Abstract
Randomly amplified polymorphic DNA analysis using primer 239 (5′ CTGAAGCGGA 3′) was performed to characterize Leuconostoc sp. strains. All the strains of Leuconostoc mesenteroides subsp. mesenteroides (with the exception of two strains), two strains formerly identified as L. gelidum, and one strain of Leuconostoc showed a common band at about 1.1 kb. This DNA fragment was cloned and sequenced in order to verify its suitability for identifying L. mesenteroides subsp. mesenteroides strains.
Leuconostoc mesenteroides subsp. mesenteroides is used industrially to produce dextrans and is the most common species isolated from fermented feed and food products (4, 9). In addition, its recognized antagonistic or synergistic properties in mixed microflora populations have led to studies on its importance in some starter cultures (8). Therefore, unequivocal criteria and reliable methods for identification and characterization of leuconostoc isolates are important requirements in industrial microbiology as well as in other fields of applied microbiology.
Randomly amplified polymorphic DNA (RAPD) analysis has been shown to be suitable for generating strain- and species-specific oligonucleotide probes and/or primers with known sequences (3, 6, 10). Therefore, the developed RAPD-PCR method was applied to find a significant RAPD fragment within L. mesenteroides subsp. mesenteroides strains. Moreover, a set of primers to use in PCR amplification was designed in order to evaluate its suitability as a specific probe for identifying L. mesenteroides subsp. mesenteroides.
A total of 66 leuconostoc strains were used in this study. The 13 reference strains from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM; Braunschweig, Germany) are listed in the legend of Fig. 1. Fifty-three leuconostocs, identified in previous works (2, 11), were as follows: 21 strains of L. mesenteroides subsp. mesenteroides, 5 strains of L. mesenteroides subsp. dextranicum, 3 strains of L. pseudomesenteroides, 3 strains of L. gelidum, 5 strains of Weissella paramesenteroides, 2 strains of Oenococcus oeni, 1 strain of L. citreum, 1 strain of L. argentinum, 1 strain of L. carnosum, and 11 Leuconostoc spp. These strains were isolated from such different sources as pizza dough, field grass, natural whey cultures, mozzarella cheese, and sausages.
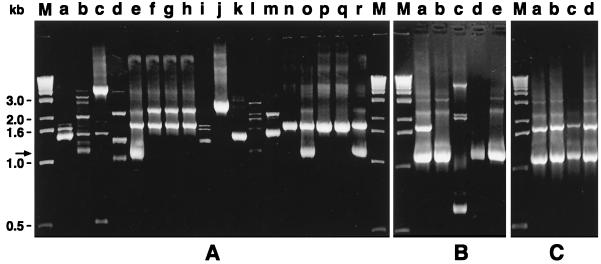
FIG. 1.
RAPD patterns of different leuconostoc species generated with primer 239 (5′ CTGAAGCGGA 3′). The bold arrows show the location of the 1.1-kb band. (A) Lane a, Leuconostoc sp. strain DSM 20186; lane b, L. citreum DSM 20188; lane c, L. lactis DSM 20202T; lane d, W. paramesenteroides DSM 20288T; lane e, L. mesenteroides subsp. mesenteroides DSM 20343T; lane f, L. mesenteroides subsp. cremoris DSM 20346T; lane g, L. mesenteroides subsp. cremoris DSM 20200; lane h, L. mesenteroides subsp. dextranicum DSM 20484T; lane i, L. fallax DSM 20189T; lane j, L. pseudomesenteroides DSM 20193T; lane k, L. carnosum DSM 5576T; lane l, L. argentinum DSM 8581T; lane m, L. gelidum DSM 5578T; lane n, L. mesenteroides subsp. mesenteroides A27; lane o, L. mesenteroides subsp. mesenteroides A52; lane p, L. gelidum A57; lane q, L. gelidum A72; lane r, L. mesenteroides subsp. mesenteroides A65. (B) Lanes a to d, L. mesenteroides subsp. mesenteroides strains DSM 20343T (lane a), 501 (lane b), 574 (lane c), 526 (lane d); lane e, Leuconostoc sp. strain 511. (C) Lanes a to d, L. mesenteroides subsp. mesenteroides strains DSM 20343T (lane a), 11X (lane b), 23X (lane c), 27X (lane d). M, 1-kb DNA ladder (GIBCO BRL) used as a molecular weight marker. The amplified products (25 μl) were resolved by electrophoresis on a 1.5% (wt/vol) agarose–Tris-borate-EDTA gel at 5 V cm−1 for 4 h.
Preparation of crude cell extracts and RAPD-PCR assays under standard conditions were carried out as previously described (5). In brief, 28 different 10-mer primers were tested initially by screening of DNA from the 13 leuconostoc strains belonging to the DSM in RAPD-PCR assays. The suitability of each primer was scored on the basis of intensity and distribution of bands. Experiments indicated that primer 239 (5′ CTGAAGCGGA 3′; Primm, Milan, Italy) allowed reproducible patterns with bands from 0.5 to 3 kb to be generated (results not shown). Its ability to generate the same pattern was evaluated by amplifying DNA from the DSM leuconostoc strains three times with the same RAPD conditions. Therefore, this primer was used to differentiate all of the 66 leuconostoc strains.
Representative RAPD profiles are shown in Fig. 1. Primer 239 produced 11 different RAPD patterns among the 13 strains of the DSM, since DSM 20346T and DSM 20200 of L. mesenteroides subsp. cremoris (Fig. 1A, lanes f and g, respectively) and L. mesenteroides subsp. dextranicum DSM 20484T (Fig. 1A, lane h) exhibited the same profile. The RAPD pattern of L. mesenteroides subsp. mesenteroides DSM 20343T (Fig. 1A, lane e) revealed two major bands of 1.1 and 1.7 kb, which also occurred in L. mesenteroides subsp. mesenteroides strains A52, A65, 11X, 23X, and 27X. All the other L. mesenteroides subsp. mesenteroides strains tested (with the exception of strains 574 and A27) and Leuconostoc sp. strain 511 exhibited RAPD profiles with only a DNA fragment amplified at about 1.1 kb. This significant band was absent in all other strains of the genus Leuconostoc.
In a previous work (11) we demonstrated that in spite of its biochemical pattern, the strain 574 from natural whey cultures exhibited a sodium dodecyl sulfate-polyacrylamide gel electrophoresis pattern as well as a ribopattern different from those of L. mesenteroides subsp. mesenteroides DSM 20343T. It can therefore represent a misidentification.
The L. mesenteroides subsp. mesenteroides A27 and L. gelidum A72 and A57 strains displayed the same RAPD pattern with only the fragment of 1.7 kb. This result could be attributed to the lack of the homology sequence with the 239 primer on the genome of these strains located after 1.1 kb from the first site of annealing. A misidentification for the A72 and A57 strains could be supposed because their RAPD patterns were strongly different from that of the type strain of L. gelidum. These two strains were dextran producers and galactose negative, and they could be strains of L. mesenteroides subsp. mesenteroides lacking galactose fermentation.
The 1.1-kb DNA fragment from DNA of L. mesenteroides subsp. mesenteroides DSM20343T that was amplified by primer 239 was cloned into the pCR-Script SK(+) cloning vector (Stratagene, La Jolla, Calif.), and the DNA sequence of the whole fragment was determined by the dideoxy chain termination method (7) by using the DNA sequencing kit (Perkin-Elmer Cetus, Emeryville, Calif.) according to the manufacturer's instructions. Research for DNA homologies performed with the GenBank and EMBL databases revealed less than 20% homology to any known sequences.
Two oligonucleotides (LMMf: 5′ CCGTTACCCCTAAATTTTGAC; LMMr: 5′ GACCAAATACAATAGGTTGCG) were chosen inside (positions 139 to 160 and 944 to 965, respectively) the 1,151-bp sequenced fragment to increase the stringency in the specific amplification of DNA from L. mesenteroides subsp. mesenteroides strains. PCR was performed at 94°C for 3 min, followed by 25 cycles of 1 min at 94°C, 45 s at 67°C, and 1 min at 72°C and a 5-min extension period at 72°C.
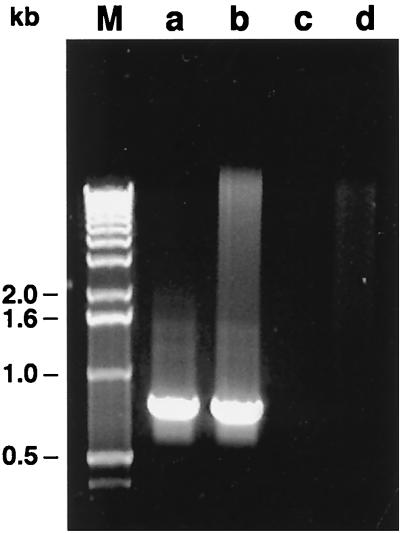
Representative results of all the strains tested for PCR specificity with the selected primers are shown in Fig. 2. Twenty-one of 22 L. mesenteroides subsp. mesenteroides strains, strains A72 and A57 (formerly reported as L. gelidum), and Leuconostoc sp. strain 511 exhibited a PCR fragment located at 825 bp. No DNA amplification was observed in strain 574, in 42 strains belonging to other leuconostoc species, or in two reference strains of the genus Lactococcus, 2 strains of Enterococcus, 5 strains of Lactobacillus, 5 strains of Streptococcus, or 5 strains of Pediococcus assayed as a negative control.
FIG. 2.
Ethidium bromide-stained 1.5% agarose gel of PCR-specific products obtained with primers LMMf and LMMr targeting the 825-bp fragment. Lane a, L. mesenteroides subsp. mesenteroides DSM 20343T; lane b, Leuconostoc sp. strain 511; lane c, L. gelidum DSM 5578T; lane d, L. mesenteroides subsp. dextranicum DSM 20484T; M, 1-kb DNA ladder (GIBCO BRL) used as a molecular weight marker.
The 825-bp PCR fragment from L. mesenteroides subsp. mesenteroides DSM 20343T was used as a probe in a DNA dot blot hybridization performed at 67°C. The hybridization signal obtained by the labelled 825-bp DNA probe with the total DNA from L. mesenteroides subsp. mesenteroides DSM 20343T was defined as the 100% intensity signal, while the intensities of the signals obtained with total DNA from the other strains tested were calculated by the program Phoretix. Calibration of values was through linear regression (1). A high hybridization response (98 to 100% homology) was obtained with every L. mesenteroides subsp. mesenteroides strain (with the exception again of strain 574) plus L. gelidum A72 and A57 and Leuconostoc sp. strain 511. A weaker signal (13 to 23%) was observed with DNA from L. mesenteroides subsp. dextranicum DSM 20484T and 2X. When the temperature of annealing was reduced from 67 to 65°C in the specific PCR amplification, a weak band appeared for the two above-mentioned strains (results not shown). This probably reflects the close phylogenetic relationship of L. mesenteroides subsp. mesenteroides and L. mesenteroides subsp. dextranicum. No hybridization signal was obtained with DNA from other Leuconostoc species or from members of the other genera that were considered.
PCR primers performed in this study were able to identify as L. mesenteroides subsp. mesenteroides the strain 511, which in a previous work (11) was not discriminated at species level.
Sources and geographical origins of L. mesenteroides subsp. mesenteroides strains used in this study were strongly different (e.g., DSM 20343T from fermenting olives, 501 from field grass, A65 from pizza dough). Thus, the 1.1-kb RAPD fragment cannot be considered a strain-specific marker.
In conclusion, the set of primers from RAPD-PCR that was developed in this study allowed identification of strains belonging to L. mesenteroides subsp. mesenteroides, and it could be used for monitoring this microorganism in mixed microflora populations that are implicated in fermented feed and food products.
Nucleotide sequence accession number.
The GenBank accession number for the sequence data reported here is AF124525.
Acknowledgments
This work was supported by a grant from MURST, Rome, Italy.
We thank Anna Calvanico and Elena Nebuloso for scientific and technical collaboration and Mark Walters for final proofreading of the manuscript.
REFERENCES
- 1.Boesch C, Trcek J, Sievers M, Teuber M. Acetobacter intermedius, sp. nov. Syst Appl Microbiol. 1998;21:220–229. doi: 10.1016/S0723-2020(98)80026-X. [DOI] [PubMed] [Google Scholar]
- 2.Coppola S, Pepe O, Masi P, Sepe M. Characterization of leavened doughs for pizza in Naples. Adv Food Sci. 1996;59:160–162. [Google Scholar]
- 3.Erlandson K, Batt C A. Strain-specific differentiation of lactococci in mixed starter culture populations using randomly amplified polymorphic DNA-derived probes. Appl Environ Microbiol. 1997;63:2702–2707. doi: 10.1128/aem.63.7.2702-2707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holzapfel W H, Schillinger V. The genus Leuconostoc. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. Vol. 2. New York, N.Y: Springer-Verlag Inc.; 1992. pp. 1508–1534. [Google Scholar]
- 5.Moschetti G, Blaiotta G, Aponte M, Catzeddu P, Villani F, Deiana P, Coppola S. Random amplified polymorphic DNA (RAPD) and amplified ribosomal DNA spacer polymorphism: powerful methods to differentiate Streptococcus thermophilus strains. J Appl Microbiol. 1998;85:25–36. doi: 10.1046/j.1365-2672.1998.00461.x. [DOI] [PubMed] [Google Scholar]
- 6.Quere F, Deschamps A, Urdaci M C. DNA probe and PCR-specific reaction for Lactobacillus plantarum. J Appl Microbiol. 1997;82:783–790. doi: 10.1046/j.1365-2672.1997.00157.x. [DOI] [PubMed] [Google Scholar]
- 7.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiles M E. Bacteriocins produced by Leuconostoc species. J Dairy Sci. 1994;77:2718–2724. doi: 10.3168/jds.S0022-0302(94)77214-3. [DOI] [PubMed] [Google Scholar]
- 9.Stiles M E, Holzapfel W H. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1994;36:1–29. doi: 10.1016/s0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 10.Tilsala-Timisjärvi A, Alatossava T. Strain-specific identification of probiotic Lactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl Environ Microbiol. 1998;64:4816–4819. doi: 10.1128/aem.64.12.4816-4819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villani F, Moschetti G, Blaiotta G, Coppola S. Characterization of strains of Leuconostoc mesenteroides by analysis of soluble whole-cell protein pattern, DNA fingerprinting and restriction of ribosomal DNA. J Appl Microbiol. 1997;82:578–588. doi: 10.1111/j.1365-2672.1997.tb03588.x. [DOI] [PubMed] [Google Scholar]