Abstract
We report a dual labeling technique involving two green fluorescent protein (GFP) variants that is compatible with confocal microscopy. Two lasers were used to obtain images of (i) mixed cultures of cells, where one species contained GFPuv and another species contained GFPmut2 or GFPmut3, and (ii) a single species containing both GFPuv and GFPmut2 in the same cell. This method shows promise for monitoring gene expression and as a nondestructive and in situ technique for confocal microscopy of multispecies biofilms.
Over the past decade, green fluorescent protein (GFP) from the jellyfish Aequorea victoria has emerged as a versatile reporter gene and in situ cell marker. Advantages such as species independence and the lack of a requirement for substrates and cofactors make GFP unique as a reporter gene (3). GFP has become an especially valuable marker for nondestructively visualizing cells, particularly in biofilms. The use of GFP in combination with confocal laser scanning microscopy (CLSM) has led to new insights into biofilm processes (6). Several GFP variants with excitation and emission properties different from those of the wild-type protein have been developed (1). One such protein, GFPuv (5) (Clontech, Palo Alto, Calif.), emits bright green light (maximum at 509 nm) when exposed to UV or blue light (395 or 470 nm). Mutant proteins GFPmut2 and GFPmut3 (4) have emission maxima of 507 and 511 nm when excited by blue light (481 and 501 nm, respectively). Unlike GFPuv, GFPmut2 and GFPmut3 are not excited by UV light, a difference that allows differential imaging of these proteins in the same sample.
A dual-labeling technique based on two GFPs has been used for epifluorescent microscopy (11). However, dual labeling with fluorescent proteins for confocal microscopy has not been developed, because of the limited selection of excitation wavelengths available with Kr/Ar lasers, which are frequently features of commercially available confocal microscopes. In this study, we used a confocal microscope equipped with Kr/Ar and UV lasers to expand the range of fluorescent proteins that could be imaged. The two-laser system was used to image (i) mixed cultures of cells, where one species contained GFPuv and another species contained GFPmut2 or GFPmut3, and (ii) a single species containing both GFPuv and GFPmut2 in the same cell.
Strains, plasmids, and media.
The strains and plasmids used are listed in Table 1. All strains were grown in minimal salts medium (MSM) (7) with 0.1% glycerol, 0.1% Casamino Acids, and 100 μg of ampicillin ml−1. Escherichia coli and Pseudomonas strains were cultured at 37 and 30°C, respectively. Cultures containing pCSAK50 were induced with 0.02% arabinose.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH10B | F−-mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK1 rpsL nupG | Life Technologies, Inc. (Gaithersburg, Md.) |
| E. coli 33456 | Wild type | 13 |
| Pseudomonas sp. strain GJ1 | 9 | |
| Plasmids | ||
| pGFPuv | pUC-based vector carrying gfpuv, Apr | Clontech |
| pGFPmut3 | pKEN containing gfpmut3, Apr | 4 |
| pCSAK50 | pBAD-based vector containing gfpuv, Apr | A. Khlebnikov and J. D. Keasling, unpublished data |
| pSMC21b | pUCP-based plasmid containing gfpmut2, Apr, Cbr, Kmr | 2 |
Apr, ampicillin resistance; Cbr, carbenicillin resistance; Kmr, kanamycin resistance.
pSMC21 is identical to pSMC2 but has an additional Kmr gene.
Bench-scale flow cell.
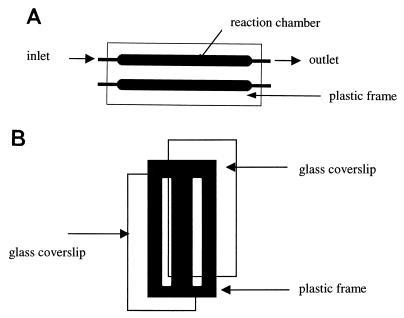
Biofilms were prepared in bench-scale parallel plate flow cells (reactor volume of 0.35 ml) (Fig. 1). A cover glass was glued to the plastic frame with silicone rubber adhesive sealant RTV 102 (GE Silicones, Waterford, N.Y.). Flow cells were operated in recirculating (start-up) or continuous modes at a constant flow rate (0.862 ml min−1).
FIG. 1.
Bench-scale flow cell. (A) Top view; (B) cross-sectional view. Chamber size: 46 by 4 by 2 mm.
Preparation of poly-l-lysine-coated cover glass.
A 1.5-ml amount of 0.025% poly-l-lysine (Sigma Diagnostics, St. Louis, Mo.) was placed on the surface of Fisherbrand microscope cover glass (type 24 × 60-1; Fisher Scientific, Pittsburgh, Pa.) (8). The poly-l-lysine was dried onto the cover glass overnight, and the glass was subsequently glued to the biofilm reactor.
Confocal microscopy.
Confocal microscopy was performed with an MRC-1024 laser scanning confocal imaging system (Bio-Rad Microsciences, Cambridge, Mass.) equipped with a Nikon Diaphot 200 inverted microscope (Nikon, Inc., Tokyo, Japan). A 60× Plano-Apo oil immersion lens with a numerical aperture of 1.4 was used for all images. The 488-nm line from a Kr/Ar laser was used to excite GFPuv, GFPmut2, and GFPmut3. The 363-nm line of an argon ion UV laser (model ENT 622; Innova Technology/Coherent Enterprises, Santa Clara, Calif.) was used to excite GFPuv. Emissions from all GFPs were detected with a standard fluorescein filter set (522 ± 35 nm) and a sequential scanning program was used; a delay of approximately 4 s occurred between scans. Images obtained by excitation with the 488-nm line of the Kr/Ar laser were directed to the red channel, while images obtained by excitation with the 363 nm line of the UV laser were directed to the green channel. The resulting red and green images were overlaid by using Confocal Assistant 4.02 (Todd Clark Brelje) and Adobe Photoshop 5.0 (Adobe Systems, Inc., San Jose, Calif.) software. Colocalization of red and green signals produced a yellow color.
Mixed cultures of cells containing different GFPs on poly-l-lysine-coated surfaces.
E. coli DH10B harboring pGFPmut3 was grown to stationary phase in a shake flask and was then recirculated over a poly-l-lysine-coated cover glass for 2 h. E. coli 33456 containing pGFPuv was grown to stationary phase and was recirculated in the same reactor for 5 h after strain DH10B had been recirculated through the flow cell. MSM (15 ml) was then pumped through each flow cell to remove cells that had not adhered to the surface of the cover glass. The attached cells were imaged immediately.
Mixed-culture biofilms containing different GFPs on bare glass.
Exponentially growing E. coli 33456 harboring pGFPuv was recirculated through a flow cell over a bare cover glass for 8 h to promote colonization of the surface. Following that, Pseudomonas sp. strain GJ1 harboring pSMC21 was recirculated through the same flow cell for 3 h. Subsequently, sterile MSM was continuously pumped through the flow cell at a flow rate of 0.063 ml min−1 for 16 h.
Two GFPs in one cell.
E. coli DH10B containing both pCSAK50 and pSMC21 was grown with and without 0.02% arabinose. Each culture was recirculated through a separate flow cell containing a poly-l-lysine-coated cover glass for 3 h. The flow cell was then rinsed and imaged as described above.
Mixed cultures of cells containing different GFPs on poly-l-lysine-coated surfaces.
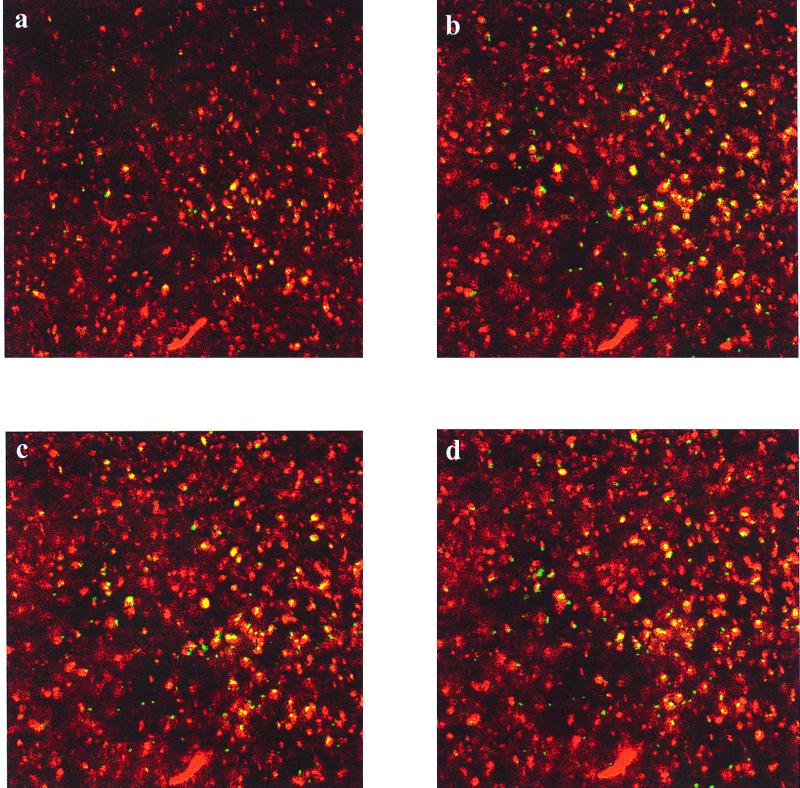
The dual-GFP technique was used to find the locations of two different E. coli strains, each with a different GFP, in a biofilm formed on poly-l-lysine-coated glass. Distinct regions of E. coli 33456(pGFPuv) and DH10B(pGFPmut3) could be observed after the cells flowed over poly-l-lysine-coated cover glass (Fig. 2). Red-colored cells represent the fraction of the population that expressed GFPmut3. Yellow-colored cells resulted from the excitation of GFPuv by both 363- and 488-nm light. No fluorescence from GFPmut3 was detected when the samples were excited with 363-nm light.
FIG. 2.
Horizontal sections of E. coli DH10B(pGFPmut3) and E. coli 33456(pGFPuv) attached to a poly-l-lysine-coated cover glass. The images represent cells that were excited by either 363 nm (green), 488 nm (red), or both (yellow). Images at 3 μm (a), 6 μm (b), 7 μm (c), and 8 μm (d) above the substratum are shown.
Mixed culture biofilms containing different GFPs on bare cover glass.
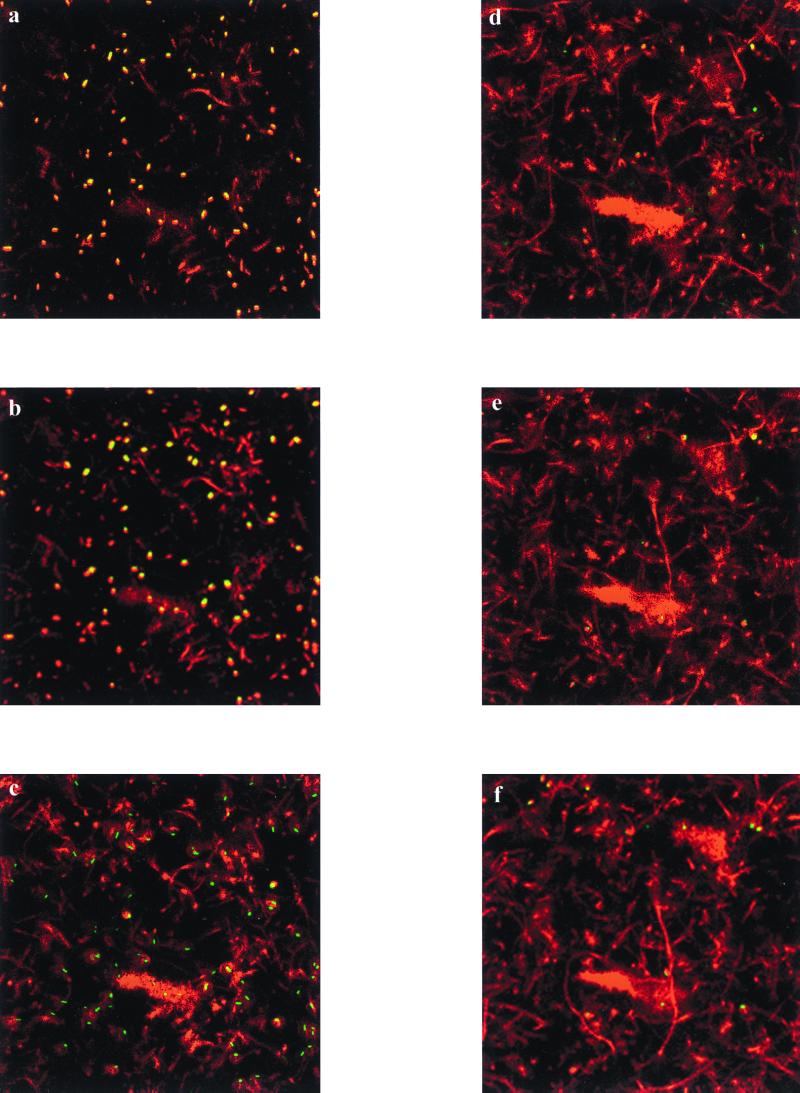
Biofilms comprised of E. coli 33456(pGFPuv) and Pseudomonas sp. strain GJ1(pSMC21) were cultured on a bare glass substratum (Fig. 3). The two species could be distinguished by color as well as morphology: E. coli cells are small and yellow (due to GFPuv), while Pseudomonas cells form long chains and are red (due to GFPmut3). A heterogeneous spatial distribution of the two species is evident in the biofilm, with the bulk of the E. coli cells concentrated near the substratum, and the Pseudomonas cells forming a thick layer above the E. coli cells. In the merged images of dual-species biofilms, we observed cells that were excited by the UV laser but not by the Kr/Ar laser (green cells in Fig. 3). Since the lasers were activated at different times, it must be that these cells are unattached and floated to a new location between laser excitations. That these cells were unattached is also supported by the fact that they were most frequently observed in voids within the biofilm. The dynamic nature of biofilm evolution through growth and cellular detachment is partially responsible for the observed heterogeneity in microbial biofilms (10, 12). An alternative explanation is that the signal is the result of UV-excitable debris in the sample. Thus, this technique may be useful for examining the dynamic nature of multispecies biofilms as well as the spatial distribution of different species.
FIG. 3.
Horizontal sections of a coculture biofilm with Pseudomonas sp. strain GJ1(pSMC21) and E. coli 33456(pGFPuv). The images represent cells that were excited by either 363 nm (green), 488 nm (red), or both (yellow). Images at 2 μm (a), 4 μm (b), 9 μm (c), 14 μm (d), 17 μm (e), and 20 μm (f) above the substratum are shown.
Two GFPs in a single cell.
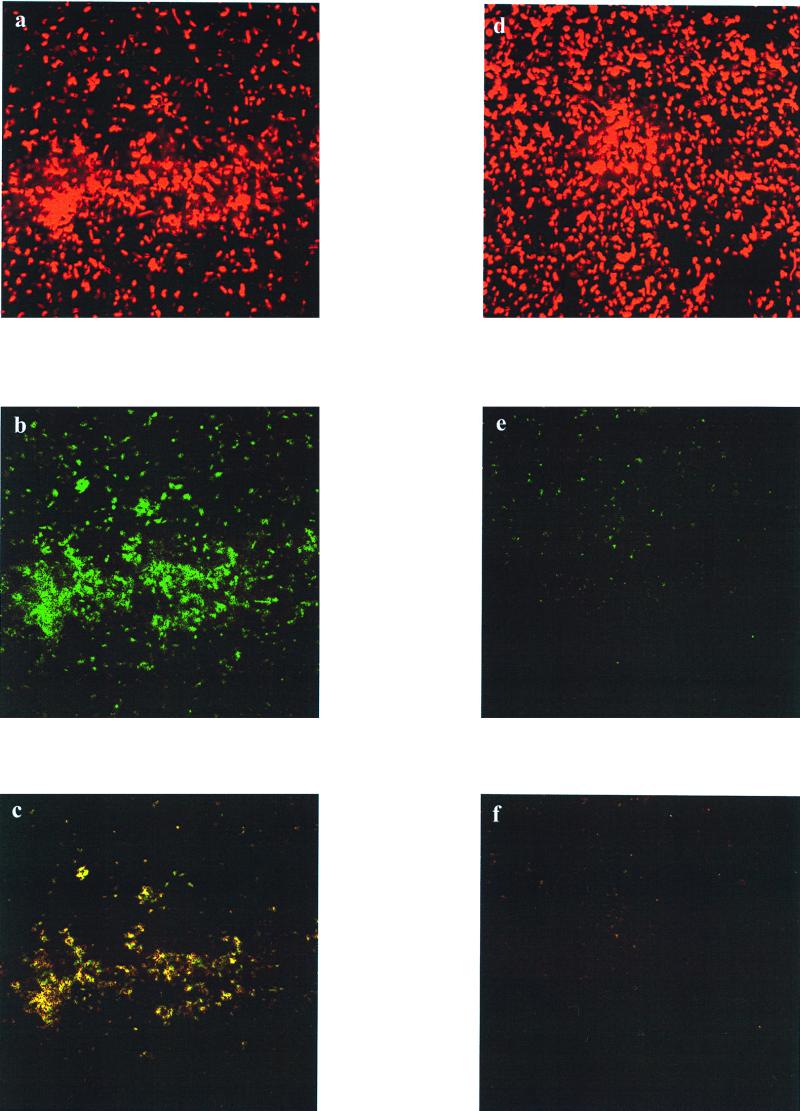
By combining the signal output from both the red and the green channels, information about the attachment and induction state of the illuminated cells was obtained. In the case of the E. coli harboring two fluorescent proteins (Fig. 4), both arabinose-induced and uninduced cells fluoresced upon illumination with the Kr/Ar laser; this was due to expression of gfpmut3 from the constitutive plasmid pSMC21 and also gfpuv from induced pCSAK50. Cells that had been induced with arabinose also produced high levels of GFPuv and fluoresced upon illumination by the UV laser (Fig. 4b), while there was only a faint signal corresponding to a basal level of expression of gfpuv in the uninduced culture (Fig. 4e). Cells that fluoresced upon illumination with both lasers (yellow color) contained both GFP variants. Images of biofilms with two GFP variants colocalized in a single cell could be beneficial for quantifying the fraction of a microbial population that is induced after exposure to an inducer or for determining expression of two different genes simultaneously.
FIG. 4.
E. coli DH10B harboring pCSAK50 and pSMC21. (a to c) Arabinose-induced cells excited at wavelengths of 488 nm (a), 363 nm (b), and 488 and 363 nm (c). (d to f) Uninduced cells excited at wavelengths of 488 nm (d), 363 nm (e), and 488 and 363 nm (f).
The dual-fluorescent-protein system described in this work is a powerful tool for nondestructive analysis of microbial communities using confocal microscopy. This technique could be particularly beneficial for investigations of multispecies biofilms, where the three-dimensional architecture of the sample is of great interest. The model systems indicate that dual labeling using GFP variants can serve as an in situ method for imaging of cocultures as well as for monitoring gene expression in biofilms. The dual-labeling technique differs from other fluorescent dual-labeling strategies in that the two GFP signals are distinguished by differential excitation rather than differential emission. Dual labeling using GFP for analysis of biofilms has been accomplished by using fluorescent stains, such as propidium iodide (14) and soluble nucleic acid dyes (K. M. Kazmerzak, R. N. Andersen, R. T. Mondshine, and P. E. Kolenbrander, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. J-8, 1999), or with fluorescently labeled oligonucleotide probes (1). A drawback of these techniques is that the sample must be sacrificed to image the biofilm. In contrast, using the labeling strategy presented here, biofilms can be imaged continuously by CLSM.
Dual labeling with green fluorescent proteins coupled with confocal microscopy is an alternative to existing methods for investigation of natural biofilms and fixed cultures. With the increasing availability of broad-host-range vectors for transformation of environmental isolates, the use of fluorescent proteins as specific markers should facilitate the study of microbial consortia and intercellular processes including quorum sensing and horizontal gene transfer.
Acknowledgments
We thank Carolyn Larabell at Lawrence Berkeley National Laboratory for use of the confocal microscope and Eric Granlund from the UC Berkeley College of Chemistry machine shop for his help in designing and constructing the flow cells. We are grateful to George O'Toole and Raphael Valdivia for their contributions of pSMC21 and pGFPmut3, respectively.
Stacie E. Cowan was supported by a National Institute of Health Training Grant. Artem Khlebnikov was supported by the Swiss National Scientific Foundation (Bourse de Relève du FNRS). This work was supported by the National Science Foundation (BES-9814088 and EEC-9731725) and the Office of Naval Research (N00014-99-1-0182).
REFERENCES
- 1.Andersen J B, Sternberg C, Kongsbak-Poulsen L, Petersen-Bjorn S, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloemberg G V, O'Toole G A, Lugtenberg B J J, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euslichen G, Ward W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 5.Crameri A, Whitehorn E A, Tate E, Stemmer W P C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 6.Eberl L, Schulze R, Ammendola A, Geisenberger O, Erhart R, Sternberg C, Molin S, Amann R. Use of green fluorescent protein as a marker for ecological studies of activated sludge communities. FEMS Microbiol Lett. 1997;149:77–83. [Google Scholar]
- 7.Gilbert E S, Crowley D E. Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Appl Environ Microbiol. 1997;63:1933–1938. doi: 10.1128/aem.63.5.1933-1938.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg S, Doyle R J, Rosenberg M. Mechanism of enhancement of microbial cell hydrophobicity by cationic polymers. J Bacteriol. 1990;172:5650–5654. doi: 10.1128/jb.172.10.5650-5654.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssen D B, Scheper A, Witholt B. Biodegradation of 2-chloroethanol and 1,2-dichloroethane by pure bacterial cultures. Prog Ind Microbiol. 1984;20:169–178. [Google Scholar]
- 10.Lawrence J R, Korber D R, Wolfaardt G M. Heterogeneity of natural biofilm communities. Cells Mater. 1996;6:175–191. [Google Scholar]
- 11.Lewis P J, Marston A L. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene. 1999;227:101–109. doi: 10.1016/s0378-1119(98)00580-0. [DOI] [PubMed] [Google Scholar]
- 12.Ornston L N, editor. Microbial biofilms. Vol. 49. Palo Alto, Calif: Annual Reviews Inc.; 1995. [Google Scholar]
- 13.Shen H, Wang Y-T. Simultaneous chromium reduction and phenol degradation in a coculture of Escherichia coli ATCC 33456 and Pseudomonas putida DMP-1. Appl Environ Microbiol. 1995;61:2754–2758. doi: 10.1128/aem.61.7.2754-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skillman L C, Sutherland I W, Jones M V, Goulsbra A. Green fluorescent protein as a novel species-specific marker in enteric dual-species biofilms. Microbiology. 1998;144:2095–2101. doi: 10.1099/00221287-144-8-2095. [DOI] [PubMed] [Google Scholar]