Abstract
Scutellarin related drugs have superior therapeutic effects on cerebrovascular and cardiovascular diseases. Here, an optimal biosynthetic pathway for scutellarin was constructed in Yarrowia lipolytica platform due to its excellent metabolic potential. By integrating multi-copies of core genes from different species, the production of scutellarin was increased from 15.11 mg/L to 94.79 mg/L and the ratio of scutellarin to the main by-product was improved about 110-fold in flask condition. Finally, the production of scutellarin was improved 23-fold and reached to 346 mg/L in fed-batch bioreactor, which was the highest reported titer for de novo production of scutellarin in microbes. Our results represent a solid basis for further production of natural products on unconventional yeasts and have a potential of industrial implementation.
Keywords: Scutellarin, Yarrowia lipolytica, Metabolic engineering, Combinatorial gene overexpression
1. Introduction
Scutellarin is the main biosynthetic component of breviscapine extracted from Erigeron breviscapus, which has anti-inflammatory, antioxidant [1], antiplatelet aggregation [2], microcirculation improvement [3], neuroprotection [4], and antitumor effects [5]. Scutellarin can be used for the treatment of diabetic complications, cardio-cerebral vascular diseases, stroke, nephropathy, and nonalcoholic fatty liver disease (NAFLD) [[6], [7], [8], [9], [10]]. Unfortunately, many of these illnesses are the major cause of permanent disability and death in the world [4,[11], [12], [13]]. Market analysis shows that the annual demand for scutellarin in China is approximately 100 tons, with over 10 million patients using scutellarin and related drugs each year [14]. However, extraction from E. breviscapus remains the main source of scutellarin, greatly depleting the wild resources of this species [15]. Chemical synthesis is overshadowed by the use of expensive starting materials and cumbersome synthesis procedures [16]. Therefore, using synthetic biology to construct cellular factory for scutellarin synthesis is an effective way to solve the shortage of E. breviscapus resources.
Unlike the traditional hosts Saccharomyces cerevisiae and Escherichia coli, Y. lipolytica stands out as a non-conventional oleaginous yeast due to its excellent metabolic potential [17]. Y. lipolytica is generally regarded as safe (GRAS) by the Food and Drug Administration (FDA). As a very specialized aerobic yeast, it is lack of Crabtree effect, which will produce ethanol under the condition of high glucose or respiratory restriction, and is rich in key respiratory intermediates (acetyl-CoA, NADPH, ATP), which could be used as the precursors for natural compounds [[18], [19], [20], [21], [22], [23], [24], [25], [26]]. Furthermore, genetic manipulation tools have been developed in Y. lipolytica through engineering DNA double-strand break (DSB) repair protein complex Ku70/Ku80 [27,28]. So far, several efforts have been made to utilize Y. lipolytica as platform to produce natural products (Supplementary Table S1) and the highest yield of resveratrol has reached to 12 g/L [[29], [30], [31], [32], [33], [34], [35]]. More importantly, naringenin and arbutin have been successfully produced, indicating sufficient precursor and abundant glycosylated donor of scutellarin in Y. lipolytica [36,37]. Inspired by these results, we considered Y. lipolytica as a potential factory for scutellarin production.
Although the biosynthesis of scutellarin was successfully realized in S. cerevisiae, the scutellarin fermentation products was not the major products (37.5%) accompanied by a large number of apigenin-7-O-glucuronide [14]. Here, we screened key enzymes from different species to construct cell factory for de novo biosynthesis of scutellarin in Y. lipolytica and improved the yield of scutellarin to 346 mg/L through increasing the copy number of key enzymes and optimizing the fermentative condition in 1.3 L controlled fed-batch bioreactor.
2. Materials and methods
2.1. Genes and strains
All metabolically modified Y. lipolytica strains were from the strain W29 carrying Cas9 on the KU70 locus, which was kindly provided by Dr. Zongjie Dai (Tianjin Institute of Industrial Biotechnology). Genes encoding AtPAL2, EbPAL, AtC4H, EbC4H, EbCHS, HaCHS, At4CL1, Eb4CL, EbFSII, EbF7GAT, EbF6H, CcF6H, CcarF6H, SbaiCYP82D1, SbaiCYP82D4, SbarCYP82D1, SbarCYP82D4, SbarCYP82D5, ATR2, ScARO4K229L, ScARO7G141S, SeACSP641L and EbUDPGDH were all codon-optimized toward Y. lipolytica by GenScript (Nanjing, China). Integration sites, YlARO4K221L, YlARO7G139S, PEX10, TEFin and GPD promoters, tlip2 and PEX20 terminators were amplified from Y. lipolytica genomic DNA. All primers and genes were listed in (Table S2, S3 and S6).
2.2. Cultivation and medium
E. coli strains were cultured using 2 YT at 37 °C with shaking at 230 rpm and Y. lipolytica strains were incubated using YNB or YPD at 30 °C and 230 rpm. Additionally, YPD solid medium with hygromycin B (250 mg/L) and nourseothricin (400 mg/L) were used to select transformed Y. lipolytica strains. After 3 days of cultivation, individual colonies of recombinant strains were picked from plates, inoculated into 3 mL YNB in 24-well plates with gas permeable sealing membrane (Sigma-Aldrich) and grown for 48 h. The cultures were then transferred to a 250 mL shake flask containing 30 mL YNB medium and cultivated for 4 days. The yeast solution was lined onto antibiotic-free YPD plates for 1 day to remove the gRNA plasmid in preparation for the next round of integration. The composition of the mineral medium (MM), YNB, YPD were shown in the supplementary material. All flask fermentation results represented the average standard deviation of three independent experiments.
2.3. Construction of plasmids and strains
All derived strains and plasmids constructed were listed in (Tables S4–S5). Amplification of target gene fragments using Phanta Max Super-Fidelity DNA polymerase (Vazyme Biotech, Nanjing, China) by polymerase chain reaction (PCR). The Zymo Fragment Recovery Kit (Zymo Research) was used for purification of PCR product, construction of recombinant plasmids by Gibson assembly [38]. The EasyPure® Plasmid MiniPrep Kit (Trans Biotech, Beijing, China) was used for plasmid isolation. All processes were carried out following the manufacturer's instructions. To construct integrative strains, a single gRNA plasmid and a linearized homologous donor plasmid were introduced by the CRISPR/Cas9 system. The integrative plasmid was constructed using pMD-19 T as the backbone, with different integration sites as described in Refs. [39,40], and single gRNA plasmids were graciously provided by Dr. Zongjie Dai (Tianjin Institute of Industrial Biotechnology). Using the lithium acetate method to construct engineered Y. lipolytica strains and specific protocol as described previously [39]. Recombinant was verified by PCR amplification. The detailed construction process of the plasmids could be found in the Supplementary Methods.
2.4. HPLC and LC-MS analysis
500 μL of fermented product was mixed with 500 μL of methanol, sonicated for 30 min, and centrifuged at 12,000 rpm for 20 min. HPLC and LC-MS assays were as before [14].
2.5. Fed-batch fermentation
The yeast cells were scribed on YPD plates and grown for two days. A single colony from the plate was transferred to 4 mL of mineral medium for 48 h and then transferred to 40 mL of mineral medium for 24 h. Fed-batch cultivations were performed in a 1.3 L parallel bioreactor (T&J-Mini Box, Shanghai T&J Bio-engineering Co., L TD) and an initial volume was 400 mL. The initial medium (MM) was prepared in a previous study [32]. Temperature was maintained at 30 °C during fermentation. PH maintained at 5.0 by automatic addition of 10% ammonia. The dissolved oxygen (DO) was set at 40% by automatically controlling agitation speed (200–1200 rpm), and the initial aeration was set at 0.5 L/min. Sterile antifoam was added automatically when foaming. The glucose in the feed medium was 15-fold and other components were 5-fold of the initial medium. After all the glucose was exhausted, the feed solution was added and the initial replenishment rate was 3 mL/h and the glucose concentration was controlled to be below 5 g/L. Fermentation was stopped when the glucose concentration in the medium was not depleted and started to rise. Samples of 5 mL culture medium were collected every 24 h to analyze cell dry weight, residual glucose content and scutellarin accumulation. Error bars represent the mean of the three data.
2.6. Detection of the biomass and sugar
500 μL of fermentation broth was washed twice to remove all remaining material in the medium prior to lyophilization, and cell dry weight (DCW) was measured by freeze-drying the cells by weighing. The residual glucose concentration was measured with an SBA-40D biosensor (Shandong Academy of Sciences, China). Erythritol was quantified by HPLC equipped with a refractive index detector and an Aminex HPX-87H column using 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 mL/min and a temperature of 40 °C.
3. Results
3.1. Modularization of scutellarin biosynthetic pathway in Y. lipolytica
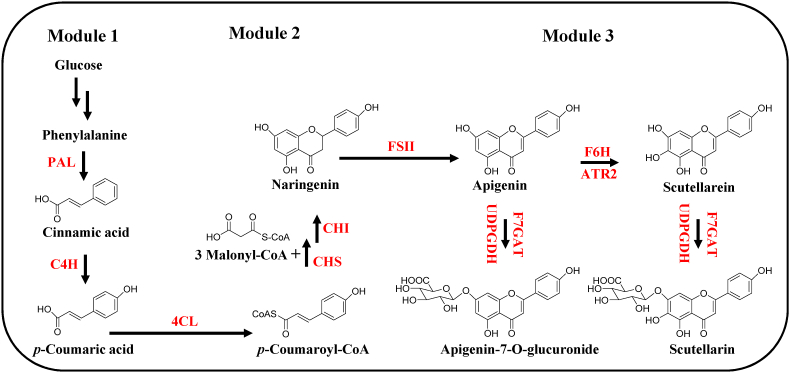
The biosynthetic pathway of scutellarin from l-phenylalanine to scutellarin has been well characterized (Fig. 1), which comprised eight consecutive steps, catalyzed by PAL, C4H, 4CL, CHS, CHI, FSII, F6H, F7GAT. Simultaneously, apigenin was hydroxylated by F6H at C6 position to produce the main precursor and also glycosylated by F7GAT at C7 position to yield the major by-product apigenin-7-O-glucuronide. In order to optimize the whole pathway stepwise, we divided it into three modules, p-coumaric acid production module (Module I), naringenin production module (Module II), and scutellarin production module (Module III). The naringenin production module catalyzed by 4CL and CHS, is the pivotal bridge between Module I and Module III and determines the realization of the whole synthesis pathway. Hence, we optimized the naringenin production module as a starting point and then engineered the other two modules with further optimization.
Fig. 1.
Metabolic pathway for scutellarin synthesis from glucose in Yarrowia lipolytica. Abbreviations: PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; FSII, flavone synthase II; F6H, flavone-6-hydroxylase; ATR2, cytochrome P450 reductase; F7GAT, flavonoid-7-O-glucuronosyltransferase; UDPGDH, UDP-glucose dehydrogenase. Multiple arrows represent multiple enzymatic steps, red genes describe the introduced artificial pathway. In this study, PAL and C4H from A. thaliana or E. breviscapus, 4CL from A. thaliana or E. breviscapus, CHS from H. androsaemum or E. breviscapus, ATR2 from A. thaliana, F6H from E. breviscapus, Scutellaria baicalensis Georgi, Scutellaria barbata D. Don, C. canadensis and C. cardunculus.
3.2. Optimization of the naringenin production module
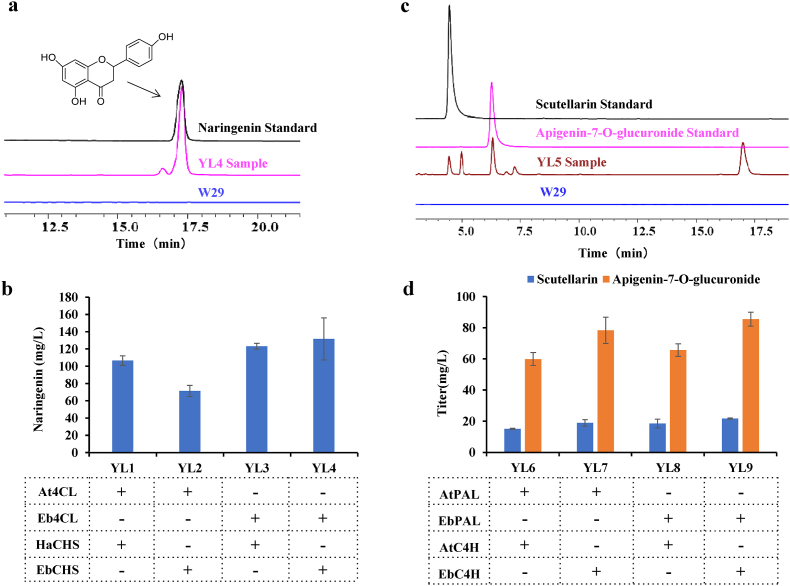
In this study, we selected 4CL from Arabidopsis thaliana or E. breviscapus and CHS from Hypericum androsaemum or E. breviscapus as the candidate genes to catalyze the p-coumaric acid into naringenin [14,41]. They were overexpressed in the p1 plasmid under the control of the strong constitutive promoters TEFin and GPD [39,42]. Then, the linearized p1-CHS-4CL was transformed into the Ku80 site of Y. lipolytica W29. YNB medium with 1 mM p-coumaric acid was used for cultivation. Under HPLC characterization, the fermentation of engineered yeast YL4 had a characteristic retention time of 17.159 min, which was the same as the naringenin standard (Fig. 2a). We observed four strains YL1-YL4 yielded ranging from 71.36 to 131.59 mg/L for the synthesis of naringenin from p-coumaric acid (Fig. 2b), and we found that the engineered strains could also produce the phloretin. Consistent with previously reported literature that HaCHS was the most efficient CHS for phloretin production [39]. The peak area of phloretin in YL3 which integrated the HaCHS was twice as large as that of YL4. Taking the above phenomena into account, we selected YL4 for further modification which integrated the EbCHS (Fig. S1).
Fig. 2.
Naringenin, apigenin-7-O-glucuronide and scutellarin produced in engineered Y. lipolytica strains. Error bars represent standard deviation of at least three biological replicates. The “-” and “+” symbols indicate lack or existence of the corresponding genetic modification, respectively. A. HPLC analysis of YL4 fermentation samples and naringenin standard. B. Naringenin production in different combinations of CHS and 4CL in shake-flask fermentation. C. HPLC analysis of the scutellarin standard, apigenin-7-O-glucuronide standard and YL5 samples. D. Scutellarin and apigenin-7-O-glucuronide production with different combinations of PAL and C4H in shake-flask fermentation.
We next sought to produce scutellarin from the optimized strain YL4. As follows, five enzymes were further integrated into the strain. Similar to p1, FSII was constructed in the p2 vector, F6H and ATR2 were constructed in the p3 vector, F7GAT and UDPGDH were constructed in the p4 vector. These linearized plasmids were integrated into strain YL4 sequentially to generate the strain YL5. Under HPLC characterization, the fermentation of engineered yeast YL5 had a characteristic retention time of 4.536 min, which was the same with the standard of scutellarin (Fig. 2c). Meanwhile the peak at 6.379 min was coincident with apigenin-7-O-glucuronide standard (Fig. 2c). LC-MS analysis results were presented in (Fig. S2). The titer of scutellarin was 15.11 mg/L, and apigenin-7-O-glucuronide was 53.63 mg/L (Fig. S3).
3.3. Bottlenecks exploration of the p-coumaric acid production module
As successful biosynthesis of scutellarin in strain YL5 from p-coumaric acid, we proposed to explore the bottleneck from phenylalanine to p-coumaric acid production module. In p-coumaric acid production module, C4H belongs to the P450 enzyme family and is usually considered to be the rate-limiting step in the biosynthesis of coumaric acid-derived compounds [43,44]. PAL and C4H were screened to determine the optimal gene combination for the highest production of scutellarin. We chose PAL and C4H from A. thaliana or E. breviscapus. Previous reports have proven that they could be functionally expressed in S. cerevisiae [41,45]. PAL and C4H were constructed in a p5 vector integrated into strain YL5 to obtain strain YL6-YL9 (Fig. 2d). The titer of scutellarin was 15.05 mg/L to 21.60 mg/L, and apigenin-7-O-glucuronide was 59.87 mg/L to 85.44 mg/L (Fig. 2d).
Several studies have demonstrated that boosting the supply of aromatic amino acid and malonyl-CoA precursors could effectively increase the production of flavonoids, whether in S. cerevisiae or in Y. lipolytica [32,36,[46], [47], [48], [49]]. Overexpression of mutants of DAHP synthase ARO4 and chorismate mutase ARO7 could avoid feedback inhibition of aromatic amino acids and thereby increase the yield of flavonoid and stilbene products [50]. However, the production did not show improvement by expressing ScARO4K229L and ScARO7G141S or YlARO4K221L and YlARO7G139S in strain YL9, respectively (Fig. S4). We also detected a decrease in OD600, presumably due to a toxic effect of p-coumaric acid accumulation in cells. In addition, we also did not observe higher scutellarin production either by overexpressing SeACSP641L [35,51] and PEX10 [49], which could increase the malonyl-CoA pool (Fig. S4). Our results suggested that the precursors (p-coumaric acid and malonyl-CoA) were not a limiting step in this background strain.
3.4. Optimizing the scutellarin production module
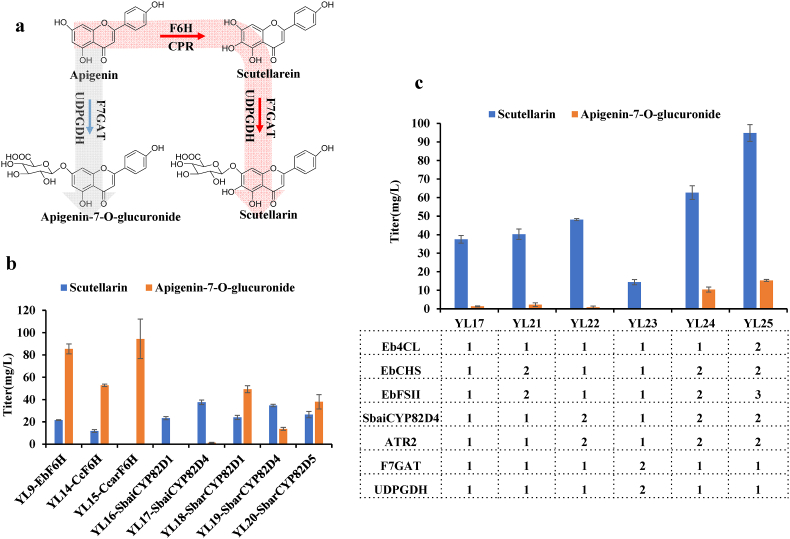
According to the national standard in China, there is more than 90% scutellarin in breviscapine oral drugs and 98% in injection drugs [52,53]. However, in our original version of engineered strains, the production of by-product apigenin-7-O-glucuronide was much higher than that of scutellarin. The glucuronosyltransferase could be competed with the P450 enzyme F6H against the substrate apigenin. It was possible to improve the yield ratio of scutellarin by employing a F6H with higher catalytic efficiency (Fig. 3a). Seven enzymes from different species (Scutellaria baicalensis Georgi, Scutellaria barbata D. Don, C. canadensis and C. cardunculus) were used in this study [54,55]. To determine whether these enzymes could alter the product ratio, we expressed them in YL9 to generate strains YL14-YL20. The results showed that CYP82D4 from S. baicalensis (YL17) presented the best catalytic activity to produce scutellarin both in terms of yield and proportion (Fig. 3b). In comparison with strain YL9, the titer of scutellarin was improved to 37.8 mg/L, the percentage of scutellarin was increased from 20.18% to 96.54% and the percentage of apigenin-7-O-glucuronide was decreased from 79.81% to 3.46% (Fig. 3b and Fig. S5), which meant that the ratio of scutellarin to the apigenin-7-O-glucuronide was improved about 110-fold in flask condition and the specific calculations could be found in the supplementary materials. These results suggested that the screening of isozymes was an efficacious tactic to improve the productivity of heterologous pathways in specific host [[56], [57], [58]].
Fig. 3.
Metabolic engineering to improve the ratio and yield of scutellarin.
A. Schematic illustration of the biosynthetic pathways leading to the production of scutellarin and the byproduct apigenin-7-O-glucuronide. B. Scutellarin and apigenin-7-O-glucuronide production in different species of F6H in shake-flask fermentation. C. Evaluation of scutellarin and apigenin-7-O-glucuronide produced by yeast strains YL17, YL21, YL22, YL23, YL24 and YL25 in shake flask.
In order to further increase the titer of scutellarin, we subsequently increased the copy number of key enzymes in the engineered strain YL17. Since accumulation of p-coumaric acid as well as naringenin was observed in YL17 (Fig. S6), we combined EbCHS and EbFSII, SbaiCYP82D4 and ATR2, F7GAT and UDPGDH respectively with additional copies in the strains YL17 to generate YL21-YL25 (Fig. 3c). Our results showed that the addition of copy numbers of EbCHS, EbFSII, SbaiCYP82D4 and ATR2 indeed significantly increased the yield of scutellarin. Therefore, combining all four genes led the strain YL24 to produce 62.66 mg/L of scutellarin (Fig. 3c). Besides, overexpression of genes F7GAT and UDPGDH did not result in a significant increase in scutellarin production (Fig. 3c). However, in strain YL24, we still observed p-coumaric acid and naringenin (Fig. S7), and pushing the accumulation of intermediates downwards might further increase the yield of scutellarin. Since increasing the copy number of CHS in strain YL21 could not effectively solved this problem. We speculated that the bottleneck might be 4CL instead of CHS, so we added one copy of 4CL to YL24. For the accumulation of naringenin, we added another copy of FSII to obtain the final strain YL25, which produced 94.79 mg/L of scutellarin (Fig. 3c), indicating that multiple copies of downstream genes were vital for directing carbon flux toward scutellarin synthesis.
3.5. Optimization of the fermentation process
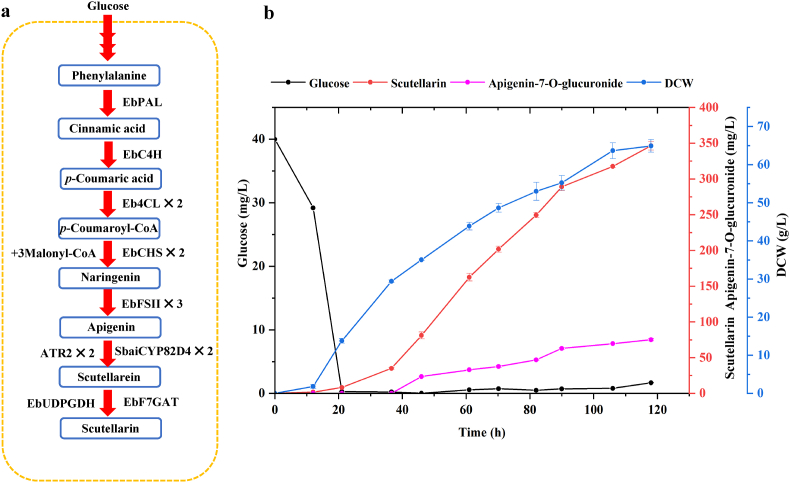
In order to further improve the production of scutellarin, we conducted a fed-batch fermentation culture of the best engineered strain YL25 using mineral medium (Fig. 4a). Since glucose concentration played a decisive role during the fermentation process, the amount of residual sugar was measured every 6 h (Fig. 4b). When the glucose exhausted in the initial medium, we started fed-batch fermentation at an initial rate of 3 mL/h at 20 h, which then was dynamically regulated to maintain the PH at 5. During the whole fermentation process, glucose was added in a continuous flow and ensured that the amount of glucose was less than 5 g/L, which did not cause high osmotic pressure and no erythritol was detected (Fig. S8). After 12 h of lag phase, the cells entered the logarithmic growth phase, and the DCW finally reached to 66.8 g/L at 118 h (Fig. 4b).
Fig. 4.
Fed-batch fermentation of the engineering strain YL25 in 1.3 L bioreactor. A. Schematic diagram of scutellarin biosynthesis process in YL25 strain. The number after the gene represents the number of copies of the gene. B. Production of scutellarin and apigenin-7-O-glucuronide by strain YL25 in fed-batch fermentation.
Considering that there were three P450 enzymes in the scutellarin synthesis pathway and that the improvement of oxygen supply could help to increase the activity of the key P450 enzymes [59], the DO was maintained at 40% by adjusting the speed and aeration. When the cells entered to the logarithmic phase, scutellarin began to accumulate. However, a small amount of by-product apigenin-7-O-glucuronide was accompanied with the production of scutellarin after 36 h. Finally, the yield of scutellarin reached to 346 mg/L at 118 h (Fig. 4b), which was the highest reported so far in microbes. The titer of apigenin-7-O-glucuronide also was improved and reached to 75.14 mg/L, accounting for 18% of the final products, which was higher than that in flask condition. The yield of scutellarin could be further optimized by the regulation of fermentation conditions in the future.
4. Discussion
It is known that the precursor limitation is one of the major obstacles to produce natural compounds in microbial cell factories [[60], [61], [62]]. However, our results indicated that precursor supply was not the major bottleneck at the initial engineering stage due to the excellent primary metabolic level of Y. lipolytica. The optimization of downstream heterogenous pathways appeared to be more important. The inherent low catalytic efficiency of enzymes for the biosynthesis of natural product is a challenge for flavonoid production, especially P450 enzymes [63,64]. The P450 enzyme F6H is regarded as the key rate-limiting enzyme in the scutellarin biosynthetic pathway. We compared F6H from seven species and found that CYP82D4 from S. baicalensis successfully enhanced the yield of scutellarin and significantly reduced the yield of the by-product apigenin-7-O-glucuronide, implying that the catalytic activity of enzymes from different sources has a significant effect on the product. In addition, the copy numbers of key downstream genes were optimized and the final flask production of scutellarin increased 4.28-fold to 94.79 mg/L. This result further confirmed that the yield of many natural products could be improved by integrating multiple copies of key enzymes in Y. lipolytica [32,35,65].
Our de novo biosynthesis system provided a concise fermentation process and achieved in a cheap mineral medium. The fed-batch fermentation of scutellarin in YL25 finally reached to 346 mg/L, which was 3.2-fold higher than that produced in S. cerevisiae [14]. Although the titers in shake flasks and fermenters for E. coli system were similar with our results, there were many expensive inducers and precursors used in E. coli production system, such as IPTG, tyrosine, and sodium malonate [66,67]. In addition, we also noticed that the percentage of apigenin-7-O-glucuronide increased from 3.46% in flask to 18% in the fermenter. A reasonable speculation was that the supply of oxygen in the fermenter could affect the expression of P450 enzymes and the fermentation process needed to be controlled more accurately.
The productivity of scutellarin in our study was only 1.83 mg/g glucose. One of important reasons could be that there were large amounts of precipitated products accumulating inside the fermenter during our fermentation process. HPLC result showed that the major constitute part of this precipitate was naringenin (Figs. S9 and S10). Naringenin is the first step in the formation of flavonoids and also an intermediate of scutellarin. The precipitating of intermediate product became an obstacle for further effective biotransformation. The copy number of FSII needed to be further increased to enhance the downstream biotransformation of naringenin and improve the productivity of the downstream compounds.
Our study also presented a robust work for further genetic engineering of the Y. lipolytica chassis. In this study, the producing of scutellarin was achieved through a modular expression, which divided of the biosynthetic pathway into three modules. A large panel of synthetic genes were recruited and the expression cassettes were integrated into specific locus using a CRISPR/Cas9 kit without markers remained. Collectively, our work shows that Y. lipolytica has great potential for the production of scutellarin and even other flavonoids.
CRediT authorship contribution statement
Yina Wang: designed and wrote the paper and conducted relevant experiments. Xiaonan Liu: designed and wrote the paper and conducted relevant experiments, revised the paper. Bihuan Chen: performed the fermenter operation. Wei Liu: performed the fermenter operation. Zhaokuan Guo: performed the fermenter operation. Xiangyu Liu: performed the fermenter operation. Xiaoxi Zhu: performed the fermenter operation. Jiayu Liu: performed the fermenter operation. Jin Zhang: performed the fermenter operation. Jing Li: performed the fermenter operation. Lei Zhang: performed the fermenter operation. Yadi Gao: performed the fermenter operation. Guanghui Zhang: revised the paper. Yan Wang: revised the paper. M. Iqbal Choudhary: revised the paper. Shengchao Yang: revised the paper. Huifeng Jiang: revised the paper.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgements
We thank Dr. Zongjie Dai (Tianjin Institute of Industrial Biotechnology) for the gift of Yarrowia lipolytica W29, YL0, and plasmid pgRNA-YL. We thank Dr. Guokun Wang (Tianjin Institute of Industrial Biotechnology) for kindly providing plasmids pSBNR-PL007, pSBNR-PL010, pSBNR-PL011, and pSBNR-PL013.
This project has received funding from the Major Science and Technique Programs in Yunnan Province (No. 2019ZF011-1), the National Natural Science Foundation of China (No. 81960689), the National Key R&D Program of China (2020YFC1316400), the Project of Young and Middle-Aged Talent of Yunnan Province (No. 2019HB019), Science and Technology Innovation team of Yunnan (No. 202105AE160011), Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (No. TSBICIP-KJGG-002-02) and the Tianjin Science Fund for Distinguished Young Scholars (No.18JCJQJC48300).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.05.009.
Contributor Information
Xiaonan Liu, Email: liu_xn@tib.cas.cn.
Shengchao Yang, Email: shengchaoyang@163.com.
Huifeng Jiang, Email: jiang_hf@tib.cas.cn.
Appendix A. Supplementary data
Supporting Information: Supporting information could be found in the online version of this article. Supplementary methods, plasmids construction. Methods for calculating the ratio of scutellarin to the main by-product apigenin-7-O-glucuronide, composition of media. Fig. S1. HPLC analysis of phloretin and naringenin standard and the fermented products of YL3 and YL4. Fig. S2, MS analysis of scutellarin and apigenin-7-O-glucuronide in the fermented products of YL5. Fig. S3, Production of scutellarin and apigenin-7-O-glucuronide with supplementation of 1 mM p-coumaric acid. Fig. S4. Evaluation of scutellarin and apigenin-7-O-glucuronide produced by yeast strains YL9, YL10, YL11, YL12, and YL13 in shake flask. Fig. S5, HPLC analysis of the YL9, YL14, YL15, YL16, YL17, YL18. YL19 and YL20 samples. Fig. S6, HPLC analysis of the p-coumaric acid standard, naringenin standard and YL17 samples. Fig. S7. HPLC analysis of the p-coumaric acid standard, naringenin standard and YL24 samples. Fig. S8. HPLC analysis of the erythritol standard, glucose standard, 12 h sample, 36 h sample, and 118 h sample. Fig. S9, Naringenin accumulated on the walls of the fermentation tank. Fig. S10, HPLC analysis of the sedimentation on the fermentation tank wall. Table S1, Microbial production of flavonoids in Y. lipolytica; Table S2, The origin of the heterologous genes and the encoded enzymes in this study. Table S3, Primers used in this study; Table S4, Plasmids used in this study; Table S5, Strains used in this study; Table S6, Codon-optimized genes for Y. lipolytica used in this study.
The following is the Supplementary data to this article:
References
- 1.Liu Y., Wen P.H., Zhang X.X., Dai Y., He Q. Breviscapine ameliorates CCl4-induced liver injury in mice through inhibiting inflammatory apoptotic response and ROS generation. Int J Mol Med. 2018;42(2):755–768. doi: 10.3892/ijmm.2018.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin X., Luo Y., Chen Y., Ma Y., Yue P., Yang M. Novel breviscapine nanocrystals modified by panax notoginseng saponins for enhancing bioavailability and synergistic anti-platelet aggregation effect. Colloids Surf B Biointerfaces. 2019;175:333–342. doi: 10.1016/j.colsurfb.2018.11.067. [DOI] [PubMed] [Google Scholar]
- 3.Gao J., Chen G., He H., Liu C., Xiong X., Li J., Wang J. Therapeutic effects of breviscapine in cardiovascular diseases: a review. Front Pharmacol. 2017;8:289. doi: 10.3389/fphar.2017.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pengyue Z., Tao G., Hongyun H., Liqiang Y., Yihao D. Breviscapine confers a neuroprotective efficacy against transient focal cerebral ischemia by attenuating neuronal and astrocytic autophagy in the penumbra. Biomed Pharmacother. 2017;90:69–76. doi: 10.1016/j.biopha.2017.03.039. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z., Li H., Yan J., Liu Y. Flavonoid compound breviscapine suppresses human osteosarcoma Saos-2 progression property and induces apoptosis by regulating mitochondria-dependent pathway. J Biochem Mol Toxicol. 2021;35(1) doi: 10.1002/jbt.22633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L., Gao Y., Zhang S., Fang Z. The effects of breviscapine injection on hypertension in hypertension-induced renal damage patients: a systematic review and a meta-analysis. Front Pharmacol. 2019;10:118. doi: 10.3389/fphar.2019.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Gui Y., Luo Y., Liu Y., Tu L., Ma Y., Yue P., Yang M. Design and evaluation of inhalable nanocrystals embedded microparticles with enhanced redispersibility and bioavailability for breviscapine. Powder Technol. 2021;377:128–138. [Google Scholar]
- 8.Song Z., Yin J., Xiao P., Chen J., Gou J., Wang Y., Zhang Y., Yin T., Tang X., He H. Improving breviscapine oral bioavailability by preparing nanosuspensions, liposomes and phospholipid complexes. Pharmaceutics. 2021;13(2):132. doi: 10.3390/pharmaceutics13020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L., Liu M., Fang Z. Combined therapy of hypertensive nephropathy with breviscapine injection and antihypertensive drugs: a systematic review and a meta-analysis. Evid Based Complement Alternat Med. 2018 doi: 10.1155/2018/2958717. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan T., Jiang S., Zhang J., Weng Q., Yu Y., Li H., et al. Breviscapine alleviates nonalcoholic steatohepatitis by inhibiting TGF-beta-activated kinase 1-dependent signaling. Hepatology. 2021:1–17. doi: 10.1002/hep.32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seshadri S., Beiser A., Kelly-Hayes M., Kase C.S., Au R., Kannel W.B., Wolf P.A. The lifetime risk of stroke: estimates from the Framingham Study. Stroke. 2006;37(2):345–350. doi: 10.1161/01.STR.0000199613.38911.b2. [DOI] [PubMed] [Google Scholar]
- 12.Wang L., Ma Q. Clinical benefits and pharmacology of scutellarin: a comprehensive review. Pharmacol Ther. 2018;190:105–127. doi: 10.1016/j.pharmthera.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Prince M.J., Wu F., Guo Y., Gutierrez Robledo L.M., O'donnell M., Sullivan R., Yusuf S. The burden of disease in older people and implications for health policy and practice. Lancet. 2015;385(9967):549–562. doi: 10.1016/S0140-6736(14)61347-7. [DOI] [PubMed] [Google Scholar]
- 14.Liu X., Cheng J., Zhang G., Ding W., Duan L., Yang J., Kui L., Cheng X., Ruan J., Fan W., Chen J., Long G., Zhao Y., Cai J., Wang W., Ma Y., Dong Y., Yang S., Jiang H. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun. 2018;9(1):1–10. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J., Zhang G., Zhang J., Liu H., Chen W., Wang X., Li Y., Dong Y., Yang S. Hybrid de novo genome assembly of the Chinese herbal fleabane Erigeron breviscapus. GigaScience. 2017;6(6):1–7. doi: 10.1093/gigascience/gix028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong Z.X., Li N.G., Zhang P.X., Gu T., Wu W.Y., Shi Z.H. An efficient chemical synthesis of scutellarein: an in vivo metabolite of scutellarin. Molecules. 2016;21(3):263. doi: 10.3390/molecules21030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeldon I., Blenner M. Yarrowia lipolytica methods and protocols: methods and protocols. Methods Mol Biol. 2021 [Google Scholar]
- 18.Wasylenko T.M., Ahn W.S., Stephanopoulos G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab Eng. 2015;30:27–39. doi: 10.1016/j.ymben.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Blank L.M., Lehmbeck F., Sauer U. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res. 2005;5(6–7):545–558. doi: 10.1016/j.femsyr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Ma J., Gu Y., Marsafari M., Xu P. Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J Ind Microbiol Biotechnol. 2020;47(9–10):845–862. doi: 10.1007/s10295-020-02290-8. [DOI] [PubMed] [Google Scholar]
- 21.Muhammad A., Feng X., Rasool A., Sun W., Li C. Production of plant natural products through engineered Yarrowia lipolytica. Biotechnol Adv. 2020;43 doi: 10.1016/j.biotechadv.2020.107555. [DOI] [PubMed] [Google Scholar]
- 22.Abghari A., Chen S. Yarrowia lipolytica as an oleaginous cell factory platform for production of fatty acid-based biofuel and bioproducts. Front Energy Res. 2014;2:21. [Google Scholar]
- 23.Abdel-Mawgoud A.M., Markham K.A., Palmer C.M., Liu N., Stephanopoulos G., Alper H.S. Metabolic engineering in the host Yarrowia lipolytica. Metab Eng. 2018;50:192–208. doi: 10.1016/j.ymben.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Wong L., Holdridge B., Engel J., Xu P. Genetic tools for streamlined and accelerated pathway engineering in Yarrowia lipolytica. Methods Mol Biol. 2019;1927:155–177. doi: 10.1007/978-1-4939-9142-6_11. [DOI] [PubMed] [Google Scholar]
- 25.Celinska E., Ledesma-Amaro R., Larroude M., Rossignol T., Pauthenier C., Nicaud J.M. Golden Gate Assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica. Microb Biotechnol. 2017;10(2):450–455. doi: 10.1111/1751-7915.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Z., Gao C., Li J., Hou J., Lin C.S.K., Qi Q. Engineering of unconventional yeast Yarrowia lipolytica for efficient succinic acid production from glycerol at low pH. Metab Eng. 2017;42:126–133. doi: 10.1016/j.ymben.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Verbeke J., Beopoulos A., Nicaud J.M. Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol Lett. 2013;35(4):571–576. doi: 10.1007/s10529-012-1107-0. [DOI] [PubMed] [Google Scholar]
- 28.Lustig A.J. The Kudos of non-homologous end-joining. Nat Genet. 1999;23(2):130–131. doi: 10.1038/13755. [DOI] [PubMed] [Google Scholar]
- 29.Wei W., Zhang P., Shang Y., Zhou Y., Ye B.C. Metabolically engineering of Yarrowia lipolytica for the biosynthesis of naringenin from a mixture of glucose and xylose. Bioresour Technol. 2020;314 doi: 10.1016/j.biortech.2020.123726. [DOI] [PubMed] [Google Scholar]
- 30.Lv Y., Gu Y., Xu J., Zhou J., Xu P. Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab Eng. 2020;61:79–88. doi: 10.1016/j.ymben.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akram M., Rasool A., An T., Feng X., Li C. Metabolic engineering of Yarrowia lipolytica for liquiritigenin production. Chem Eng Sci. 2020;230 [Google Scholar]
- 32.Sáez-Sáez J., Wang G., Marella E.R., Sudarsan S., Cernuda Pastor M., Borodina I. Engineering the oleaginous yeast Yarrowia lipolytica for high-level resveratrol production. Metab Eng. 2020;62:51–61. doi: 10.1016/j.ymben.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu Y., Ma J.B., Zhu Y.L., Ding X.Y., Xu P. Engineering Yarrowia lipolytica as a chassis for de novo synthesis of five aromatic-derived natural products and chemicals. ACS Synth Biol. 2020;9(8):2096–2106. doi: 10.1021/acssynbio.0c00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Q., Szczepańska P., Yuzbashev T., Lazar Z., Ledesma-Amaro R. De novo production of resveratrol from glycerol by engineering different metabolic pathways in Yarrowia lipolytica. Metab Eng Commun. 2020;11 doi: 10.1016/j.mec.2020.e00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv Y., Marsafari M., Koffas M., Zhou J., Xu P. Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth Biol. 2019;8(11):2514–2523. doi: 10.1021/acssynbio.9b00193. [DOI] [PubMed] [Google Scholar]
- 36.Palmer C.M., Miller K.K., Nguyen A., Alper H.S. Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a beta-oxidation mediated strategy. Metab Eng. 2020;57:174–181. doi: 10.1016/j.ymben.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Wei W., Zhang P., Shang Y., Zhou Y., Ye B.C. Metabolically engineering of Yarrowia lipolytica for the biosynthesis of naringenin from a mixture of glucose and xylose. Bioresour Technol. 2020;314 doi: 10.1016/j.biortech.2020.123726. [DOI] [PubMed] [Google Scholar]
- 38.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 39.Holkenbrink C., Dam M.I., Kildegaard K.R., Beder J., Dahlin J., Domenech Belda D., Borodina I. EasyCloneYALI: CRISPR/Cas9-Based synthetic toolbox for engineering of the yeast Yarrowia lipolytica. Biotechnol J. 2018;13(9) doi: 10.1002/biot.201700543. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz C., Shabbir-Hussain M., Frogue K., Blenner M., Wheeldon I. Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica. ACS Synth Biol. 2017;6(3):402–409. doi: 10.1021/acssynbio.6b00285. [DOI] [PubMed] [Google Scholar]
- 41.Eichenberger M., Lehka B.J., Folly C., Fischer D., Martens S., Simon E., Naesby M. Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab Eng. 2017;39:80–89. doi: 10.1016/j.ymben.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tai M., Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Trantas E., Panopoulos N., Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng. 2009;11(6):355–366. doi: 10.1016/j.ymben.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Yan Y., Kohli A., Koffas M.A. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl Environ Microbiol. 2005;71(9):5610–5613. doi: 10.1128/AEM.71.9.5610-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehka B.J., Eichenberger M., Bjorn-Yoshimoto W.E., Vanegas K.G., Buijs N., Jensen N.B., Dyekjaer J.D., Jenssen H., Simon E., Naesby M. Improving heterologous production of phenylpropanoids in Saccharomyces cerevisiae by tackling an unwanted side reaction of Tsc13, an endogenous double-bond reductase. FEMS Yeast Res. 2017;17(1):fox004. doi: 10.1093/femsyr/fox004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M., Kildegaard K.R., Chen Y., Rodriguez A., Borodina I., Nielsen J. De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab Eng. 2015;32:1–11. doi: 10.1016/j.ymben.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Liu Q., Yu T., Li X., Chen Y., Campbell K., Nielsen J., Chen Y. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat Commun. 2019;10(1):1–13. doi: 10.1038/s41467-019-12961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li G.J., Li H.B., Lyu Y.B., Zeng W.Z., Zhou J.W. Enhanced biosynthesis of dihydromyricetin in Saccharomyces cerevisiae by coexpression of multiple hydroxylases. J Agric Food Chem. 2020;68(48):14221–14229. doi: 10.1021/acs.jafc.0c05261. [DOI] [PubMed] [Google Scholar]
- 49.Markham K.A., Palmer C.M., Chwatko M., Wagner J.M., Murray C., Vazquez S., Swaminathan A., Chakravarty I., Lynd N.A., Alper H.S. Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation. Proc Natl Acad Sci U S A. 2018;115(9):2096–2101. doi: 10.1073/pnas.1721203115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luttik M., Vuralhan Z., Suir E., Braus G., Pronk J., Daran J. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab Eng. 2008;10(3–4):141–153. doi: 10.1016/j.ymben.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Gallego-Jara J., Terol G.L., Ecija Conesa A., Zambelli B., Canovas Diaz M., De Diego Puente T. Characterization of acetyl-CoA synthetase kinetics and ATP-binding. Biochim Biophys Acta Gen Subj. 2019;1863(6):1040–1049. doi: 10.1016/j.bbagen.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 52.Zhang W., Chen W., Wang Y., Yang G., Kong D., Li H. Studies on the flavone glycosides from the extract of Erigeron breviscapus. Zhong Cao Yao. 2000;31(8):565–566. [Google Scholar]
- 53.Commission C.P. vol. 1. China Medical Science Press; Beijing, China: 2015. pp. 191–193. (Chinese pharmacopoeia). [Google Scholar]
- 54.Liu X., Cheng J., Zhu X., Zhang G., Yang S., Guo X., Jiang H., Ma Y. De novo biosynthesis of multiple pinocembrin derivatives in Saccharomyces cerevisiae. ACS Synth Biol. 2020;9(11):3042–3051. doi: 10.1021/acssynbio.0c00289. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Q., Cui M.Y., Levsh O., Yang D., Liu J., Li J., Hill L., Yang L., Hu Y., Weng J.K., Chen X.Y., Martin C. Two CYP82D enzymes function as flavone hydroxylases in the biosynthesis of root-specific 4'-deoxyflavones in Scutellaria baicalensis. Mol Plant. 2018;11(1):135–148. doi: 10.1016/j.molp.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y., Xiao W., Wang Y., Liu H., Li X., Yuan Y. Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb Cell Factories. 2016;15(1):1–13. doi: 10.1186/s12934-016-0509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma T., Zhou Y., Li X., Zhu F., Cheng Y., Liu Y., Deng Z., Liu T. Genome mining of astaxanthin biosynthetic genes from Sphingomonas sp. ATCC 55669 for heterologous overproduction in Escherichia coli. Biotechnol J. 2016;11(2):228–237. doi: 10.1002/biot.201400827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu T., Zhou J., Tong Y., Su P., Li X., Liu Y., Liu N., Wu X., Zhang Y., Wang J., Gao L., Tu L., Lu Y., Jiang Z., Zhou Y.J., Gao W., Huang L. Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metab Eng. 2020;60:87–96. doi: 10.1016/j.ymben.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Xu L., Wang D., Chen J., Li B., Li Q., Liu P., Qin Y., Dai Z., Fan F., Zhang X. Metabolic engineering of Saccharomyces cerevisiae for gram-scale diosgenin production. Metab Eng. 2022;70:115–128. doi: 10.1016/j.ymben.2022.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Xu P., Ranganathan S., Fowler Z.L., Maranas C.D., Koffas M.A. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng. 2011;13(5):578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 61.Zha W., Rubin-Pitel S.B., Shao Z., Zhao H. Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab Eng. 2009;11(3):192–198. doi: 10.1016/j.ymben.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 62.Johnson A.O., Gonzalez-Villanueva M., Wong L., Steinbuchel A., Tee K.L., Xu P., Wong T.S. Design and application of genetically-encoded malonyl-CoA biosensors for metabolic engineering of microbial cell factories. Metab Eng. 2017;44:253–264. doi: 10.1016/j.ymben.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 63.Bar-Even A., Salah Tawfik D. Engineering specialized metabolic pathways--is there a room for enzyme improvements? Curr Opin Biotechnol. 2013;24(2):310–319. doi: 10.1016/j.copbio.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Shang Y., Huang S. Engineering plant cytochrome P450s for enhanced synthesis of natural products: past achievements and future perspectives. Plant Commun. 2020;1(1) doi: 10.1016/j.xplc.2019.100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marella E.R., Dahlin J., Dam M.I., Ter Horst J., Christensen H.B., Sudarsan S., Wang G., Holkenbrink C., Borodina I. A single-host fermentation process for the production of flavor lactones from non-hydroxylated fatty acids. Metab Eng. 2020;61:427–436. doi: 10.1016/j.ymben.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 66.Li J., Tian C., Xia Y., Mutanda I., Wang K., Wang Y. Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab Eng. 2019;52:124–133. doi: 10.1016/j.ymben.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 67.Ji D., Li J., Xu F., Ren Y., Wang Y. Improve the biosynthesis of baicalein and scutellarein via manufacturing self-assembly enzyme reactor in vivo. ACS Synth Biol. 2021;10(5):1087–1094. doi: 10.1021/acssynbio.0c00606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.