Abstract
A homolog of the major eubacterial cold shock gene cspA was identified in Sinorhizobium meliloti RM1021 by luxAB reporter transposon mutagenesis. Here we further characterize the organization and regulation of this locus. DNA sequence analysis indicated that the locus includes three open reading frames (ORFs) encoding homologs corresponding to CspA, a novel 10.6-kDa polypeptide designated ORF2, and a homolog of the Escherichia coli ribosomal protein S21. Transcription analysis indicated that this locus produced two different-sized cspA-hybridizing transcripts upon cold shock, a 400-nucleotide (nt) RNA encoding cspA alone and a 1,000-nt transcript encoding cspA-ORF2-rpsU. The sizes of the transcripts agreed with the location of the transcription start site determined by primer extension and the locations of two putative transcriptional terminators. The promoter of the cspA-ORF2-rpsU locus had −10 and −35 elements similar to the E. coli ς70 consensus promoter and, like the cspA locus of E. coli, included an AT-rich region upstream of the −35 hexamer. The promoter of the S. meliloti cspA locus was found to impart cold shock-induced mRNA accumulation. In addition, the 5′-untranslated region (5′ UTR) was found to increase the fold induction of cspA transcripts after cold shock and depressed the level of luxAB mRNA prior to cold shock, another feature similar to cspA regulation in E. coli. No “cold box” was identified upstream of the S. meliloti cspA gene, however, and there was no other obvious sequence identity between the S. meliloti 5′ UTR and that of E. coli. DNA hybridization analysis indicated that outside the cspA-ORF2-rpsU cold shock locus there are several additional cspA-like genes and a second rpsU homolog.
The cold shock response in eubacteria includes a number of adaptive changes ranging from alterations in membrane composition to changes in nucleoid structure (19, 37). Some of these changes involve the induction of cold shock genes. Indeed, a highly conserved feature of the cold shock response is the induction of one or more homologs of the major cold shock gene of Escherichia coli, cspA (25). CspA proteins are thought to act as RNA chaperones that bind to mRNAs and prevent the formation of secondary structures that prohibit their translation at low temperatures (23). Low-temperature regulation of the E. coli cspA gene has been shown to involve both transcriptional (18) and posttranscriptional (11, 22) mechanisms. Gene fusion studies indicate that the E. coli cspA promoter alone is responsive to cold shock, increasing transcription approximately sevenfold after temperature downshift (18). In addition, the 5′-untranslated region (5′ UTR) imparts dramatic temperature regulation of cspA mRNA accumulation which appears to result from temperature-influenced changes in mRNA secondary structure affecting mRNA stability (11, 22).
Sinorhizobium meliloti is a ubiquitous soil bacterium that forms nitrogen-fixing nodules on alfalfa and related plants (14). The ability of S. meliloti to form effective nodules, however, is adversely affected by a number of environmental conditions, including low temperature. This led Cloutier et al. (8) to initiate studies on the cold shock response in S. meliloti (and other temperate rhizobia) and also in arctic Rhizobium species. Their results established that rhizobia, like other bacteria, alter gene expression in response to low temperature. Beyond this, however, nothing is known about the function and regulation of cold shock genes in rhizobia. Thus, as a first step toward a better understanding of the cold shock response in S. meliloti, we conducted transposon mutagenesis by using a luxAB reporter gene to identify cold shock loci (31). Several luxAB reporter transposon recipients displayed higher LuxAB activity after cold shock. Two transposons were found to have been inserted near a homolog of the E. coli cspA gene. Here we further characterize the organization and regulation of this locus.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
S. meliloti strains (Table 1) were grown in tryptone-yeast extract (TY) broth medium (6) at 30°C. Cultures were maintained on solid TY medium containing 1.5% agar or frozen to −80°C in TY broth containing 10% glycerol. Cultures of E. coli were grown at 37°C in Luria-Bertani (LB) broth (33) and maintained on solid LB medium containing 1.5% agar. Antibiotics for S. meliloti were added to solid medium at the following concentrations: streptomycin (SM), 200 μg/ml (50 μg/ml in broth); spectinomycin (SP), 50 μg/ml; gentamicin (GM), 25 μg/ml; kanamycin (KM), 200 μg/ml (50 μg/ml in broth). KM and ampicillin (AP) were added to media for E. coli at 50 μg/ml. Tetracycline (TC) was added to media at a concentration of 10 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| Sinorhizobium meliloti | ||
| RM1021 | Wild type; Strr | F. de Bruijn |
| RM509 | RM1021 containing Tn5-1062 between cspA and ORF2 | 31 |
| RM11 | RM1021; ORF2::Tn5-1062 | 31 |
| RM37 | Ino−; RM1021 with cspA promoter-luxAB fusion replacing part of the ino locus | This study |
| RM39 | Ino−; RM1021 with cspA promoter-UTR-luxAB fusion replacing part of the ino locus | This study |
| Escherichia coli DH5α | supE44 hsdR17 recA1 thi-1 lacU169(φ80lacZM15) endA1 gyrA96 relA1 | 20 |
| Plasmids | ||
| pBluescript KS(−) Apr | Stratagene | |
| pMW193 | Tcr SP-Strr; pRK290-based vector for replacing the ino locus of RM1021 with promoter-reporter gene fusions | 7 |
| pRK2013 | Kmr Tra+; mob; helper plasmid for triparental matings | 13 |
| pJB251 | Gmr; incompatible with pRK290-based vectors | 7 |
| pRL1062a | Tcr Kmr; vector for Tn5-1062 mutagenesis and source of 2.4-kb XbaI luxAB fragment | 9, 41 |
| pH2 | Kmr; template for amplification of cspA promoter and coding region | 31 |
| pEco1 | Kmr; template for amplification of rpsU | 31 |
| pJJ23 | Kmr; template for amplification of ORF2 | 31 |
| pKO11 | 2.4-kb XbaI fragment with promoterless luxAB genes in pBluescript KS(−) | This study |
| pKO23 | cspA promoter (−188 to +1) on SacI-NotI fragment in pKO11 | This study |
| pKO26 | cspA promoter and 5′ UTR (−188 to +110) on SacI-NotI fragment in pKO11 | This study |
| pKO37 | cspA promoter-luxAB fusion from pKO23 cloned into KpnI site of pMW193 | This study |
| pKO39 | cspA promoter-UTR-luxAB fusion from pKO26 cloned into KpnI site of pMW193 | This study |
DNA cloning, sequencing, and hybridization analysis.
Cloned DNA was prepared for sequencing from cells of E. coli by using Qiagen Maxi-Columns and cut with restriction enzymes according to the manufacturer's instructions (New England Biolabs). Manual double-stranded DNA sequencing reactions were performed with the Sequenase 2.0 Kit (Amersham) and [35S]dATP. Oligonucleotide primers were synthesized by the Macromolecular Synthesis Facility, Michigan State University. Automated cycle sequencing reactions (fluorescent dye-terminator) were performed at the Michigan State University DNA Sequencing Facility by using an ABI Catalyst 8000 Molecular Workstation (Applied Biosystems, Inc.). Additional sequencing and primer synthesis were performed at the Biotechnology Resource Laboratory, Yale University. Total genomic DNA was isolated from S. meliloti essentially as described earlier (3), omitting the NaCl-CTAB (cetyltrimethylammonium bromide) extraction step. Probe DNA was amplified from cspA, ORF2, and rpsU sequences by standard methods (21) by using the templates and primers listed in Table 1. Labelling of probe DNA with [α-32P]dCTP, agarose gel electrophoresis, Southern blotting, and autoradiography were performed as described elsewhere (33).
Construction and integration of promoter-luxAB fusions.
Promoterless luxAB genes were excised from pRL1062 on a 2.4-kb XbaI fragment and cloned into the XbaI site of pBluescript KS(−), generating plasmid pKO11. Fragments of DNA encoding the cspA promoter alone (−188 to +1) or the cspA promoter and 5′ UTR (+188 to +110) were amplified by standard methods (21) by using plasmid pH2 as template and the primers listed in Table 1. The promoter fragments (including linkers added during primer synthesis) were cut with SacI and NotI and ligated into the SacI and NotI sites of pKO11 to generate plasmids pKO23 and pKO26. The pcspA-luxAB fusion (in pKO23) and the pcspA-UTR-luxAB fusion (in pKO26) were then excised with KpnI and ligated into the unique KpnI site of pMW193, creating plasmids pKO37 and pKO39, respectively. Plasmid pMW193 is designed to allow replacement of the inositol utilization locus (ino) with cloned novel DNA by homologous recombination, while isolating the integrated DNA from transcriptional activity in the S. meliloti chromosome (7).
To integrate the promoter-reporter fusions into the ino locus in RM1021, plasmids pKO37 and pKO39 were mobilized separately into S. meliloti RM1021 from E. coli DH5α by triparental mating. Plasmid pJB251 (Gmr), which is incompatible with pKO37 and pKO39, was then introduced, and transconjugants were selected for Tcr, Spr, and Gmr. The resulting strains, containing single recombinations between ino DNA on pKO37 or pKO39 and the chromosomal ino locus, were cultured in TY medium containing GM and SP but not TC. In subsequent screening we obtained isolates that were Gmr and Spr but Tcs and lacked the ability to use inositol as a sole carbon source. Replacement of chromosomal ino DNA with the promoter-reporter fusions, giving rise to strains RM37 and RM39, was confirmed by hybridization (not shown).
RNA isolation and analysis.
Total RNA was isolated from bacteria essentially as described earlier (3), except that rifamicin (150 μg/ml) was added to the bacteria immediately upon harvesting to prevent further transcription. RNA was precipitated with 2 volumes of cold 100% ethanol, washed twice with 70% ethanol, dried, resuspended in water, and quantified spectrophotometrically. Northern and slot blot hybridization experiments were performed as described elsewhere (3, 33). The intensity of radioactive bands on slot blot filters was quantified with a Molecular Dynamics PhosphorImager and analyzed with ImageQuant 3.3 software. The fold induction of mRNAs was calculated by using the 0-h (30°C, pre-cold shock) level as the reference level. Primer extension experiments were carried out as described previously (33) with enzymes supplied with the Primer Extension System (Promega) and with 10 μg of S. meliloti RNA per reaction. The sequences of the oligonucleotide primers are given in Table 2.
TABLE 2.
Oligonucleotides
| Oligonucleotide | Sequence | Function and features |
|---|---|---|
| MT171 | 5′-CCGGAATTCAAACGCTCCCTGCCAGTACATCCG-3′ | Amplification of cspA DNA; EcoRI |
| MT172 | 5′-CCGGAATTCCGTCCGGGAACTCTAGCGATTGAA-3′ | Amplification of cspA DNA; EcoRI |
| MT173 | 5′-CCGGAATTCGACCAGGCTCTTCGCGTTCTCAAG-3′ | Amplification of rpsU DNA; EcoRI |
| MT174 | 5′-CCGGAATTCCTACAGCGGCAGACGTGCAAAGA-3′ | Amplification of rpsU DNA; EcoRI |
| MT176 | 5′-ATTTCGGCGTCCTGATGCTCAAGG-3′ | Amplification of ORF2 DNA |
| MT177 | 5′-GGGTGGCATGGACGAAGGAGATGG-3′ | Amplification of ORF2 DNA |
| MT183 | 5′-CAATCGCTAGAGTTCCCG-3′ | Primer extension |
| MT184 | 5′-TGAATGAAGCCGAAGCCC-3′ | Primer extension |
| MT169 | 5′-TTTGAGCTCGGTACCAAGCCGGAAGAACCTCAGCTCGTCC-3′ | 5′ primer for promoter amplification; SacI and KpnI sites |
| MT170 | 5′-TTTGCGGCCGCTTCAATCGCTAGAGTTCCCGGACG-3′ | 3′ primer for amplifying bases −188 to +110; NotI site |
| MT208 | 5′-TTTGCGGCCGCTGAGCTATATATAAACGCTGCTTCGAGAT-3′ | 3′ primer for amplifying bases −188 to +1; NotI site |
Sequence analysis and accession number.
Sequence data were compared to known nucleotide and protein sequences by using the BLAST e-mail server (National Center for Biotechnology Information, Bethesda, Md. [1]). Additional analyses were performed by using the GCG (10) and DNASTAR sequence analysis packages. Potential open reading frames (ORFs) were identified, and codon usage was assessed with CodonUse 3.1 software (window size, 33; logarithmic range, 3), written by Conrad Halling, University of Chicago. The sequence reported here has been assigned GenBank accession number AF030523.
RESULTS
Sequence analysis of the cspA cold shock locus of S. meliloti.
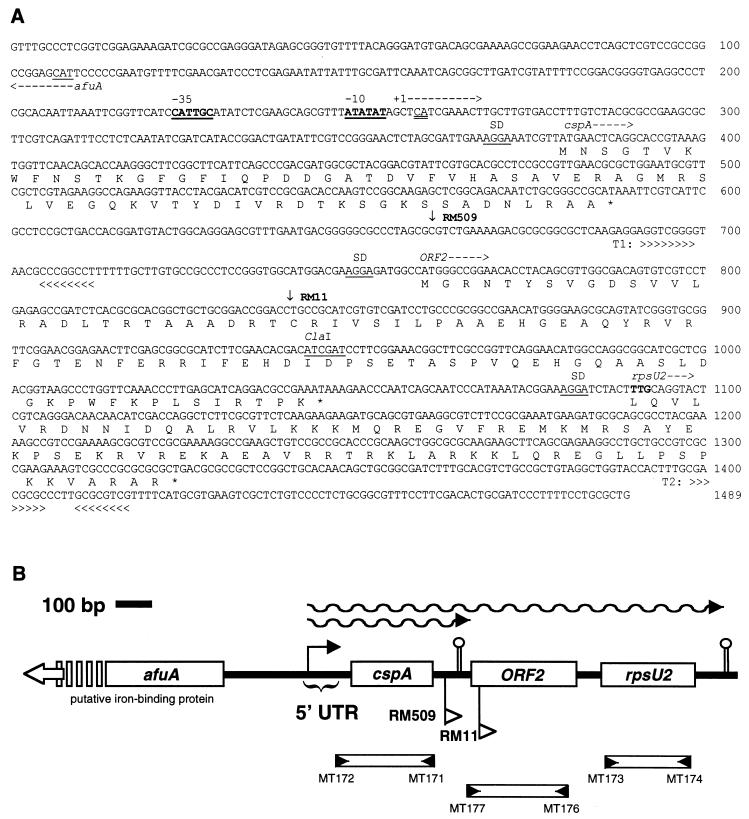
The transposon insertions in two of these mutants, RM509 and RM11, were determined to be located downstream of a homolog of cspA. DNA sequencing of this region (Fig. 1) (GenBank accession number AF030523) revealed the presence of three tandem ORFs with codon usage typical for S. meliloti proteins. The 5′ ORF encodes a polypeptide of 69 amino acids with a predicted molecular size of 7.4 kDa and an isoelectric point of 8.6. A BLAST search of the GenBank-EMBL database revealed significant similarity between the 5′ ORF and the major cold shock protein of E. coli, CspA (Fig. 2A) (25). The best match, however, was with a putative CspA homolog encoded by an ORF on the recently sequenced symbiotic plasmid of Rhizobium sp. strain NGR234 (Fig. 2B) (16). The predicted S. meliloti CspA protein contains the consensus cold shock domain amino acids conserved among CspA homologs, the eukaryotic Y-box-binding proteins and certain glycine-rich proteins of Arabidopsis thaliana (40). Based on sequence similarity and induction by cold shock, we designated the 5′ ORF as the cspA gene of S. meliloti.
FIG. 1.
Cold shock operon in S. meliloti RM1021. (A) Nucleotide sequence of the cold shock operon (bases 1001 to 2489 of accession number AF030523). The deduced amino acid sequences and putative gene identifications are given underneath the nucleotide sequence for ORFs which display codon usage typical of S. meliloti. Promoter sequences are printed in boldface and are underlined, and the location of the transcriptional start sites as determined by primer extension are double-underlined. Putative ribosome-binding sites (SD) are underlined. Rho-independent transcriptional terminators (T1 and T2) are indicated by chevrons. Inverted arrows indicate the location of transposon Tn5-1062 insertions in mutants RM11 and RM509. (B) Diagram of cold shock gene region in S. meliloti. The bent arrow indicates the transcriptional start site as determined by primer extension. Flags below the map indicate the location of Tn5-1062 insertions and the direction of transcription of the luxAB reporter genes. Bars below the map represent the amplified fragments used as probes in Southern and Northern blot experiments. Small arrows in the bars represent the oligonucleotide primers listed in Table 2. The complement of the start codon of the putative afuA homolog (bases 107 to 109) is underlined, and the direction of transcription (divergent from cspA) is indicated by the arrow. Part of the afuA sequence is omitted for brevity; however, it is included in GenBank in its entirety.
FIG. 2.
Similarities between the predicted CspA amino acid sequence from S. meliloti and CspA sequences from other bacteria by using the BESTFIT program (10). Asterisks indicate the conserved cold shock domain residues, as listed in PROSITE (5): [FY]-GFI-X(6,7)-[DER]-[LIVM]-FXHX-[STK]-X-[LIVMFY]. (A) Comparison with the CspA protein from E. coli. (B) Comparison with the closest known match, a predicted cold shock protein from the symbiotic plasmid of Rhizobium sp. strain NGR234 (16).
Just downstream of cspA was a second ORF that was preceded by a consensus ribosome binding site (Fig. 1A). The ORF was deduced to encode a polypeptide of 95 amino acids having a molecular size of 10.6 kDa and an isoelectric point of 7.4 (Fig. 1). The predicted polypeptide displayed no significant similarities with previously described polypeptides, nor did it contain sequences similar to known functional motifs. The putative cold shock gene encoding the novel polypeptide was designated ORF2.
A third ORF located just downstream of ORF2 (Fig. 1) was deduced to encode a polypeptide of 78 amino acids with a predicted molecular size of 9.3 kDa and an isoelectric point of 11.6. The predicted polypeptide was similar to ribosomal protein S21 from several species of bacteria, including that encoded by E. coli rpsU (Fig. 3). Thus, the ORF was designated rpsU. Translation of the S. meliloti rpsU homolog presumably begins at the unusual initiation codon TTG since the first in-frame ATG codon is downstream of several codons that specify amino acids that are conserved among S21 proteins (Fig. 1A). The use of TTG as an initiation codon has been documented in other bacteria, including members of the Rhizobiaceae (2). Also consistent with the TTG serving as the initiation codon is that immediately upstream of it there is a canonical ribosome-binding site.
FIG. 3.
Comparison of the predicted ribosomal protein S21 amino acid sequence from S. meliloti with S21 proteins from other prokaryotes made by using the BESTFIT program (10). Amino acid residues identical to the predicted S. meliloti protein are shaded. The last nine residues of the S. meliloti S21 sequence (Fig. 1) are omitted for brevity and display no matches to the other sequences. Rme, S. meliloti; Eco, E. coli; Hin, Haemophilus influenzae; Bst, Bacillus stearothermophilus; Mxa, Myxococcus xanthus; Ava, Anabaena variabilis.
Sequencing of the region upstream of cspA revealed the presence of a divergently oriented ORF that encodes a protein similar to AfuA, a putative iron-binding transport protein (39). The sequence of the entire ORF was determined (GenBank accession number AF030523) but was not studied further.
Transcription of the cspA cold shock locus.
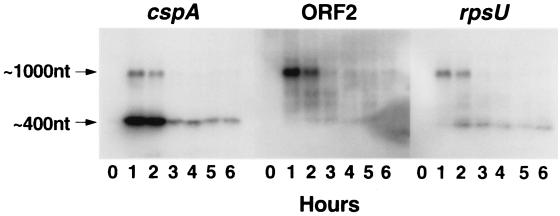
An inspection of the region upstream of the cspA gene indicated that there was a sequence resembling an E. coli ς70 consensus promoter and an AT-rich region similar to that found in the cspA promoter of E. coli (36). In addition, analysis of the entire cspA locus by using the TERMINATOR (10) program revealed two putative rho-independent termination signals, T1 and T2, downstream of cspA (Fig. 1). T1 and T2 were located, respectively, just upstream of ORF2 and just downstream of rpsU. The locations of the putative promoter and terminator sequences suggested that S. meliloti might produce two cold shock-inducible cspA mRNAs, one approximately 400 nucleotides (nt) long encoding cspA alone and another one approximately 1,000 nt long encoding cspA, ORF2, and rspU (Fig. 1B). To determine whether this was the case, total RNA was isolated from strain RM1021 before and after a temperature downshift from 30 to 15°C, and Northern blots were prepared and hybridized with probes for cspA, ORF2, and rpsU. The results indicated that cspA-hybridizing transcripts of approximately 400 and 1,000 nt did indeed accumulate upon a cold shock of 1 h (Fig. 4). Moreover, the results indicated that the 1,000-nt transcripts hybridized with the ORF2 and rpsU gene probes but that the 400-nt transcripts did not. Thus, it appears that the cspA, ORF2 and rspU genes are organized into an operon that produces transcripts of two different lengths as diagrammed in Fig. 1B.
FIG. 4.
Northern blot analysis of S. meliloti RM1021 total RNA isolated before and at 1-h intervals after a cold shock from 30 to 15°C. The filters were probed, from left to right, with the amplified cspA, ORF2, and rpsU genes shown in Fig. 1B.
The results presented in Fig. 4 indicate that expression of the cspA operon was transient. At 1 h after temperature downshift, the 400- and 1,000-nt transcripts were approximately 20- and 125-fold, respectively, higher in abundance than before the cold shock (Fig. 4, left panel [phosphorimager quantification not shown]). By 3 h, however, the 1,000-nt transcripts had returned to preshock levels. At this time transcripts of 400 nt were present a low levels that hybridized with cspA. In addition, transcripts of 400 nt were present that hybridized with the ORF2 and rpsU probes. Presumably, the faint hybridization observed with the ORF2 probe was due to degradation products generated from the 1,000-nt transcript. This could also be the case with the rpsU probes. Alternatively, this species (∼400 nt, right panel) may represent the cold shock induction of another rpsU-like gene (see below).
Identification of the cspA promoter.
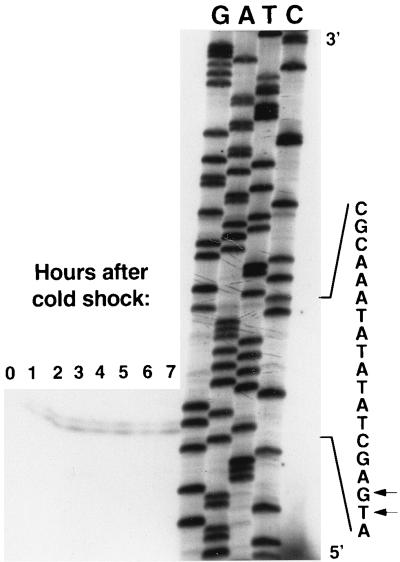
We located the 5′ end of the cspA mRNAs and confirmed the cold shock induction of cspA mRNA by primer extension analysis (Fig. 5). Cold shock induced the accumulation of mRNA that hybridized to oligomer MT183, which is complementary to sequences 5′ to the cspA ORF (Table 2). In this experiment the increase in cspA mRNA after cold shock was more gradual than in the Northern blot experiments. While there was a significant increase in cspA mRNA, the intensity of the signal increased to a higher steady-state level without a large transient increase. We believe this pattern of increased cspA expression was likely due to the use of a larger volume of culture for RNA isolation in this experiment (680- versus 220-ml cultures used to harvest RNA for Northern blots), resulting in a more gradual cooling of the cells and therefore a more gradual accumulation of cspA mRNA. The 5′ end of cspA mRNAs terminate at one of two adjacent bases, 120 and 121 nucleotides 5′ to the beginning of the cspA start codon (Fig. 5). As previously mentioned, upstream of the start of transcription is a sequence resembling an E. coli ς70 consensus promoter; there are several overlapping TATA boxes ca. 10 bp upstream of +1, preceded by a hexamer that resembles the E. coli ς70 −35 consensus sequence. The distances between the 5′ end of the cspA transcripts and the two predicted terminators agree well with the sizes of the 400- and 1,000-nt transcripts determined by Northern hybridization, providing additional support for the proposed operon structure. Primer extension experiments with oligonucleotide MT184, which binds inside the cspA ORF, gave an identical location for the 5′ end of the cspA mRNA (data not shown). Upstream of the −35 hexamer there is an AT-rich region that may serve as a UP element (32). The region surrounding +1 includes the sequence GCTCATCG, which is similar to the GCACATCA sequence that is found in the cold shock-stimulated P1 promoter of phage lambda (17). No CCAAT box was found in the entire region sequenced. We also did not find a region similar to the “cold box,” an 11-bp motif that is conserved in the 5′ UTR of cspA, cspB, and csdA of E. coli and is proposed to play a negative regulatory role in the transcription of cspA (12).
FIG. 5.
Primer extension mapping of the 5′ end of S. meliloti cspA mRNA. Total RNA from RM1021 cells was isolated before and at several 1-h intervals after a cold shock from 30 to 15°C and then hybridized to oligonucleotide MT183. Arrows indicate the 5′ ends of cspA transcripts.
Regulation of the cspA operon.
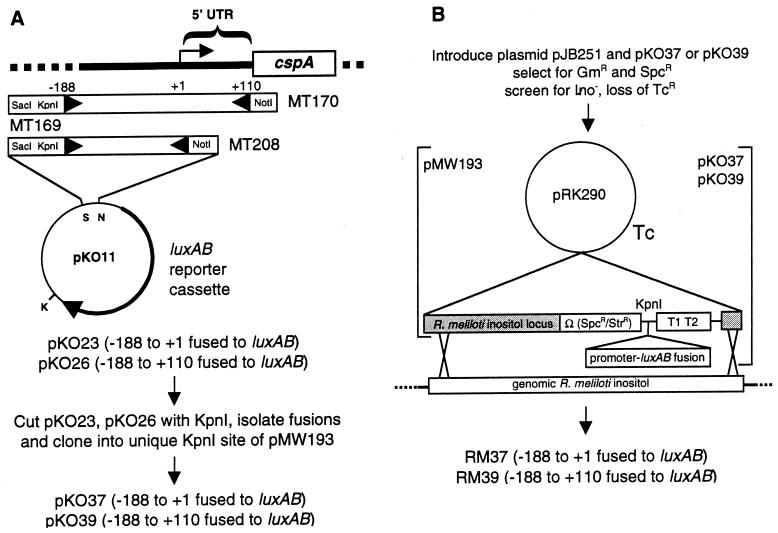
Two reporter gene constructs were made to determine whether the cspA promoter and/or the 5′ UTR were involved in regulating the accumulation of the cspA transcripts in response to cold shock. One construct, pKO23, had the promoter region of cspA—nt −188 to +1—fused to the luxAB reporter genes (Fig. 6A). The other, pKO26, had both the promoter region and most of the 5′ UTR of cspA—nt −188 to +110—fused to the luxAB reporter gene (Fig. 6A). To avoid potential high background luminescence caused by multiple copies of the fusions in plasmid vectors, the constructs were cloned into the KpnI site of pMW193, a vector designed to allow the integration of single copies of genes into the chromosome of S. meliloti. The fusions were then recombined into the inositol utilization (ino) locus (Fig. 6B), generating strains RM37 and RM39. Insertion of the promoter fusions into the ino locus was verified by Southern hybridization analysis (not shown).
FIG. 6.
Construction of pcspA-luxAB fusions and integration into the S. meliloti genome. (A) Amplification and fusion of pcspA with or without UTR sequences to promoterless luxAB. Promoter fragments were amplified by using the primers shown (boxes with arrowheads), with plasmid pH2 as template. Primer sequences are given in Table 2. SacI, KpnI, and NotI sites were added during primer synthesis. (B) Integration of promoter-luxAB fusions into the genomic inositol utilization (ino) locus of S. meliloti. The ino sequences in pMW193 flank the cloning site, omega fragment, and rrnB terminators, allowing replacement of the genomic ino locus with the promoter-luxAB fusion. Symbols: K, KpnI; N, NotI; T1 T2, tandem E. coli rrnB terminators in pMW193.
The levels of luxAB mRNA in strains RM37 (cspA promoter alone fused to luxAB) and RM39 (cspA promoter and 5′ UTR fused to luxAB) before and after a temperature downshift from 30 to 15°C were determined by slot blot hybridization (Fig. 7). Strain RM509, which contains a luxAB reporter transposon (Tn5-1062) insertion between cspA and terminator T1 (Fig. 1) was used as a reference strain. In cells grown at 30°C, the level of luxAB mRNA in strain RM37 was approximately sixfold higher than in strain RM39 (Fig. 7). Therefore, the presence of the 5′ UTR upstream of luxAB in strain RM39 resulted in a lower steady-state level of luxAB mRNA prior to cold shock. The pre-cold shock level of luxAB mRNA in strain RM509, in which the intact 5′ UTR and the entire cspA ORF are present upstream of luxAB on the mRNA molecule (Fig. 1B) (30) was comparable to the pre-cold shock luxAB mRNA level in strain RM39.
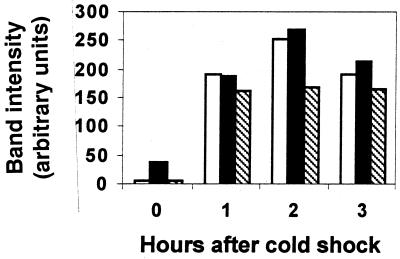
FIG. 7.
Induction of pcspA-luxAB fusions by cold shock. Total RNA from S. meliloti strains RM37, RM39, and RM509 was isolated before and at 1-h intervals after cold shock and analyzed by slot blot hybridization to luxAB mRNA. The intensity of the bands was determined with a phosphorimager as described in Materials and Methods. White columns, RM509; black columns, RM37; hatched columns, RM39. Data are representative of two experiments.
Upon cold shock (2 to 4 h), the level of luxAB mRNA increased approximately sevenfold in strain RM37 (Fig. 7), suggesting that part of the cold shock accumulation of cspA mRNA is due to an increase in promoter activity. At this same time, the level of luxAB mRNA in strain RM39 increased approximately 25-fold and, in absolute levels, approached that attained in RM37. Thus, the negative effect on transcript accumulation caused by the 5′ UTR of the cspA operon appeared to be relieved by cold shock. Upon cold shock, the level of luxAB mRNA in RM509 was induced nearly 50-fold and was about the same as that observed in RM37.
Other sequences similar to cspA and rpsU are present in S. meliloti.
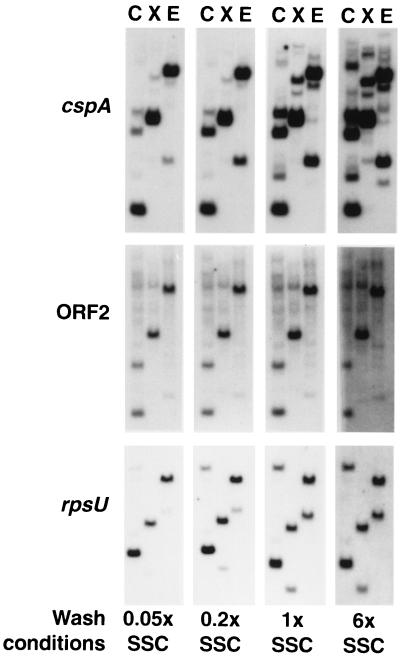
Many species of bacteria contain several genes homologous to cspA. E. coli contains nine copies of cspA-like genes, only some of which are induced by cold shock (26, 29, 42). To determine whether S. meliloti also contains multiple homologs of cspA, the amplified cspA DNA used as probe in the Northern blot experiments was hybridized to genomic DNA of S. meliloti digested with ClaI, XhoI, or EcoRI. Even under stringent conditions (0.05× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]), multiple sequences that hybridize to the amplified S. meliloti cspA were detected (Fig. 8, top panels). Washing under less-stringent conditions (0.2× to 6× SSC) revealed even more sequences that hybridized with the cspA probe, suggesting that cspA is part of a multigene family in S. meliloti with perhaps as many as five members. Similar experiments with the amplified ORF2 sequence as a probe indicated that only one sequence hybridized strongly to the ORF2 probe under the least stringent condition (6× SSC), suggesting that a single copy of ORF2 is present in S. meliloti (Fig. 8, center panels).
FIG. 8.
Southern blots of S. meliloti RM1021 genomic DNA cut with ClaI (C), XhoI (X), or EcoRI (E). Filters were probed with the amplified S. meliloti cspA, ORF2, or rpsU DNA fragments shown in Fig. 1B. One filter from each hybridization was washed in one of four aqueous SSC solutions (all include 0.1% sodium dodecyl sulfate) at 60°C and then exposed to film. EcoRI and XhoI do not cleave within the cold shock operon; ClaI cleaves ORF2 once (Fig. 1).
Duplicate Southern blots of S. meliloti genomic DNA were also probed with a DNA fragment amplified from the rpsU homolog (Fig. 8, bottom panels). The Southern blot washed at high stringency (0.05× SSC) showed the cloned rpsU homolog and another faintly hybridizing signal, based on the sizes of the restriction fragments. Washing identical blots under less-stringent conditions allowed stronger hybridization of the second sequence to the rpsU probe. As mentioned above, we have not ruled out the possibility that this other rpsU-like gene may be the source of the smaller rpsU-like transcript observed after cold shock (Fig. 4, right panel). Because both genes are visible under the most stringent wash condition (0.05× SSC), it is possible that the amplified rpsU DNA might have hybridized to transcripts of both rpsU-like genes under our standard Northern blot wash conditions (0.25× SSC).
DISCUSSION
We have identified a cold shock operon in S. meliloti that encodes a homolog of the E. coli CspA major cold shock protein, a small novel polypeptide (ORF2), and a homolog of E. coli ribosomal protein S21. To our knowledge, this is the first example of a cspA gene being organized into an operon with other cold shock genes. The significance of this grouping of genes is uncertain. The role of the S. meliloti CspA protein in cold shock is presumably the same as that proposed for other CspA proteins: to act as an RNA chaperone that aids in the translation of mRNAs at low temperature. The roles of the other two genes in the cold shock response, however, are less clear. Indeed, the ORF2 polypeptide has no sequence homology with any previously described protein or functional motif, and a transposon insertion mutation within the ORF2 coding sequence does not have any obvious effect on the growth of cells upon shift to low temperature (31). Thus, at this point, all that can be concluded is that the ORF2 gene appears to be a novel single-copy gene that is transiently induced upon cold shock but does not have an essential role in low temperature growth.
As for the rpsU gene, in vitro studies of E. coli ribosome function suggest that the rpsU-encoded ribosomal protein S21 is required for initiation of translation (38); it possibly facilitates the binding of mRNAs to the ribosome (4) or stimulates the association of ribosomal subunits (27). Thus, increased synthesis of S21-like proteins in S. meliloti might function to alleviate an inhibition of translation initiation caused by low temperatures, as other known cold shock proteins, such as RbfA, are believed to do (24). Interestingly, a homolog of rpsU in Anabaena variabilis has also been shown to be induced by cold shock (34, 35), suggesting that the induction of rpsU homologs by cold shock is an important adaptive response in some bacteria. Whether this is the case in S. meliloti remains to be determined. Mutants RM509 and RM11, which have transposon insertions that are likely to exert polar effects and block the expression of the rpsU gene, do not display any obvious cold-sensitive phenotype when grown at 15°C (31). These results suggest that the rpsU gene is not required for cold shock adaptation. However, our Southern analysis indicates that S. meliloti has a second rpsU homolog (Fig. 8). Expression of this gene could mask a role of the cold shock-inducible rpsU gene in cold shock adaptation under the conditions tested. Of course, it is also possible that protein S21 is not required for ribosomal function under some conditions; at least one prokaryote, Mycoplasma genitalium, lacks this gene entirely (15).
Our results indicate that there are a number of similarities in the regulation of the S. meliloti cspA cold shock operon and the E. coli cspA cold shock gene. The promoter regions of the cspA-ORF2-rpsU operon and the E. coli cspA gene both have −10 and −35 elements similar to the E. coli ς70 consensus promoter and have AT-rich regions upstream of the −35 hexamer (this study; see also reference 36). One study found that the presence of the AT-rich region upstream of the cspA gene of E. coli is required for the expression of cspA (28); other workers have seen a positive but less dramatic effect (18). The role of this sequence in the expression of the S. meliloti cspA gene has not been determined. Our gene fusion studies indicate that the promoter of the S. meliloti cspA operon is induced approximately sevenfold in response to cold shock (Fig. 7), which is similar to that reported for the promoter of the E. coli cspA gene (18). Moreover, our results indicate that the 5′ UTR of the S. meliloti cspA operon, like the 5′ UTR of the E. coli cspA gene (11), has a substantial effect on the accumulation of transcripts. We found that at non-cold shock temperatures, the 5′ UTR of the S. meliloti cspA operon decreases the level of transcript accumulation by about sixfold; this effect seems likely to be due to the sequence causing an increased rate of degradation of the transcripts at the non-cold shock temperature. Upon cold shock, however, the effect of the 5′ UTR appeared to be eliminated. In E. coli, the 5′ UTR of the cspA gene is sufficient to regulate much of the accumulation of cspA mRNA in response to cold shock (11, 22). The current model posits that the presence of the 5′ UTR causes cspA mRNA to be rapidly degraded at 37°C but that it assumes a secondary structure at lower temperatures that facilitates its translation when other mRNAs are inaccessible. This is borne out by a study in which mutations of the E. coli 5′ UTR which alter its secondary structure cause cspA mRNA to accumulate at 37°C (11). This effect is at least partly due to the influence of RNase E (see below). The presence of a “cold box” (a sequence in the 5′ end of the UTR that influences cspA transcription) (12) in S. meliloti was not confirmed, since only one such gene was analyzed; however, no sequence in the S. meliloti UTR matches the cold box consensus of E. coli. This may simply indicate that S. meliloti either does not have such an element or that its sequence is different, since the entire UTR seems to have little similarity to those of other species.
Lastly, there is preliminary evidence that an RNase E-like enzyme also regulates cspA accumulation in S. meliloti. In some Northern blot experiments we have observed an RNA species of approximately 250 nt that hybridizes to a cspA probe, appearing approximately 1 h after accumulation of the 400- and 1,000-nt transcripts after cold shock (unpublished data). A prediction of the secondary structure of the S. meliloti 400-nt cspA transcript (bases +1 to +458) by the MFOLD program (10) revealed two putative RNase E recognition sites (single-stranded A/U-rich regions flanked by stable secondary structures [30]) spaced about 250 nt apart. The first sequence, AUAUU (nucleotides +82 to +85), is located in the loop of a potential hairpin (bases +73 to +92; part of a larger stem interrupted by an unpaired region of about eight bases on each side). Bases +324 to +332 (CGCAUAAA) form a second potential RNase E site between two stem-loops (+302 to +323 and +333 to +388). Cleavage of these potential RNase E sites would produce an RNA species of approximately 250 bases. Fang et al. (11) found that eliminating a putative RNase E recognition site in the E. coli cspA transcript (+135 to +146) lengthens the half-life of the transcript and that the half-life of cspA mRNA is longer in a conditional rne mutant at the nonpermissive temperature. Thus, it is possible that an RNase E-like enzyme might play a role in the regulation of cspA mRNA stability in S. meliloti. Mutants with transposon insertions in an rne-like gene were recently isolated (A. M. Gustafson and M. F. Thomashow, unpublished data), and their analysis may reveal whether RNase E plays a role in regulating the accumulation of the S. meliloti cspA transcript.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation STC grant DEB9120006 in the Center for Microbial Ecology and by the Michigan Agricultural Experiment Station.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aoyama T, Hirayama T, Tamamoto S, Oka A. A putative start codon TTG for the regulatory protein VirG of the hairy-root-inducing plasmid pRiA4. Gene. 1989;78:173–178. doi: 10.1016/0378-1119(89)90325-9. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Backendorf C, Ravensbergen C J, Van der Plas J, van Boom J H, Veeneman G, Van Duin J. Basepairing potential of the 3′ terminus of 16S RNA: dependence on the functional state of the 30S subunit and the presence of protein S21. Nucleic Acids Res. 1981;9:1425–1444. doi: 10.1093/nar/9.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1996;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berenger F J. R-factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 7.Bosworth A H, Williams M K, Albrecht K A, Kwiatkowski R, Beynon J, Hankinson T R, Ronson C W, Cannon F, Wacek T J, Triplett E W. Alfalfa yield response to inoculation with recombinant strains of Rhizobium meliloti with an extra copy of dctABD and/or modified nifA expression. Appl Environ Microbiol. 1994;60:3815–3832. doi: 10.1128/aem.60.10.3815-3832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloutier J, Prévost D, Nadeau P, Antoun H. Heat and cold shock protein synthesis in arctic and temperate strains of rhizobia. Appl Environ Microbiol. 1992;58:2846–2853. doi: 10.1128/aem.58.9.2846-2853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen M F, Meeks J C, Cai A Y A, Wolk C P. Transposon mutagenesis of heterocyst-forming filamentous cyanobacteria. Methods Enzymol. 1998;297:3–17. [Google Scholar]
- 10.Devereux J, Haeberli P, Smythies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 12.Fang L, Hou Y, Inouye M. Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J Bacteriol. 1998;180:90–95. doi: 10.1128/jb.180.1.90-95.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 is dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer H M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 16.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 17.Giladi H, Goldenberg D, Koby S, Oppenheim A B. Enhanced activity of the bacteriophage lambda PL promoter at low temperature. FEMS Microbiol Rev. 1995;17:135–140. doi: 10.1111/j.1574-6976.1995.tb00195.x. [DOI] [PubMed] [Google Scholar]
- 18.Goldenberg D, Azar I, Oppenheim A B, Brandi A, Pon C L, Gualerzi C O. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol Gen Genet. 1997;256:282–290. doi: 10.1007/s004380050571. [DOI] [PubMed] [Google Scholar]
- 19.Graumann P, Marahiel M A. Some like it cold: response of microorganisms to cold shock. Arch Microbiol. 1996;166:293–300. doi: 10.1007/s002030050386. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on the transformation of Escherichia coli with plasmids. J Mol Biol. 1983;66:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Innis M A, Gelfand D H. Optimization of PCRs. In: Innis M A, et al., editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 3–12. [Google Scholar]
- 22.Jiang W, Fang L, Inouye M. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J Bacteriol. 1996;178:4919–4925. doi: 10.1128/jb.178.16.4919-4925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 24.Jones P G, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 25.Jones P G, Inouye M. The cold shock response—a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee S J, Xie A, Jiang W, Etchegaray J-P, Jones P G, Inouye M. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol Microbiol. 1994;11:833–839. doi: 10.1111/j.1365-2958.1994.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 27.Melançon P, Leclerc D, Destroismaisons N, Brakier-Gingras L. The anti-Shine-Dalgarno region in Escherichia coli 16S ribosomal RNA is not essential for the correct selection of translational starts. Biochemistry. 1990;29:3402–3407. doi: 10.1021/bi00465a037. [DOI] [PubMed] [Google Scholar]
- 28.Mitta M, Fang L, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima K, Kanamaru K, Mizuno T, Horikoshi K. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J Bacteriol. 1996;178:2994–2997. doi: 10.1128/jb.178.10.2994-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naureckiene S, Uhlin B E. In vitro analysis of mRNA processing by RNase E in the pap operon of Escherichia coli. Mol Microbiol. 1996;21:55–68. doi: 10.1046/j.1365-2958.1996.6121101.x. [DOI] [PubMed] [Google Scholar]
- 31.O'Connell K P, Gustafson A M, Lehmann M D, Thomashow M F. Identification of cold shock gene loci in Sinorhizobium meliloti by using a luxAB reporter transposon. Appl Environ Microbiol. 2000;66:401–405. doi: 10.1128/aem.66.1.401-405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross W, Aiyar S E, Salomon J, Gourse R L. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sato N. A cold-regulated cyanobacterial gene cluster encodes RNA-binding protein and ribosomal protein S21. Plant Mol Biol. 1994;24:819–823. doi: 10.1007/BF00029864. [DOI] [PubMed] [Google Scholar]
- 35.Sato N, Tachikawa T, Wada A, Tanaka A. Temperature-dependent regulation of the ribosomal small-subunit protein S21 in the cyanobacterium Anabaena variabilis M3. J Bacteriol. 1997;179:7063–7071. doi: 10.1128/jb.179.22.7063-7071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe H, Goldstein J, Yang M, Inouye M. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J Bacteriol. 1992;174:3867–3873. doi: 10.1128/jb.174.12.3867-3873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theiringer H A, Jones P G, Inouye M. Cold shock and adaptation. Bioessays. 1998;20:49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 38.Van Duin J, Wijnands R. The function of ribosomal protein S21 in protein synthesis. Eur J Biochem. 1981;118:615–619. doi: 10.1111/j.1432-1033.1981.tb05563.x. [DOI] [PubMed] [Google Scholar]
- 39.Willemsen P T, Vulto I, Boxem M, de Graaff J. Characterization of a periplasmic protein involved in iron utilization of Actinobacillus actinomycetemcomitans. J Bacteriol. 1997;179:4949–4952. doi: 10.1128/jb.179.15.4949-4952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolffe A P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1993;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 41.Wolk C P, Cai Y, Panoff J M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci USA. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamanaka K, Inouye M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J Bacteriol. 1997;179:5126–5130. doi: 10.1128/jb.179.16.5126-5130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]