Abstract
The lactic acid bacteria (LAB) are safe microorganisms which are mainly used for the preparation of fermented foods and for probiotic applications. The potential of LAB as live vehicles for the production and delivery of therapeutic molecules such as antigens is also being actively investigated today. However, very little is known about the fate of live LAB when administered in vivo and about the interaction of these microorganisms with the nasal or gastrointestinal ecosystem. For future applications, it is essential to be able to discriminate the biotherapeutic strain from the endogenous microflora and to unravel the mechanisms underlying the postulated health-beneficial effect. We therefore started to investigate both aspects in a mouse model with two LAB species presently under development as live vaccine vectors, i.e., Lactococcus lactis and Lactobacillus plantarum. We have constructed different expression vectors carrying the gfp (green fluorescent protein [GFP]) gene from the jellyfish Aequoria victoria, and we found that this visible marker was best expressed when placed under the control of the inducible strong nisA promoter from L. lactis. Notably, a threshold amount of GFP was necessary to obtain a bright fluorescent phenotype. We further demonstrated that fluorescent L. plantarum NCIMB8826 can be enumerated and sorted by flow cytometry. Moreover, tagging of this strain with GFP allowed us to visualize its phagocytosis by macrophages in vitro and ex vivo and to trace it in the gastrointestinal tract of mice upon oral administration.
The lactic acid bacteria (LAB) constitute a family of gram-positive bacteria which are well known for their use in industrial food fermentations and for their probiotic properties (27). During the past 15 years, the characterization of LAB has considerably evolved, and a variety of molecular biology tools have been developed for these microorganisms. Several reporter genes such as those encoding chloramphenicol acetyltransferase (cat-86 from Bacillus pumilus or cat-194 from Staphylococcus aureus) (1, 11), the Escherichia coli β-glucoronidase gene (30), the Leuconostoc mesenteroides β-galactosidase gene (20), the Bacillus licheniformis α-amylase gene (18), the Vibrio fischeri luciferase gene (13), and the S. aureus nuclease gene (31) have been used for LAB mainly to isolate functional expression or targeting signals. The lux system has also been applied to study lactococcal promoter strength in the digestive tract of mice (6). The phenotypic tests linked to these systems require the addition of exogenous substrates for the detection of recombinant strains expressing the reporter genes. As such, they may present limitations for in vivo studies. To circumvent this drawback, an original reporter system based on the green fluorescent protein (GFP) from the jellyfish Aequorea victoria has been developed (5) and used successfully with a variety of bacteria such as gram-negative bacteria (5), Mycobacterium bovis (12, 23), and, very recently, two LAB, Streptococcus thermophilus (34) and Lactococcus lactis (33). GFP is a protein of 238 amino acids which spontaneously emits green light at 508 nm when excited with blue light at 395 nm in the presence of O2. Its major advantage results from its intrinsic property of fluorescing in the absence of any added cofactor or substrate, thus allowing nondestructive detection of recombinant cells expressing this reporter gene (5, 36). GFP is very stable and photobleaches very slowly even after repeated observations under the epifluorescence microscope. Moreover, mutant GFPs have been generated to improve detection and expression of the fluorescent protein in prokaryotic cells. These mutants generally absorb light of a longer wavelength (>396 nm) with little change in the emission spectrum compared to that of wild-type GFP and lead to improved fluorescence over that of the wild type due to increased solubility of the protein (8, 16, 17, 36).
Our laboratory is mainly interested in potential health applications of LAB such as their use for in vivo production and delivery of biologically active molecules. Dietary LAB have been consumed since times immemorial and are thus designated “generally recognized as safe” (2), which represents an important advantage for their potential use as live therapeutic vehicles (7, 28, 38). Nevertheless, little is known about the fate of LAB administered in vivo and their interaction with either the immune system of the host or its endogenous microflora, which we started to investigate in a mouse model.
In the present report, we describe the implementation of a GFP variant optimized for bacterial expression (GFPuv [8]) as a marker for Lactobacillus plantarum NCIMB8826 and L. lactis NZ9800, two LAB species presently under study as potential live vaccine vehicles (19, 28, 29, 38). We tested expression of the gfp gene under the control of promoters of different strengths and verified whether fluorescent lactobacilli can be enumerated by flow cytometry and traced in vivo. We specifically analyzed the interaction of GFP+ recombinant L. plantarum strains with macrophages which are actively phagocytic antigen-presenting cells that play an essential role in the induction of immune responses.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
gfp expression experiments were performed with L. plantarum NCIMB8826, a human saliva isolate, and L. lactis NZ9800 (Table 1). All cloning steps were done with E. coli MC1061 (Table 1).
TABLE 1.
Plasmids and bacterial strains
| Plasmid or strain | Relevant characteristics | Antibiotic resistance | Reference or source |
|---|---|---|---|
| Plasmids | |||
| pGIT032 | L. hilgardii pLAB1000 replicon; expression vector containing the constitutive ldhL promoter from L. plantarum | Apr Emr | 15 |
| pTG2247 | L. lactis pSH71 replicon (pCK17 derivative); expression vector containing P25 promoter from S. thermophilus followed by the ldhD RBS from L. plantarum and terminator signal T1T2 from E. coli | Kmr Cmr | 19 |
| pNZ8037 | L. lactis pSH71 replicon; pNZ8032 derivative; expression vector containing nisA promoter from L. lactis NZ9800 | Cmr | 9 |
| pNZ8037mod | pNZ8037 carrying an additional KpnI site | Cmr | This study |
| pBSmod | pBluescript derivative containing an additional SphI site | Apr | P. Chagnaud (unpublished data) |
| pBAD-GFPuv | pBR322 derivative containing gfp under the control of araC promoter | Apr | Clontech |
| pMEC7 | pBSmod containing the P25-RBS-T1T2 cassette from pTG2247 | Apr | This study |
| pMEC12 | pMEC7 derivative containing gfp | Apr | This study |
| pMEC17 | pTG2247 derivative containing the P25-RBS-gfp-T1T2 cassette from pMEC12 | Kmr Cmr | This study |
| pMEC30 | pGIT032 derivative containing gfp | Apr Emr | This study |
| pMEC45 | pNZ8037 derivative containing gfp | Cmr | This study |
| Bacterial strains | |||
| E. coli MC1061 | araD139 Δ(ara-leu)7696 lacX74 galV galK hsr-hsm rpsL | 4 | |
| L. plantarum NCIMB8826 | Isolated from human saliva | NCIMBa | |
| L. plantarum NCIMB8826 Int-1 | Integrant carrying the nisRK genes in the tRNASer chromosomal locus | S. Pavan et al. (unpublished data) | |
| L. lactis NZ9800 | NZ9700 derivative, ΔnisA, carrying nisRK on the chromosome; non-nisin producer | 9 |
NCIMB, National Collection of Industrial and Marine Bacteria, Aberdeen, United Kingdom.
L. plantarum strains were cultured in MRS broth (Difco) at 37°C without shaking. L. lactis strains were grown without shaking in M17 broth (Difco) containing 0.5% (wt/vol) glucose at 30°C. E. coli strains were grown in Luria-Bertani medium at 37°C (32) under aeration. When appropriate, antibiotics were added to the culture medium. For LAB strains, chloramphenicol and erythromycin were used at final concentrations of 10 and 5 μg/ml, respectively. Ampicillin was supplied at a concentration of 100 μg/ml in the case of E. coli.
Expression of the gfp gene placed under the control of the nisin promoter was induced as follows: an overnight culture of L. plantarum NCIMB8826 was used to inoculate fresh medium at a dilution of 1:50. After 1 h of incubation, different amounts (2.5, 10, and 25 ng/ml) of nisin (Sigma) were added to the culture, which was further incubated for 3 to 4 h. For L. lactis, nisin induction was performed as described previously (10). GFP+ cells were observed by UV illumination or epifluorescence microscopy. The bacteria were washed once with phosphate-buffered saline (PBS) (Gibco) and concentrated 10- or 100-fold in PBS for in vitro or in vivo (intranasal administration) experiments, respectively. For feeding experiments, bacteria were resuspended in a 1/100 volume of gavage buffer (0.25 M sodium bicarbonate, 0.6% casein, 0.5% glucose). For bacterial enumeration on agar plates, washed cells were diluted and 100 μl of adequate dilutions was plated on selective medium. CFU were determined after 48 h of growth at 37°C.
DNA manipulation and transformation.
Plasmid DNA was purified from E. coli by the alkaline lysis method (32) and was isolated from L. plantarum and L. lactis as described previously (19, 37). Restriction endonucleases, T4 DNA ligase, and Taq polymerase were purchased from Boehringer Mannheim and used according to the recommendations of the manufacturer. Electroporation of L. plantarum NCIMB8826 and L. lactis NZ9800 was performed according to the methods of Josson et al. (21) and Wells et al. (37), respectively.
Construction of expression plasmids carrying the gfp gene.
The expression plasmids pTG2247, pGIT032, and pNZ8037mod are described in Table 1. They allow cloning of the gene of interest behind the S. thermophilus P25 (pTG2247), the L. plantarum ldhL (pGIT032), or the L. lactis nisA inducible (pNZ8037) promoter, respectively, leading to transcriptional fusions in all cases.
(i) Cloning of gfp gene under constitutive promoters.
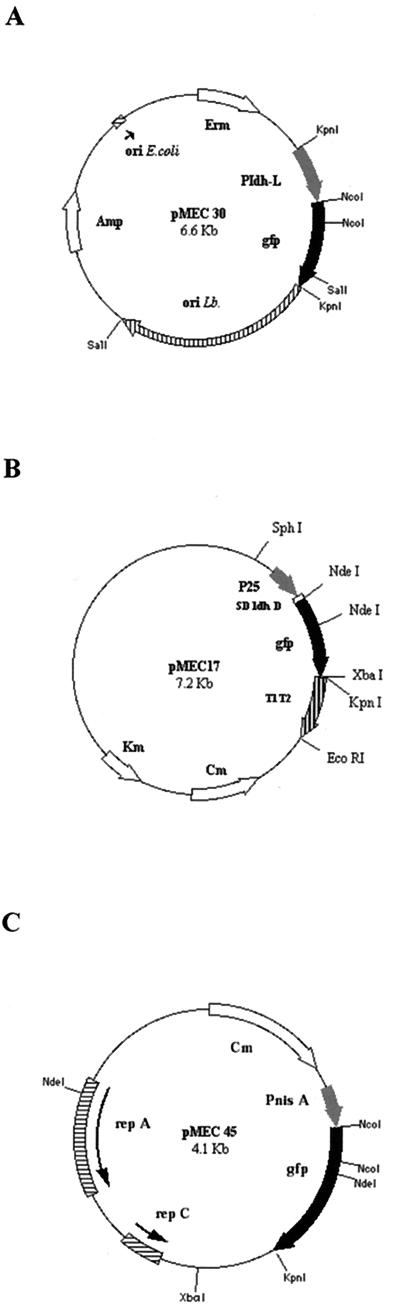
The ldhL promoter from pGIT032 and the P25 promoter from pTG2247 were first chosen to drive the expression of the gfp gene. The latter was amplified from pBAD-GFPuv (carrying the GFPuv variant optimized for bacterial expression [Clontech]) by PCR with two oligonucleotides with the following sequences: CAT GCA TGC CAT GGC TAG CAA AGG AGA AGA AC (primer 1) and CCG GGT ACC GAG CTC GAA TTC (primer 2). The first one contained a NcoI site (underlined) which included the ATG initiation codon, and the second one included a KpnI site (underlined). The 758-bp PCR product was first restricted partially by NcoI and then by KpnI and cloned into NcoI-KpnI-restricted pGIT032 (partial restriction by KpnI), giving rise to pMEC30 (Fig. 1A). In this construction, GFP is fused to the first 25 amino acids of lactate dehydrogenase (LDH), giving rise to a hybrid protein with a calculated molecular weight of 29,000.
FIG. 1.
Plasmids pMEC30 (A), pMEC17 (B), and pMEC45 (C) carrying the gfp gene under the control of the L. plantarum ldhL, the S. thermophilus P25, or the L. lactis inducible nisA promoter, respectively.
Two intermediate steps were carried out to bring the gfp gene under the control of the P25 promoter. First, the SphI-EcoRI fragment of pTG2247, which contains the P25 promoter, the ldhD ribosome binding site (RBS), and the T1T2 termination signal was cloned into the SphI-EcoRI-restricted pBSmod vector (Table 1). This intermediate plasmid, which replicates only in E. coli, was named pMEC7. pBAD-GFPuv was first restricted partially by NdeI and then by XbaI. The resulting 750-bp fragment containing gfp was then cloned into NdeI-XbaI-restricted pMEC7, thereby giving rise to pMEC12. Finally, a recombinant shuttle plasmid was obtained by inserting the SphI-KpnI fragment of pMEC12 into SphI-KpnI-restricted pTG2247. The resulting plasmid, pMEC17, thus carries the gfp gene under the control of the P25 promoter (Fig. 1B).
(ii) Cloning of gfp under the nisin-inducible promoter.
The nisin-inducible promoter from pNZ8037 (9) was used to drive the expression of gfp. The 758-bp PCR-amplified gene was restricted partially by NcoI and then by KpnI and cloned into NcoI-KpnI-restricted pNZ8037mod, which contains the nisA promoter and translational initiation region, giving rise to pMEC45 (Fig. 1C).
Western blotting.
Total protein extracts were prepared from exponentially growing cultures. The bacteria were harvested by centrifugation (3,000 × g, 10 min, 4°C), washed with PBS, resuspended in 1 ml of 10 mM Tris-HCl (pH 7.5), and disrupted with a French press (Bioritech). The cell suspension was centrifuged (10,000 × g, 10 min, 4°C) to remove cell debris. The protein concentration was determined with the Bio-Rad protein assay kit (Bio-Rad). The samples were boiled in Laemmli buffer (26) and subjected to sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis. The proteins were transferred onto nitrocellulose membranes (Optitran BA-S85; Schleicher & Schuell) with a Bio-Rad electroblotter. The blots were blocked for 2 h to overnight with 5% dried milk in blocking buffer (0.1% Tween 20, 0.5 M NaCl, 10 mM Tris-HCl, pH 8.2) and incubated for 1 h at 25°C with rabbit anti-GFP antiserum (Clontech) diluted 1/2,000 in blocking buffer. After three washes in blocking buffer, the membranes were incubated for 1 h at 25°C with alkaline-phosphatase-conjugated anti-rabbit antisera (Sigma) diluted 1/7,000 in blocking buffer. After three washes in blocking buffer and one wash in developing buffer (5 mM MgCl2, 100 mM NaCl, 50 mM Tris-HCl, pH 9.5), the blots were developed with 5.0 mg of BCIP (5-bromo-4-chloro-3-indolylphosphate) per ml and 10 mg of nitroblue tetrazolium per ml in developing buffer.
Uptake of L. plantarum GFP+ strain by macrophages.
The mouse monocyte-macrophage cell line J774A.1 (ATCC TIB67) was maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle medium (DMEM; Gibco), supplemented with 10% decomplemented fetal calf serum (Gibco). Macrophages were seeded into 24-well tissue culture plates (Lab-Tek; Nunc) at a concentration of 104 cells per chamber and were grown overnight. Prior to incubation with the L. plantarum strains, the adherent macrophage monolayer was washed with DMEM. L. plantarum GFP+ (nisin-induced culture of NCIMB8826/pMEC45) bacteria were added at a multiplicity of 1 to 5 CFU/cell. After 3 h or overnight incubation at 37°C and 5% CO2, 50 nM acidotropic probe (LysoTracker Red DND-99; Molecular Probes) was added to each chamber, and incubation was continued for 1 h. The macrophages were then washed three times with DMEM to remove noningested bacteria, fixed with 4% paraformaldehyde, and examined by epifluorescence microscopy. Alternatively, the macrophage suspension was analyzed by flow cytometry after incubation with bacteria (see below).
Administration of L. plantarum GFP+ cells to mice and histological studies.
For nasal administration, four BALB/c mice were given 10 μl (i.e., 108 CFU) of either L. plantarum GFP+ (nisin-induced culture of NCIMB8826 Int-1/pMEC45) or L. plantarum GFP− (noninduced culture of NCIMB8826 Int-1/pMEC45) bacteria in one nostril. Four hours later, the mice were sacrified and a bronchoalveolar wash was performed on each animal. The cells contained in the lavage suspension were harvested by centrifugation (1,000 × g, 10 min, 4°C), washed twice with PBS, and resuspended in 1 ml of PBS. Half of the suspension was stained with the acidotropic probe as described above and examined by epifluorescence microscopy. The other half was analyzed by flow cytometry (see below).
For feeding experiments, four BALB/c mice received 109 CFU of L. plantarum GFP+ bacteria by intragastric gavage. Mice were sacrified 90 min postadministration. The Peyer's patches and flanking intestinal segments were removed, fixed in 10% formalin, and embedded in paraffin. Sequential thin sections (10 μm) were cut, deparaffinized, and mounted in Mowiol 4-88 (Calbiochem) for direct observation by epifluorescence microscopy.
Flow cytometry analysis.
Samples were analyzed on a Coulter EPICS ELITE flow cytometer with an air-cooled 488-nm argon ion laser operated at 14 W and 6 A of power. Fluorescein isothiocyanate fluorescence was collected through a 525-nm dichroic band-pass filter after being reflected by a 550-nm dichroic long pass filter. Data were collected on 1.5 × 104 or 2 × 104 individual particles per sample. Before each analysis, 3- and 6-μm green latex beads (Coulter Corporation) were used to calibrate the light scatter and fluorescence parameters. For analysis of bacterial suspensions, exponentially growing L. plantarum GFP+ or L. plantarum GFP− bacteria were harvested by centrifugation (3,000 × g, 10 min, 4°C), washed twice with PBS, resuspended thoroughly in 1 ml of PBS, and mixed with a known concentration of fluorescent beads in order to allow enumeration of viable cells. For examination of macrophages incubated with L. plantarum GFP+ or L. plantarum GFP− bacteria, the monolayers were washed with DMEM and harvested by scraping at 4 h postincubation. The macrophages were collected by centrifugation (1,000 × g, 10 min, 4°C), washed twice with PBS, and centrifuged again. The final pellet was resuspended in 1 ml of PBS. Bronchoalveolar lavage samples were prepared as described above. Detection of a fluorescent signal by flow cytometry was always confirmed by epifluorescence microscopy.
Epifluorescence microscopy.
GFP production was examined in bacterial suspensions, macrophage cultures, or tissues by epifluorescence microscopy with a Zeiss Axiophot plan2 microscope equipped with a modular filter cube with band-pass excitation filter BP450-490 and barrier emission filter BA515-IF. Photographs were taken with a MOT DX 35 camera with Provia Fujichrome 1600 films.
RESULTS
Expression of the gfp gene in LAB.
To attempt gfp expression in LAB, we chose to clone a variant of the GFP cDNA from A. victoria (i.e., GFPuv) into the vectors pGIT032 and pTG2247, which carry constitutive expression cassettes (Table 1). pGIT032 contains strong expression signals derived from the L. plantarum ldhL gene and is a shuttle vector based on a Lactobacillus hilgardii origin of replication (limited host range). pTG2247 is based on the broad-host-range L. lactis pSH71 replicon and carries a mosaic expression cassette including the P25 promoter from S. thermophilus followed by the ldhD RBS from Lactobacillus pentosus.
The GFPuv variant was amplified by PCR from pBAD-GFPuv (Table 1) and cloned under the control of the ldhL expression signals into pGIT032 or of the P25 expression cassette into pTG2247, giving rise to pMEC30 and pMEC17, respectively. The resulting plasmids were introduced into E. coli, and all individual colonies of the recombinant E. coli MC1061/pMEC30 and MC1061/pMEC17 were found to be fluorescent upon UV illumination. However, when pMEC17 was transferred by electroporation into L. lactis or L. plantarum, no fluorescence was detected upon UV illumination or by epifluorescence microscopy (Fig. 2). Surprisingly, even though the ldhL promoter had been used previously to successfully drive high expression of foreign genes in L. plantarum (see reference 28), individual colonies of NCIMB8826/pMEC30 were found to exhibit fluorescence only transiently. To check the integrity of the plasmid constructs carried by the transformants, pMEC30 and pMEC17 were extracted from L. lactis and L. plantarum and reelectroporated into E. coli. All transformants were fluorescent upon UV illumination, and DNA analysis showed no sign of structural instability (data not shown). As the lack of a fluorescent phenotype in the pMEC30- or pMEC17-containing L. lactis or L. plantarum transformants could be linked to a low GFP synthesis, we decided to use a controlled gene expression system allowing induction of the synthesis of foreign proteins in a dose-dependent manner and the attainment of high production levels upon full induction. The nisin-inducible system, originally developed with L. lactis (9, 25), is based on signal transduction by the two-component regulatory system consisting of the response-regulator protein NisR and the sensor NisK, found in the nisin gene cluster of L. lactis (14, 24). To implement the nisin system in L. plantarum NCIMB8826, it was necessary to integrate nisRK into the chromosome of this host, generating the NCIMB8826 Int-1 strain (S. Pavan et al., unpublished data). The latter was electroporated with a plasmid containing a reporter gene (gusA) under the control of the nisA promoter (pNZ8032 [9]) in order to verify that addition of nisin to the culture medium activates transcription of the β-glucuronidase gene, which was found to be the case. The GFP-encoding sequence amplified by PCR was then cloned under the control of the nisA promoter into pNZ8037mod (Table 1), giving rise to pMEC45 (Fig. 1C). This plasmid was electroporated into L. plantarum NCIMB8826 Int-1 and into L. lactis NZ9800. Chloramphenicol-resistant transformants were obtained in both cases, and upon full induction by nisin, all colonies or individual cells of L. plantarum and L. lactis exhibited fluorescence as observed by UV illumination or epifluorescence microscopy (Fig. 2). As no fluorescence was detected in the absence of nisin, noninduced bacterial cells were used as negative controls in further experiments.
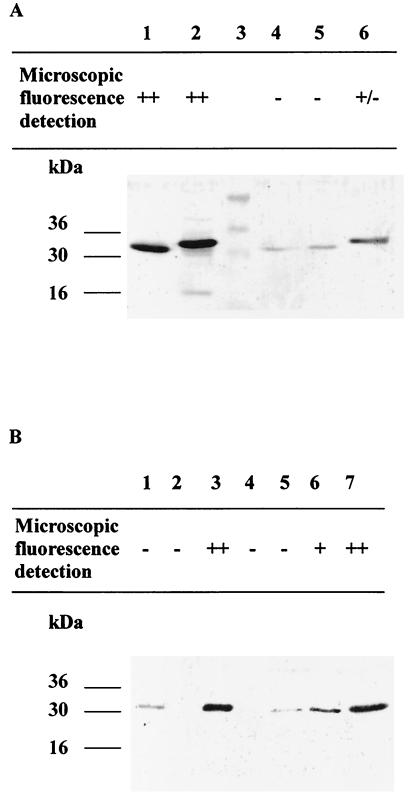
FIG. 2.
Immunoblotting of whole-cell extracts of recombinant LAB strains carrying gfp under the control of constitutive promoters (A) and the inducible nisA promoter (B). (A) Lane 1, E. coli MC1061/pMEC17; lane 2, E. coli MC1061/pMEC30; lane 3, molecular mass markers; lane 4, L. lactis NZ9800/pMEC17; lane 5, L. plantarum NCIMB8826/pMEC17; lane 6, L. plantarum NCIMB8826/pMEC30. (B) Lane 1, L. lactis NZ9800/pMEC17; lanes 2 and 3, L. lactis NZ9800/pMEC45, noninduced and induced with 5 ng of nisin per ml, respectively; lane 4, L. plantarum NCIMB8826 Int-1/pMEC45, noninduced; lanes 5, 6, and 7, L. plantarum NCIMB8826 Int-1/pMEC45 induced with 2.5, 10, or 25 ng of nisin per ml, respectively. Microscopic observations (epifluorescence) of each sample used before Western blotting are shown at the top (−, no fluorescence; +/−, transient fluorescence; ++, bright fluorescence).
The GFP production levels were examined in all recombinant L. plantarum and L. lactis strains, as well as in E. coli carrying pMEC17 or pMEC30. Total cell extracts were prepared, and equal amounts of protein were analyzed by Western blotting with polyclonal anti-GFP serum. As illustrated in Fig. 2A, GFP was present at low levels in extracts prepared from L. lactis NZ9800/pMEC17, L. plantarum NCIMB8826/pMEC17, and L. plantarum NCIMB8826/pMEC30. The LDH-GFP hybrid protein produced by the latter strain was of the expected size and did not seem to be degraded in the cell extracts. A strong signal was observed in the two recombinant E. coli strains and in fully nisin-induced L. plantarum NCIMB8826 Int-1/pMEC45 (Fig. 2B, lane 7) and L. lactis NZ9800/pMEC45 (Fig. 2B, lane 3). As these results pointed to a correlation between the fluorescent phenotype and the amount of GFP synthesized by the recombinant strains, we performed a dose-dependent nisin induction experiment. Exponentially growing cultures of L. plantarum NCIMB8826 Int-1/pMEC45 were induced with 0, 2.5, 10, or 25 ng of nisin per ml, which led to increasing levels of GFP (Fig. 2B, lanes 4 to 7). Bright fluorescence was observed only in the case of fully induced cells (Fig. 2B). Further experiments were thus performed under these conditions.
Detection and enumeration of GFP+ lactobacilli by flow cytometry.
In addition to microscopic observations of GFPuv expression by epifluorescence, we verified if L. plantarum cells producing GFPuv could be enumerated and sorted by flow cytometry. For this purpose, exponentially growing L. plantarum GFP+ or GFP− (i.e., nisin-induced or noninduced culture of NCIMB8826 Int-1/pMEC45) bacteria were monitored by cell sorting based on fluorescence intensity. As expected, fluorescent lactobacilli can easily be discriminated from their nonfluorescent counterparts by this technique (data not shown). The same suspensions were analyzed by classical counts on agar plates. As shown in Table 2, the bacterial counts determined by both techniques were in excellent agreement. It was also verified that GFPuv-producing L. plantarum cells can be enumerated in a mixed population containing both fluorescent and nonfluorescent bacteria (Table 2).
TABLE 2.
Enumeration of L. plantarum GFP+ bacteria by flow cytometry or plate counts
| Type of count | Bacterial count of L. plantarum GFP+ bacteria at dilutionc:
|
|||
|---|---|---|---|---|
| 10−3 | 10−4 | 10−5 | 10−6 | |
| Plate count (CFU/ml) | 1.1 × 106 | 1.3 × 105 | 1.6 × 104 | 1.0 × 103 |
| FACS count (no. of events/ml)a | 1.6 × 106 | 2.0 × 105 | 1.6 × 104 | 1.2 × 103 |
| FACS count in a mixed population (no. of events/ml)b | 1.9 × 106 | 1.8 × 105 | 1.8 × 104 | 1.7 × 103 |
One hundred microliters of L. plantarum GFP+ bacteria was resuspended in 900 μl of PBS, and serial dilutions were analyzed by fluorescence-activated cell sorting (FACS).
One hundred microliters of L. plantarum GFP+ and 100 μl of L. plantarum GFP− bacteria were added to 800 μl of PBS. Serial dilutions of this mixed population were analyzed by fluorescence-activated cell sorting.
The numbers in the table represent the mean values of three independent experiments.
Uptake of L. plantarum GFP+ bacteria by macrophages: microscopic and flow cytometric analysis.
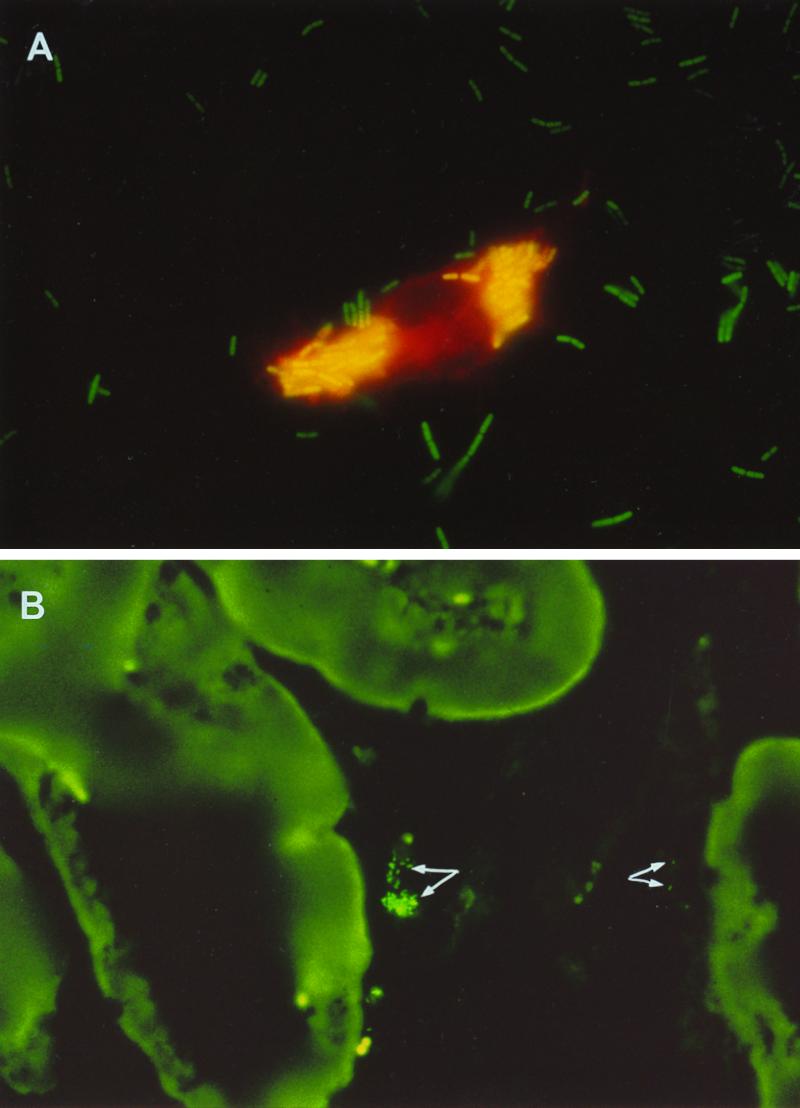
To examine whether the GFP marker could be used to visualize the interaction of fluorescent lactobacilli with specific immune cells, the murine macrophage cell line J774 was incubated in the presence of L. plantarum GFP+ bacteria at 37°C and observed by epifluorescence microscopy 4 h postincubation. The acidic compartments of the macrophage were stained with a red acidotropic probe (LysoTracker Red). The internalized lactobacilli appeared as bright yellow bacteria in contrast with the green fluorescent ones, which adhered to the surface of the macrophages or remained free in the culture medium. As shown in Fig. 3A, the NCIMB8826 strain was actively phagocytosed by the macrophages. As a control, the same experiment was conducted at 4°C, a temperature preventing activation of the macrophages. In this case, no bacteria were detected inside the macrophages (data not shown).
FIG. 3.
(A) J774 macrophages after 4 h of incubation with L. plantarum GFP+ bacteria (nisin-induced cells of L. plantarum NCIMB8826 Int-1/pMEC45). Magnification, ×1,000. (B) Thin sections of intestine segments from mice fed L. plantarum GFP+ bacteria, showing fluorescent cells (indicated by arrows) embedded in the mucus. Magnification, ×400.
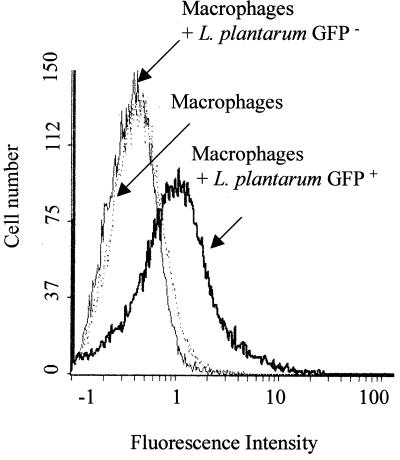
We further investigated whether macrophages that contain L. plantarum GFP+ bacteria could be separated by flow cytometry from macrophages containing nonfluorescent lactobacilli or Lactobacillus-free macrophages. J774 cultures were therefore incubated for 4 h at a cell-to-bacterium ratio of 1:1 to 5 with either L. plantarum GFP+ or L. plantarum GFP− bacteria and then processed for flow cytometry analysis. As shown in Fig. 4, the macrophages that had taken up fluorescent lactobacilli were easily distinguished from free macrophages or from macrophages containing L. plantarum GFP− bacteria. This result was confirmed by observations with epifluorescence microscopy.
FIG. 4.
Flow cytometric analysis of GFP+ and GFP− L. plantarum strains phagocytosed by J774 macrophage cell lines. Fluorescence data were gated by forward-angle light scatter. Fluorescence intensities are presented on the x axis, and cell counts are presented on the y axis. The cytometric analysis was performed on 2 × 104 events.
GFP as an in vivo and ex vivo marker for L. plantarum.
To test whether GFP can be used to monitor the fate of lactobacilli in vivo, BALB/c mice were fed with one dose of 109 CFU of fluorescent L. plantarum. Intestinal specimens consisting of Peyer's patches and flanking segments were removed and examined by fluorescence microscopy upon sacrifice of the mice. Fluorescent lactobacilli could readily be detected in the intestinal lumen, mostly embedded in the mucus, while some bacteria were found associated with the epithelial cell surface (Fig. 3B). No bacteria were detected inside Peyer's patches, probably due to the high dilution of the bacterial sample in vivo.
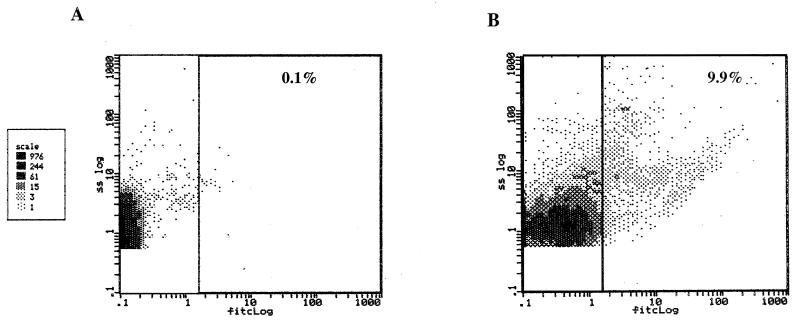
Moreover, mice were given L. plantarum GFP+ or GFP− bacteria intranasally, and after 4 h animals were killed to obtain bronchoalveolar lavage samples that were analyzed by epifluorescence microscopy and flow cytometry. Consistent with the results obtained with J774 cultured cells, L. plantarum NCIMB8826 was found to be ingested by the bronchoalveolar macrophages. The proportion of macrophages having phagocytosed lactobacilli was estimated by flow cytometry to reach 10% of the total bronchoalveolar macrophage population (Fig. 5).
FIG. 5.
Flow cytometric analysis of macrophages of bronchoalveolar lavage samples of mice after nasal administration of GFP+ or GFP− L. plantarum strains. The results are shown as the relative amounts of macrophages having taken up nonfluorescent (A) or fluorescent (B) lactobacilli against the log10 unit of fluorescence. The percentages indicate the proportions of fluorescent cells. The cytometric analysis was performed on 1.5 × 104 events. fitc, fluorescein isothiocyanate.
DISCUSSION
The advantages of using GFP compared to other reporter proteins are now well established, especially for in vivo studies. The GFP expression plasmids constructed in this study were tested in LAB strains belonging to the species L. lactis and L. plantarum, which are presently under development as live biotherapeutic agents (7, 28, 38). Although the production of GFP could be detected by Western blotting in all recombinant strains, only those synthesizing the highest level of GFP (Fig. 2) exhibited strong and consistent fluorescence. This phenotype thus seemed essentially correlated with the amount of GFP produced. Very strong fluorescence was observed for E. coli strains transformed with the plasmids carrying gfp under the control of constitutive promoters. In contrast, when pMEC17 or pMEC30 was introduced into L. lactis or L. plantarum, no fluorescence was observed and the amount of GFP produced in the recombinant LAB was much lower than that in their E. coli counterparts. In L. plantarum, pMEC30 leads to a hybrid protein of the expected size between GFP and the first 25 amino acids of the l-LDH which was produced at a much lower level than expected from previous work (see reference 28). Notably, this strain fluoresced only transiently, even though we observed no toxicity of GFP for the bacterial hosts and no protein degradation in cell extracts or structural instability of the plasmid. Not surprisingly, when the corresponding gfp expression cassette was integrated as a single copy in the chromosome of L. plantarum NCIMB8826, no fluorescence was observed (data not shown).
To increase the production level of the reporter protein, we next decided to use the lactococcal nisin-controlled expression system (9, 25). Plasmid pMEC45, carrying the gfp gene under the control of the nisA promoter, was introduced into the appropriate recipient strains, i.e., L. lactis NZ9800 and L. plantarum NCIMB8826 Int-1. Upon full induction by nisin, the corresponding transformants produced high amounts of GFP as evaluated by Western blotting. Consistently, they exhibited a strong fluorescence detectable both by UV illumination and by epifluorescence microscopy as long as the bacteria did not lyse. The hypothesis that a threshold amount of GFP is necessary to obtain bright fluorescence is supported by the following experiment. Nisin was added at increasing concentrations to exponentially growing cells of NCIMB8826 Int-1/pMEC45, leading to the progressive induction of GFP synthesis as shown by immunoblotting. An intense fluorescent signal was obtained only for bacterial cells induced with the highest amount of nisin. Different authors have mentioned the necessity of individually evaluating and adapting the gfp expression conditions for different bacterial systems (see, for example, references 3 and 35). The pMEC45 expression vector that we describe in this paper may be considered a transferable gfp expression system that should function in at least all lactic acid bacterial strains for which the nisin system has successfully been used (22). We indeed demonstrated that it was working equally well in L. lactis and in L. plantarum. As our laboratory is mostly interested in health applications of LAB, the major aim of the present study was to assess the validity of GFP as a marker to visualize the interaction of these microorganisms with specific immune cells and to monitor their fate in vivo. We have demonstrated that GFP constitutes an adequate reporter for both applications, focusing on our main model strain L. plantarum NCIMB8826. Nisin-induced NCIMB8826 Int-1/pMEC45 bacteria could easily be enumerated and discriminated from their nonfluorescent counterparts by flow cytometry, thus opening the way to quantitative detection of these bacteria in complex microbial communities. By use of an acidic probe to stain macrophage lysosomes, phagocytosis of lactobacilli by these cells could clearly be shown. Macrophages that had taken up GFP+ lactobacilli could also be analyzed and counted by flow cytometry. This was performed in vitro or on macrophages collected from bronchoalveolar lavage fluids of mice that had received fluorescent lactobacilli intranasally. The observation that L. plantarum cells are actively taken up by antigen-presenting cells is in complete agreement with the fact that recombinant lactobacilli can be used as live vaccine vehicles by the nasal route (28). Moreover, direct observation by epifluorescence microscopy allowed us to trace bacteria in intestinal sections of mice fed with nisin-induced NCIMB8826 Int-1/pMEC45. Fluorescent lactobacilli were found mostly embedded in intestinal mucus or free in the lumen, even though some bacteria seemed to be closely associated with epithelial cells. We further plan to analyze the interaction of L. plantarum with Peyer's patches by using a ligated-intestinal-loop system to avoid in vivo dilution of the sample. Preliminary studies have shown that analysis of bacterial translocation in mouse models (D. Dombrowicz, P. Desreumaux, C. Neut, F. Bouzahzah, J. P. Papin, J. F. Colombel, and M. Capron, Abstr. Keystone Symposia on Experimental Models of Immune Dysregulation and Mucosal Inflammation, abstr. 206, p. 53, 1999) is also greatly facilitated by using fluorescent bacteria, as they allow workers to easily distinguish the strain under study from the endogenous lactobacilli (data not shown).
The system that we describe relies on the in vitro induction of GFP synthesis, which alleviates potential problems of oxygen limitation that could interfere with the development of fluorescence (33). As photobleaching of GFP is very slow (36), the preloaded fluorescent bacteria can further be used for a variety of in vitro and in vivo experiments including their visualization in the gastrointestinal tract.
In summary, we have shown that GFP can be used as a useful marker in LAB to monitor their fate when administered to animals or to analyze their interactions with different cell types, both aspects being critical in the case of vaccine and probiotic applications of these bacteria. GFP+ strains will moreover facilitate the study of their survival in the environment and could be used as a tool in monitoring the risk of DNA transfer among the intestinal microflora.
ACKNOWLEDGMENTS
This work was supported by the EU BIO4-CT96-0542 grant and FEDER funds.
We are grateful to C. Grangette for her skillful help with animal experiments. The pBluescript modified vector was kindly supplied by P. Chagnaud. We thank P. Hols and C. Locht for critical reading of the manuscript and A. Veithen for helpful suggestions.
REFERENCES
- 1.Achen M G, Davidson B E, Hillier A J. Construction of plasmid vectors for the detection of streptococcal promoters. Gene. 1986;45:45–49. doi: 10.1016/0378-1119(86)90130-7. [DOI] [PubMed] [Google Scholar]
- 2.Adams M R, Marteau P. On the safety of lactic acid bacteria. Int J Food Microbiol. 1995;27:263–264. doi: 10.1016/0168-1605(95)00067-t. [DOI] [PubMed] [Google Scholar]
- 3.Bloemberg G V, O'Toole G A, Lugtenberg B J J, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadaban M J, Cohen S. Analysis of gene control signals by DNA fusion cloning in E. coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 6.Corthier G, Delorme C, Erhlich S D, Renault P. Use of luciferase genes as biosensors to study bacterial physiology in the digestive tract. Appl Environ Microbiol. 1998;64:2721–2722. doi: 10.1128/aem.64.7.2721-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corthier G, Renault P. Future directions for research on biotherapeutic agents: contribution of genetic approaches on lactic acid bacteria. In: Elmer G W, editor. Biotherapeutic agents and infectious diseases. Totowa, N.J: Humana Press; 1998. pp. 269–304. [Google Scholar]
- 8.Crameri A, Whitheorn E A, Tate E, Stemmer W P C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 9.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, Alen-Boerrigter I, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vos W M. Gene cloning and expression in lactic streptococci. FEMS Microbiol Rev. 1987;46:281–295. [Google Scholar]
- 12.Dhandayuthapani S, Via L E, Thomas C A, Horowitz P M, Deretic D, Deretic V. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 13.Eaton T J, Shearman C A, Gasson M J. The use of bacterial luciferase genes as reporter genes in Lactococcus: regulation of the Lactococcus lactis subsp. lactis lactose genes. J Gen Microbiol. 1993;139:1495–1501. doi: 10.1099/00221287-139-7-1495. [DOI] [PubMed] [Google Scholar]
- 14.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian K D. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol. 1994;60:814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferain T, Garmyn D, Bernard N, Hols P, Delcour J. Lactococcus plantarum ldhL gene: overexpression and deletion. J Bacteriol. 1994;176:596–601. doi: 10.1128/jb.176.3.596-601.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 17.Heim R, Prasher D C, Tsien R Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hols P, Baulard A, Garmyn D, Delplace B, Hogan S, Delcour J. Isolation and characterization of genetic expression and secretion signals from Enterococcus faecalis through the use of broad-host-range α-amylase probe vectors. Gene. 1992;118:21–30. doi: 10.1016/0378-1119(92)90244-j. [DOI] [PubMed] [Google Scholar]
- 19.Hols P, Slos P, Dutot P, Reymund J, Chabot P, Delplace B, Delcour J, Mercenier A. Efficient secretion of the model antigen M6-gp41E in Lactobacillus plantarum NCIMB8826. Microbiology. 1997;143:2733–2741. doi: 10.1099/00221287-143-8-2733. [DOI] [PubMed] [Google Scholar]
- 20.Israelsen H, Madsen S M, Vrang A, Hansen E B, Johansen E. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl Environ Microbiol. 1995;61:2540–2547. doi: 10.1128/aem.61.7.2540-2547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josson K, Scheirlinck T, Michiels F, Plateeuw C, Stanssens P, Joos H, Dhaese P, Zabeau M, Mahillon J. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid. 1989;21:9–20. doi: 10.1016/0147-619x(89)90082-6. [DOI] [PubMed] [Google Scholar]
- 22.Kleerebezem M, Beerthuyzen M M, Vaughan E E, de Vos W M, Kuipers O P. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kremer L, Baulard A, Estaquier J, Poulain-Godefroy O, Locht C. Green fluorescent protein as a new expression marker in mycobacteria. Mol Microbiol. 1995;17:913–922. doi: 10.1111/j.1365-2958.1995.mmi_17050913.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuipers O P, Beerthuyzen M M, de Ruyter P G G A, Wesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers O P, de Ruyter P G G A, Kleerebezem M, de Vos W M. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 1997;15:135–140. doi: 10.1016/S0167-7799(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Marteau P, Rambaud J C. Potential of using lactic acid bacteria for therapy and immunomodulation in man. FEMS Microbiol Rev. 1993;12:207–222. doi: 10.1111/j.1574-6976.1993.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 28.Mercenier A. Lactic acid bacteria as live vaccines. In: Tannock G M, editor. Probiotics: a critical review. Wymondham, Norfolk, United Kingdom: Horizon Scientific Press; 1999. pp. 113–127. [Google Scholar]
- 29.Mercenier A, Dutot P, Kleinpeter P, Aguirre M, Paris P, Reymund J, Slos P. Development of lactic acid bacteria as live vectors for oral or local vaccines. Adv Food Sci. 1996;18:73–77. [Google Scholar]
- 30.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucoronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poquet I, Ehrlich S D, Gruss A. An export-specific reporter designed for gram positive bacteria: application to Lactococcus lactis. J Bacteriol. 1998;180:1904–1912. doi: 10.1128/jb.180.7.1904-1912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Scott K P, Mercer D K, Glover L A, Flint H J. The green fluorescent protein as a visible marker for lactic acid bacteria in complex ecosystems. FEMS Microbiol Ecol. 1998;26:219–230. [Google Scholar]
- 34.Solaiman D K Y, Somkuti G A. Construction of a green-fluorescent protein-based, insertion-inactivation shuttle vector for lactic acid bacteria and Escherichia coli. Biotechnol Lett. 1997;19:1175–1179. [Google Scholar]
- 35.Stretton S, Techkarnjanaruk S, McLennan A M, Goodman A E. Use of green fluorescent protein to tag and investigate gene expression in marine bacteria. Appl Environ Microbiol. 1998;64:2554–2559. doi: 10.1128/aem.64.7.2554-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsien R Y. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 37.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;154:1–9. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 38.Wells J M, Wilson P W, Norton P M, Gasson M J, Le Page R W F. Lactococcus lactis: a high level expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol. 1993;8:1155–1162. doi: 10.1111/j.1365-2958.1993.tb01660.x. [DOI] [PubMed] [Google Scholar]