Abstract
Ampelomyces and Phoma species are frequently confused with each other. Isolates previously attributed to the genus Ampelomyces were shown to be Phoma isolates through studies of their morphology and life cycle and ribosomal DNA internal transcribed spacer region 1 sequence analysis. Phoma glomerata can colonize and suppress development of powdery mildew on oak and may have utility as a mycoparasitic agent.
Powdery mildews are widespread plant pathogens that are conspicuous by their white mycelia and powder-like conidia (20). The fungus Ampelomyces quisqualis Ces. is the only fungus that has been demonstrated to be generally effective as a biocontrol agent of powdery mildew (4, 9, 16). Many morphologically similar species may be confused with A. quisqualis (10). To evaluate this possibility, we examined and identified Ampelomyces-like fungi isolated from powdery mildew and compared these cultures with isolates identified as Ampelomyces in culture collections.
Isolation and growth.
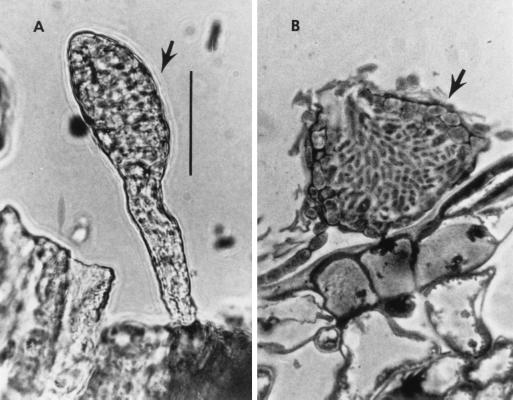
Leaves of sycamore trees (Platanus occidentalis L.) bearing infections of powdery mildew (Microsphaera penicillata (Wallr.:Fr.) Lèv.) were located in South River, New Jersey, in July 1998. Microscopic examination of the leaves revealed two types of pycnidia: stipitate pycnidia, typical of A. quisqualis, and sessile pycnidia, typical of the genus Phoma (Fig. 1) (17). Both types of pycnidia were removed from leaves with fine needles and placed on potato dextrose agar (Difco, Inc., Detroit, Mich.) containing the antibiotics gentamicin (40 mg/liter), streptomycin (40 mg/liter), and penicillin (20 mg/liter) (PDA + 3). Two different fungi were consistently recovered. The stipitate pycnidia developed into slow-growing colonies whose characteristics corresponded to those expected for A. quisqualis (5, 11). The sessile pycnidia developed into rapidly growing colonies whose characteristics corresponded to those of Phoma glomerata (Cda) Wollenw. (2, 19).
FIG. 1.
(A) Stipitate pycnidium of A. quisqualis (arrow). (B) Section of sessile pycnidium of P. glomerata on oak leaf (arrow). Scale bar = 20 μm.
Agar plugs (6 mm in diameter) of mycelia cut from the margins of rapidly growing colonies of both the South River Ampelomyces and South River P. glomerata isolates were transferred to five plates each of PDA+3 and incubated at room temperature (21 to 22°C) for 3 weeks to measure growth rates. We measured an average growth of 8 ± 1 mm/day for the P. glomerata isolates and an average growth of 0.8 ± 0.1 mm/day for the Ampelomyces isolates. With age, cultures of P. glomerata produced alternarioid dictyochlamydospores measuring 41 ± 7.5 × 12 ± 1.4 μm.
Inoculation experiments.
Koch's postulates (1, 3) were used to establish the pathogenicity of P. glomerata to powdery mildew. A suspension of P. glomerata conidia from cultures grown on PDA+3 was made in sterile water (∼8 × 106 conidia/ml). The conidial suspension was used to inoculate epiphyllous mycelia of the powdery mildew Phyllactinia guttata (Wallr.:Fr.) Lèv. on intact (left on the tree) leaves of oak (Quercus coccinea Münch.) by moistening an approximately 15-mm2 region on the upper surface of the leaves. Controls were repeats of this process with sterile water. Ten replicates of both the treatment and control were made, and the sites of inoculation were marked by placing white tape on the reverse of the leaves at the inoculation sites. The leaves were monitored for 30 days. During this time, control leaves developed powdery mildew cleistothecia while all leaves treated with P. glomerata conidia developed abundant pycnidia in and around the inoculation sites but did not produce powdery mildew cleistothecia. None of the control leaves showed development of P. glomerata pycnidia, and cleistothecia developed normally. To fulfill Koch's postulates, pycnidia were removed from treated leaves with fine needles and plated on PDA+3 medium to recover P. glomerata. Colonies that developed were confirmed to be P. glomerata by observation of dictyochlamydospores, pycnidia, and subsequent sequence analysis.
Phylogenetic analysis.
The nuclear ribosomal DNA internal transcribed spacer region 1 (ITS1) from P. glomerata, several Ampelomyces spp., and several Phoma spp. were sequenced. The South River P. glomerata and A. quisqualis, as well as American Type Culture Collection (ATCC) cultures of Ampelomyces heraclei (Dejeva) Rudakov (ATCC 36804) and A. quisqualis (ATCC 200245), were grown on PDA+3. DNA extraction, amplification, and sequencing were accomplished as described previously (14).
Several additional ITS1 sequences identified as Ampelomyces spp., including Ampelomyces humuli (Fautr.) Rudakov, Ampelomyces quercinus (Syd.) Rudakov, Phaeosphaeria avenaria (Weber) Eriksson, and Leptosphaeria microscopica P. Karst. were obtained from GenBank (Fig. 2).
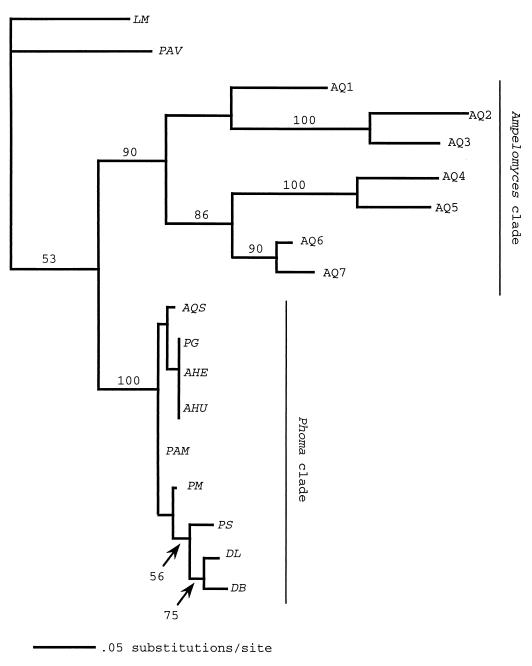
FIG. 2.
Maximum likelihood phylogram based on ITS region sequences between the 18S and 5.8S ribosomal DNA (ITS1). Bootstrap confidence levels (percent) of branches are shown. The scale bar is based on a total tree length of 204. Taxa included in the analysis, GenBank numbers (if known), and their sources are as follows: LM, L. microscopica (LMU04234); PAV, P. avenaria (PAU77357); AQ1, A. quisqualis (AQU82451, DSM [Deutsche Sammlung von Mikrooganismen und Zellkulturen GmbH, Braunschweig, Germany] 2223); AQ2, A. quisqualis (AF126818, South River); AQ3, A. quisqualis (AF126817, ATCC 200245); AQ4, A. quisqualis (AF035782); AQ5, A. quisqualis (AQU82449, CBS [Centraalbureau voor Schimmelcultures, Baarn, The Netherlands] 130.79); AQ6, A. quisqualis (AF035783, Ecogen AQ10); AQ7, A. quisqualis (AF035781, CBS 131.31); AQS, A. quercinus (AF035778, ATCC 36786); PG, P. glomerata (AF126816, South River); AHE, A. heraclei (AF126819, ATCC 36804); AHU, A. humuli (AF035779, ATCC 38616); PAM, P. americana Morgan-Jones et White (AF046016); PM, P. macrostoma Mont. (AF046020); PS, P. sorghina (Sacc.) Boerema (AF046022); DL, D. lycopersici Klebahn (AF046015); and DB, D. bryoniae (Auersw.) Rehm (AF046014).
The SeqLab interface for the Wisconsin Package Version 9.1 (Genetics Computer Group, Madison, Wis.) was used to generate alignments and make manual adjustments. PAUP version 4.0b2 for Macintosh (17) was used for phylogenetic analysis. Heuristic searches were performed by using maximum parsimony (14). Bootstrapping, using the same criteria with 400 replicates, was performed to determine the confidence levels of the inferred phylogenies. Trees found by maximum parsimony were subjected to heuristic searches by using maximum likelihood criteria by the Hasegawa-Kishino-Yano model (6) to find the most likely tree (See Treebase [http://herbaria.harvard.edu/treebase] submission no. SN145 for alignment and tree construction details).
Maximum parsimony analysis resulted in ten trees. Maximum likelihood analysis of these trees resulted in one tree (−ln L = 1200, tree length = 204, consistency index = 0.78, homoplasy index = 0.22, retention index = 0.79). The South River P. glomerata is identical to A. heraclei and A. humuli, and they group together with A. quercinus in the Didymella/Phoma clade (Fig. 2). Our South River A. quisqualis isolate grouped in the Ampelomyces clade with Ecogen's A. quisqualis AQ10. There was strong bootstrap support for the Ampelomyces (90%) and Phoma (100%) clades.
Distinguishing Phoma from Ampelomyces.
Stipitate pycnidia developing into slow-growing colonies characterize A. quisqualis (5, 11). Sessile pycnidia developing into rapidly growing colonies characterize P. glomerata (2, 19). The cultures identified as A. heraclei, A. humuli, and A. quercinus are typical representatives of their species.
The process of pycnidium formation in association with the powdery mildew is also different between the two genera. Ampelomyces spp. infect conidiogenous cells of powdery mildew, internally colonizing and forming pycnidia within the conidiophore; the pycnidia appear stipitate (Fig. 1). Phoma sp. does not appear to internally infect conidiogenous cells, and pycnidia are formed directly on the leaf surface; they are sessile (Fig. 1). While many species of Phoma are plant pathogens (17), P. glomerata is not. However, Phoma can grow saprophytically in tissues of plants and is known to be a secondary invader of diseased tissues, perhaps feeding on fungal saprophytes or pathogens of diseased tissues (12, 17, 19). P. glomerata has been isolated from species of the powdery mildew genus Microsphaera in the United States (this study) and Russia (as A. quercinus) and from species of the genus Sphaerotheca (as A. humuli) in Russia (15). It also has been isolated from the downy mildew of grapes (Plasmopara viticola (Berk. et Curt.) Berl. et De Toni) in Russia (as A. heraclei) (15). It is apparent that P. glomerata has a widespread distribution.
Potential new mycoparasitic agent.
Currently a single strain of fungus, A. quisqualis AQ10 Biofungicide, is in commercial use for biocontrol of powdery mildew on grapes and other crops (7). This strain grouped within the divergent Ampelomyces clade and appears to correctly represent a species of that genus. A few reports have identified Phoma species that are antagonistic to fungal plant pathogens. A Phoma sp. (P66A) significantly reduced conidial germination of an apple scab (Venturia inaequalis (Cooke) Wint.) (13), and Phoma etheridgei Hutch. & Hirat. produced antifungal compounds inhibitory to the tree pathogen Phellinus tremulae (Bond) Bond et Borisov (8).
Our results suggest that P. glomerata is frequently misidentified as A. quisqualis or other species of Ampelomyces. Additionally, P. glomerata often may inhabit powdery mildew infections and may be an important component of a hyperparasitic guild of fungi that naturally infect powdery mildews. Further study is warranted to evaluate the effectiveness of P. glomerata in the hyperparasitic control of fungal plant pathogens.
Nucleotide sequence accession numbers.
The following sequences were deposited in GenBank: A. quisqualis ATCC 200245 and South River, P. glomerata South River, A. heraclei ATCC 36804. Their accession numbers are listed in the legend for Fig. 2.
REFERENCES
- 1.Agrios G N. Plant Pathology. 3rd ed. San Diego, Calif: Academic Press, Inc.; 1988. [Google Scholar]
- 2.Boerema G H, Dorenbosch M M J, van Kesteren H A. Remarks on species of Phoma referred to Peyronellaea. Persoonia. 1965;4:47–68. [Google Scholar]
- 3.Brock T D. Robert Koch, a life in medicine and bacteriology. Madison, Wis: Science Tech Publishers; 1988. [DOI] [PubMed] [Google Scholar]
- 4.Falk S P, Gadoury D M, Cortesi P, Pearson R C, Seem R C. Partial control of grape powdery mildew by the mycoparasite Ampelomyces quisqualis. Plant Dis. 1995;79:483–490. [Google Scholar]
- 5.Galper S, Sztejnberg A, Lisker N. Scanning electron microscopy of the ontogeny of Ampelomyces quisqualis pycnidia. Can J Microbiol. 1985;31:961–964. [Google Scholar]
- 6.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 7.Hofstein R, Fridlender B. Brighton Crop Protection Conference—pests and diseases. Vol. 3. Farnham, United Kingdom: British Crop Protection Council; 1994. Development of production, formulation and delivery systems; pp. 1273–1280. [Google Scholar]
- 8.Hutchinson L J, Chakravarty P, Kawchuck L M, Hirasuka Y. Phoma etheridgei sp. nov. from black galls and cankers of trembling aspen (Populus tremuloides) and its potential role as a bioprotectant against the aspen decay pathogen Phellinus tremulae. Can J Bot. 1994;72:1424–1431. [Google Scholar]
- 9.Jarvis W R, Slingsby K. The control of powdery mildew of greenhouse cucumber by water spray and Ampelomyces quisqualis. Plant Dis Rep. 1977;61:728–730. [Google Scholar]
- 10.Kiss L. Genetic diversity in Ampelomyces isolates, hyperparasites of powdery mildew fungi, inferred from RFLP analysis of the rDNA ITS region. Mycol Res. 1997;101:1073–1080. [Google Scholar]
- 11.Kiss L, Nakasone K K. Ribosomal DNA internal transcribed spacer sequences do not support the species status of Ampelomyces quisqualis, a hyperparasite of powdery mildew fungi. Curr Genet. 1998;33:362–367. doi: 10.1007/s002940050348. [DOI] [PubMed] [Google Scholar]
- 12.Morgan-Jones G. Phoma glomerata. In CMI descriptions of pathogenic fungi and bacteria no. 134. Kew, United Kingdom: Commonwealth Mycological Institute; 1967. [Google Scholar]
- 13.Ouimet A, Carisse O, Neumann P. Environmental and nutritional factors affecting the in vitro inhibition of the vegetative growth of Venturia inaequalis by five antagonistic fungi. Can J Bot. 1997;75:632–639. [Google Scholar]
- 14.Reddy P V, Patel R, White J F., Jr Phylogenetic and developmental evidence supporting reclassification of cruciferous pathogens Phoma lingham and P. wasabiae in genus Plenodomus. Can J Bot. 1998;76:1916–1922. [Google Scholar]
- 15.Rudakov O L. Fungi of the genus Ampelomyces Ces. Ex. Schlect. Mikologiya i Fitopatologiya. 1979;13:104–110. [Google Scholar]
- 16.Sundheim L, Tronsmo A. Hyperparasites in biological control. In: Mukerji K G, Garg K L, editors. Biocontrol of plant diseases. Boca Raton, Fla: CRC Press; 1988. pp. 53–70. [Google Scholar]
- 17.Sutton B C. The coelomycetes. Kew, United Kingdom: Commonwealth Mycological Institute; 1980. [Google Scholar]
- 18.Swofford D L. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 19.White J F, Jr, Morgan-Jones G. Studies in the genus Phoma. VII. Concerning Phoma glomerata. Mycotaxon. 1987;28:437–445. [Google Scholar]
- 20.Yarwood C E. History and taxonomy of powdery mildews. In: Spencer D M, editor. The powdery mildews. London, United Kingdom: Academic Press, Inc.; 1978. [Google Scholar]