., Mocarski E., Moore P.S., Roizman B., Whitley R., Yamanishi K., editors. 2007. New approaches to antiviral drug discovery (genomics/proteomics) Cambridge. [PubMed] [Google Scholar]
- The rapidly emerging antiviral drug development initiative (READDI) [WWW Document] 2021. https://www.readdi.org/ 9.6.21, URL.
- Ricagno S., Egloff M.-P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. Unit. States Am. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Brunzelle J., Satchell K.J.F. High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 2020;13 doi: 10.1126/SCISIGNAL.ABE1202/SUPPL_FILE/ABE1202_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J.-F. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antivir. Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., el Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. 2020. Activity Profiling and Structures of Inhibitor-Bound SARS-CoV-2-PLpro Protease Provides a Framework for Anti-COVID-19 Drug Design. bioRxiv : the Preprint Server for Biology 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo M., Morikawa S., Fukushi S., Mizutani T., Hasegawa H., Nagata N., Iwata N., Kurane I. Inhibitory effect of mizoribine and ribavirin on the replication of severe acute respiratory syndrome (SARS)-associated coronavirus. Antivir. Res. 2005;66:159–163. doi: 10.1016/j.antiviral.2005.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladino V., Algeri D., Auriemma V. The psychological and social impact of Covid-19: new perspectives of well-being. Front. Psychol. 2020;11:1–6. doi: 10.3389/fpsyg.2020.577684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif-Yakan A., Kanj S.S. Emergence of MERS-CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:1–6. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillover [WWW Document], n.d. URL https://spillover.global/ranking-comparison/(accessed 8.12.21).
- Swaim C.D., Perng Y.-C., Zhao X., Canadeo L.A., Harastani H.H., Darling T.L., Boon A.C.M., Lenschow D.J., Huibregtse J.M. 6-Thioguanine blocks SARS-CoV-2 replication by inhibition of PLpro protease activities. bioRxiv. 2020 doi: 10.1101/2020.07.01.183020. 2020.07.01.183020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.P. 2022. Pfizer, in a Rare COVID-19 Setback, Dumps Paxlovid's Intravenous Sibling in Further Blow to ACTIV-3 [WWW Document]https://www.fiercebiotech.com/biotech/pfizer-a-rare-covid-19-setback-dumps-paxlovid-s-intravenous-sibling-to-leave-activ-3-future 3.6.22. URL. [Google Scholar]
- Theerawatanasirikul S., Kuo C.J., Phetcharat N., Lekcharoensuk P. In silico and in vitro analysis of small molecules and natural compounds targeting the 3CL protease of feline infectious peritonitis virus. Antivir. Res. 2020;174 doi: 10.1016/j.antiviral.2019.104697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilocca B., Soggiu A., Sanguinetti M., Musella V., Britti D., Bonizzi L., Urbani A., Roncada P. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microb. Infect. 2020;22:188–194. doi: 10.1016/j.micinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Bavari S. Broad-spectrum coronavirus antiviral drug discovery. Expet Opin. Drug Discov. 2019;14:397–412. doi: 10.1080/17460441.2019.1581171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigant F., Santos N.C., Lee B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol. 2015;13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020:2019–2021. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO MERS situation update, February 2022 [WWW Document] 2022. http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html 3.25.22, URL.
- WHO SARS [WWW Document] 2022. http://www.emro.who.int/health-topics/severe-acute-respiratory-syndrome/introduction.html 3.25.22, URL.
- WHO coronavirus (COVID-19) dashboard [WWW Document] 2022. https://covid19.who.int/ 3.25.22, URL.
- Willyard C. How antiviral pill molnupiravir shot ahead in the COVID drug hunt. Nature. 2021 doi: 10.1038/d41586-021-02783-1. [DOI] [PubMed] [Google Scholar]
- Xu Z., Yao H., Shen J., Wu N., Xu Y., Lu X., Zhu W., Li L.-J. 2020. Nelfinavir Is Active against SARS-CoV-2 in Vero E6 Cells. ChemRxiv : the Preprint Server for Chemistryiv. [DOI] [Google Scholar]
- Xu J., Xue Y., Zhou R., Shi P.-Y., Li H., Zhou J. Drug repurposing approach to combating coronavirus: potential drugs and drug targets. Med. Res. Rev. 2021;41:1375–1426. doi: 10.1002/med.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Norio, Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J., Rabenau H., Doerr H.W., Hunsmann G., Otaka A., Tamamura H., Fujii N., Yamamoto Naoki. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004;318:719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the achilles heel of coronaviruses. Curr. Pharmaceut. Des. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- Yazdani S., Maio N. De, Ding Y., Shahani V., Goldman N., Schapira M. Genetic variability of the SARS-CoV-2 pocketome. J. Proteome Res. 2021;20:4215. doi: 10.1021/ACS.JPROTEOME.1C00206. [DOI] [PubMed] [Google Scholar]
- Yoon J., Kim G., Jarhad D.B., Kim H.-R., Shin Y.-S., Qu S., Sahu P.K., Kim H.O., Lee H.W., Wang S. Bin, Kong Y.J., Chang T.-S., Ogando N.S., Kovacikova K., Snijder E.J., Posthuma C.C., van Hemert M.J., Jeong L.S. Design, synthesis, and anti-RNA virus activity of 6′-fluorinated-aristeromycin analogues. J. Med. Chem. 2019;62:6346–6362. doi: 10.1021/acs.jmedchem.9b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Guo S., Yi D., Li Q., Ma L., Zhang Y., Wang J., Li X., Guo F., Lin R., Liang C., Liu Z., Cen S. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antivir. Res. 2021;190 doi: 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.