Abstract
The big-data use is becoming a standard practice in the neuroimaging field through data-sharing initiatives. It is important for the community to realize that such open science effort must protect personal, especially facial information when raw neuroimaging data are shared. An ideal tool for the face anonymization should not disturb subsequent brain tissue extraction and further morphological measurements. Using the high-resolution head images from magnetic resonance imaging (MRI) of 215 healthy Chinese, we discovered and validated a template effect on the face anonymization. Improved facial anonymization was achieved when the Chinese head templates but not the Western templates were applied to obscure the faces of Chinese brain images. This finding has critical implications for international brain imaging data-sharing. To facilitate the further investigation of potential culture-related impacts on and increase diversity of data-sharing for the human brain mapping, we released the 215 Chinese multi-modal MRI data into a database for imaging Chinese young brains, namely’I See your Brains (ISYB)’, to the public via the Science Data Bank (10.11922/sciencedb.00740).
Subject terms: Cognitive neuroscience, Psychology
| Measurement(s) | brain imaging measurements |
| Technology Type(s) | magnetic resonance imaging |
| Factor Type(s) | multimodal neuroimaging metrics |
| Sample Characteristic - Organism | Homo |
| Sample Characteristic - Environment | magnetic |
| Sample Characteristic - Location | North China |
Background & Summary
With the worldwide increase in the acquisition of neuroimaging data using magnetic resonance imaging (MRI), the use of big data is becoming a standard practice in the field through international data-sharing initiatives1–4. Data sharing can bring many benefits to neuroscience research and increase sample sizes, which allow for greater precision and improve the statistical power to detect smaller effects, although smaller effects may be associated with either smaller biological effects or potentially confounding factors5. This has accelerated many fields into their new stages or even entirely new field, such as the developmental population neuroscience6,7 or statistical connectomics8. On the one hand, it can facilitate researchers to conduct test-retest studies when more neuroimaging public databases are available and improve the reproducibility of research results9,10. Data reuse can also make peer review better9,11. On the other hand, it encourages cooperation and communication between institutions and organizations, and thus creates a win-win situation for both researchers giving and receiving data12.
As the concept of sharing data continues to grow, more and more public databases are emerging to support researchers such as the Human Connectome Project (HCP)13 and its derivatives across human lifespan14 (e.g., the Adolescent Brain Cognition Development, ABCD15), UK Biobank16, the Autism Brain Imaging Data Exchange (ABIDE)17,18, the 1000 Functional Connectomes Project19. These efforts on big-data sharing has propelled the cognitive neurology field forward (see more data projects from our recent reviews20,21). However, most of the existing databases are based on Caucasian populations. This preventing us from identifying ethnic or culture-related differences in neuroscience associations. Moreover, China has the largest population in the world and there is still a need to expand and improve the public database of the Chinese brain mapping (few exceptions, e.g., the Chinese Color Nest Project20, the REST-meta-MDD project22, the Chinese Human Connectome Project23 and the Consortium for Reliability and Reproducibility9).
Much like the genetics field24, data-sharing faces many challenges in the neuroscience field. One of the challenges is to ensure that personal information, especially facial information, remains protected when raw MRI data are shared. This is especially challenging for high-resolution T1-weighted anatomical images, in which facial information has been fully embedded. The higher the image resolution is, the clearer the facial information becomes. It has been possible to restore subject’s facial information through the combination of existing facial recognition and reconstruction technologies. As a result, personal information can easily be disclosed. Therefore, we must first remove or obscure the facial features in brain images, especially in high-resolution structural MRI images. In response to this, the Health Insurance Portability and Accountability Act (HIPAA) of the United States specifically stipulates that the subject’s private photos or equivalent images must be removed from the medical images to protect their privacy25. The European Union has general data protection, which requires research institutions to strengthen the privacy protection of personal data. Therefore, data protection should be considered and implemented during the design phase of scientific studies24,26.
In order to ensure participant’s private security, all the aforementioned face masking algorithms are involved in taking a standard brain template as references, so as to blur the facial information in the template space. However, to date, previous studies have not dealt with whether a different facial mask will be generated due to a different ethnic template used during anonymization. This may reflect the situation that the existing public brain imaging databases rarely contain non-western samples. In this data descriptor, we release the’I See Your Brains’ database for imaging Chinese young brains and demonstrate the template effects on face masking using head templates of different races. Specifically, ethnically diverse brain templates are employed to anonymize Chinese multimodal MRI data independently. The discrepancy between Chinese brain images anonymized using different templates of race is quantitatively examined. We hypothesized that the use of different (Chinese versus Western) templates will cause significant differences in the face masking performance. Use of the corresponding ethnic template will achieve a better performance in face anonymization.
Methods
Participant recruitment
The database, Imaging Chinese Young Brains (namely, “I See Your Brains”, ISYB), is generated by the Institute of Psychology, Chinese Academy of Sciences (IPCAS), which is one contributing site of the Chinese imaging genetics cohort (CHIMGEN)27 to enhance cross-ethnic and cross-geographic brain research. It is the largest prospective neuroimaging genetic cohort for Chinese Han adults with natural and socioeconomic measurements obtained from remote sensing. In accordance with CHIMGEN, all participants in ISYB were recruited by advertisements posted in colleges and communities. Participants were excluded if they met any of the following criteria: regular smoker, pregnancy, abnormal color discrimination, a history of alcohol or drug abuse, currently any medication, MRI contraindications, neuropsychiatric or severe somatic disorder and sedative-hypnotic medication within a month or any medication for major neuropsychiatric disorders. The ISYB study was approved by the IPCAS ethical committee and written informed consent was obtained from each participant.
Image acquisition
ISYB contains multi-modal neuroimaging data from 241 right-handed Chinese healthy volunteers. Each participant received MRI scans of the brain images at a 3.0 Tesla MRI scanner (GE MR750) at the IPCAS MRI Research Center including: (1) the high-resolution T1-weighted structural MRI (sMRI, matrix = 256 × 256, number of slices = 188, field of view = 256 × 256 mm, repetition time = 8.16 ms, echo time = 3.18 ms, flip angle = 12°, inversion time = 450 ms), (2) resting-state functional MRI (rfMRI, matrix = 64 × 64, number of slices = 36, field of view = 220 × 220 mm, repetition time = 2000ms, echo time = 30 ms, flip angle = 90°), (3) diffusion tensor MRI (dMRI, matrix = 128 × 128, number of slices = 50, field of view = 256 × 256 mm, repetition time = 6000 ms, echo time = 65 ms, flip angle = 90°, 64 diffusion-sensitisation directions at b = 1000 with b = 0), and (4) arterial spin labeling MRI (aMRI, matrix = 128 × 128, number of slices = 50, field of view = 240 × 240 mm, repetition time = 5046 ms, echo time = 11.09 ms, flip angle = 111°, inversion time = 2025 ms).
Quality assurance
All participants were included in the first-step quality assessments of the neuroimaging data. Their anatomical T1 sMRI images were visually inspected to ensure no substantial head motion and structural abnormalities. Two participants were excluded for excessive head motion. In the second-step quality assessments, the T1 sMRI and rfMRI image quality of ISYB datasets were examined using the MRIQC toolkit (https://github.com/poldracklab/mriqc), which incorporated a series of quantitative metrics as in the following list. More details of these metrics can be found in the previous work28,29.
Spatial Metrics (sMRI, rfMRI):
Signal-to-Noise Ratio (SNR)30: The mean value of gray matter divided by the standard deviation (SD) of the air.
Foreground to Background Energy Ratio (FBER): Mean energy of image values (i.e., mean of squares) within the head relative to outside the head.
Contrast-to-Noise Ratio (CNR) (only sMRI)30: Calculated as the mean of the gray matter values minus the mean of the white matter values, divided by the standard deviation of the air values.
Entropy Focus Criteria (EFC)31: Shannon’s entropy is used to summarize the principal directions distribution.
Smoothness of Voxels32: The full-width half maximum (FWHM) of the spatial distribution of image intensities.
- Temporal Metrics (rfMRI):
- Mean Framewise Displacement (FD)33: A measure of subject head motion, which compares the motion between the current and previous volumes. This is calculated by summing the absolute value of displacement changes in the x, y and z directions and rotational changes about those three axes. The rotational changes are given distance values based on the changes across the surface of a 50 mm radius sphere.
- Standardized DVARS33: The spatial standard deviation of the temporal derivative of the data (D referring to temporal derivative of time series, VARS referring to root-mean-square variance over voxels), normalized by the temporal standard deviation and temporal auto-correlation.
- Global Correlation (GCOR)34: The average of the entire brain correlation matrix, which is computed as the brain-wide average time series correlation over all possible combinations of voxels.
The mean and SD were calculated for each of the eight (five spatial metrics and three temporal metrics) to remove extreme values or outliers which is 3-SD above or below the mean. Taking the effects of head movement in rfMRI into consideration, the data with average FD greater than 0.20 mm33 were further excluded. We note that there were 26 participants excluded by the quality assessment procedure, remaining 215 participants included for the subsequent analyses (156 females; 18–30 years old, mean = 22.55 yrs, SD = 2.68 yrs).
The assessments on the results of the quality assurance of structural MRI images and functional MRI images are summarized in Figs. 1 and 2, respectively. Details of individual quality reports are presented in the supplementary figures (SFig. 1 for sMRI and SFig. 2 for rfMRI).
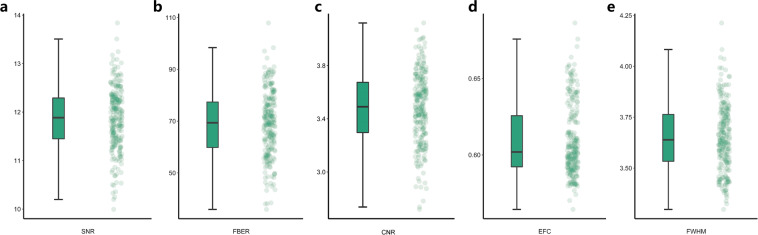
Fig. 1.
Quality assessments on the ISYB sMRI data using both bar (the left column) and scatter (the right column) plots of the following quality metrics: (a) Signal-to-Noise Ratio (SNR) (b) Foreground to Background Energy Ratio (FBER) (c) Contrast-to-Noise Ratio (CNR) (d) Entropy Focus Criteria (EFC) (e) Full-Width Half Maximum (FWHM).
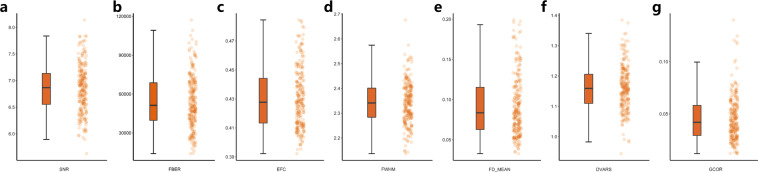
Fig. 2.
Quality assessments on the ISYB rfMRI data using both bar (the left column) and scatter (the right column) plots of the following quality metrics: (a) Signal-to-Noise Ratio (SNR) (b) Foreground to Background Energy Ratio (FBER) (c) Entropy Focus Criteria (EFC) (d) Full-Width Half Maximum (FWHM), (e) Mean Framewise Displacement (FD_MEAN), (f) Derivative of time series and root-mean-square VARiance over voxelS (DVARS), (g) Global Correlation (GCOR).
Standard brain templates
Taking into consideration on the global scope of the open science community and data-sharing initiatives, it is critical to realize that the face masking procedure initially uses Western head template as reference. Extensive research has shown that the race factor played a major role when comparing the group differences in brain morphology metrics. For example, previous studies have indicated significant morphological differences between Chinese and Western brains in size35, shape36, and anatomical volume37. Caucasian brains are significantly longer but present a smaller width and a decreased height compared with Chinese brains23,38. Thus, taking templates of different races as reference during registration exerts a significant impact on the results of the registration, which may lead to systematic differences in subsequent morphological measurements39. Selecting ethnicity-matched templates to analyze structural MRI data may be preferable, which could improve the accuracy of the registration and obtain robust morphological results40. This is attributed to the differences in the brain structure between China and the West population, which cause some systematic bias in the registration between individual images and template images and reduce the accuracy in morphological measurements. Such bias has been reproduced in diverse samples across imaging protocols and lifespan studies41–44.
Ethnicity-specific brain templates are discrepant regarding the diversity in the brain structures between the East and the West. To test the template effects on face masking, we chose two Chinese brain templates (CN200 and Chinese2020): (1) CN200 is a head template assembled by MRI T1 images using the 3 T Siemens Prisma scanner from 200 healthy Chinese Han participants from the Chinese Human Connectome Project23; (2) Chinese2020 is a head template generated based on the MRI T1 images under multiple 1.5 T magnets collected from more than 2,000 healthy participants across multiple centers and regions45. The MNI152 head template was used as the Western template, which is the default head template as in the face masking toolkit.
Face anonymization
In order to effectively blur the part of the face in images and protect privacy as much as possible, researchers have developed several toolkits to remove facial information. For example, Quickshear segments brain images into facial features and brain, and configures the voxels of the facial features to 0, thus resulting in identifiable facial features cut effectively25. FreeSurfer determines the location of facial information and creates a brain template after manually separating brain tissue from non-brain tissue. In the subsequent images, the voxel position of non-brain tissue is obtained through the established linear transformation relationship and the voxel value of the facial information position is eliminated46. Although these two methods effectively remove facial privacy information, they also lose some areas of the original image at the same time (e.g., intracranial volume and cerebrospinal fluid volume), which may cause the problem of lacking important anatomical features when calculating certain structural metrics. To address this privacy concern, Milchenko and colleagues have developed a deface toolkit, namely face masking, which extracts the anatomical surface of the face in the volume space and then obscures it47. The recognition rate of the image is reduced by changing the image resolution, leading to ideal anonymization of the data without disturbing reliability and validity of subsequent tissue extraction and measurements of brain morphology48–51. This procedure has become a built-in function of the Extensible Neuroimaging Archive Toolkit52 for facial anonymization and been incorporated into the Human Connectome Project53. For these reasons mentioned above, when data in the ISYB database were anonymized for facial information, we adopted the CN200 Chinese template. The purpose of which is to remove more privacy and obtain a better anonymization effect for facial information in Chinese brain data. The results and discussion of data anonymization based on different ethnic brain templates are included in Usage Notes session. The ISYB data are processed by the face masking toolkit.
Data Records
As supports of the open sciences, we have released the multimodal neuroimaging data of the 215 participants in the ISYB database for both research and education in human brain mapping as well as culture association studies. Specifically, as demonstrated in the previous sections, we first applied the face masking pipeline based on CN200 brain template to anonymize the MRI data by blurring facial information while all the participants agreed to share their anonymized data to the public. We then organized the data according to the Brain Imaging Data Structure (BIDS: https://bids.neuroimaging.io)54 and employed the MRIQC toolkit for monitoring the quality of all structural and functional images. This toolkit generated comprehensive reports on the data quality, which were presented as in the Supplementary Fig. 1 and Supplementary Fig. 2 at group level. All the BIDS-formatted data and related files on the QC reports have been publicly shared through the Science Data Bank (SciDB) at the National Basic Science Data Center (10.11922/sciencedb.00740)55.
The ISYB_DATA folder contains 215 folders labeled with participant ID numbers (e.g., sub-0002). Each participant’s folder contains subfolders labeled func containing the rfMRI images (.nii.gz), anat containing defaced T1-weighted sMRI images (.nii.gz), perf containing aMRI images (.nii.gz), dwi containing dMRI images (.nii.gz,.bval,.bvec). The ISYB_QC folder contains QC reports generated by MRIQC toolkit for each participant. All QC results were integrated into webpages, and linked in an overview page on group analysis. Of note, the current ISYB release contained the demographic information (age and sex) but did not include any behavioral and clinical measurements.
Technical Validation
As in the Consortium for Reliability and Reproducibility (CoRR)9, we demonstrate the utility of the released ISYB multi-modal MRI data. All the anonymized and quality-controlled data were preprocessed using the Connectome Computation System (CCS56, https://github.com/zuoxinian/CCS) with the most recent updates57. This pipeline integrates multiple analytical software packages to achieve imaging processing of multi-modal MRI data. Considering the advantages of surface-based functional brain mapping58, we reconstructed cortical surface models to generate NIFTI file for structural and functional metrics on the cortical surface. The structural image went through the following preprocessing steps: (1) spatially adaptive non-local means denoising, (2) rough inhomogeneity correction, (3) spatial normalization into the MNI standard brain space, (4) inhomogeneity correction, (5) intensity normalization, (6) brain extraction by non-local intracranial cavity extraction (NICE), and (7) gray and white matter segmentation, surface reconstruction. The rfMRI image preprocessing included (1) dropping off the first 5 EPI volumes, (2) removing and interpolating temporal spikes, (3) correcting acquisition timing among image slices and head motion among image volumes, (4) normalizing the 4D global mean intensity to 10,000, (5) regressing out head motion artifacts and other spurious noise by using ICA-AROMA59,60, and (6) removing linear and quadratic trends from the rfMRI signals to mitigate the scanner-related influences56.
The preprocessed NIFTI images were converted into the GIFTI format using the Ciftify toolbox61. The sMRI images were further preprocessed using the custom pipeline to generate highly refined cortical surface meshes which were spatially normalized to the MNI space and re-sampled to have 32k vertices, i.e., the fsaverage_LR32 cortical surface. Individual dMRI and aMRI images were preprocessed in individual volume space and then transformed onto the fsaverage_LR32 space. Individual surface-mapped rfMRI signals were then brought into the register across participants using a multi-modal surface matching algorithm to the and vectorized into the CIFTI format. This maps each surface vertex to an index in a vector by the Ciftify toolbox. A group-level surface mask was finally established by including every vertex where all the 215 participants obtained the full rfMRI signals. Based on the surface-based data, we derived two sets of brain imaging metrics using CCS for both structural and functional characteristics, respectively. Specifically, following derivatives were calculated:
Cortical Thickness (CT)62: The average distance between the gray/white boundary and the pial surface within each ROI.
Surface Area (SA)62: The sum of the areas of each tessellation falling within each ROI.
Gray Matter Volume (GMV): The measure of the density of brain cell body in a particular region.
Local Gyrification Index (LGI)63: The ratio of a regional surface area for the pial surface to a smooth surface contour estimated to wraps around the pial surface.
Fractional Anisotropy (FA)64: An index for the amount of diffusion asymmetry within a voxel, defined in terms of its eigenvalues.
Mean Diffusivity (MD)64: The mean amount of diffusion in each of the principal directions calculated in the tensor.
Regional Homogeneity (ReHo)58: The synchronicity of a voxel’s time series and that of its nearest neighbors based on Kendall’s coefficient of concordance to measure the local brain functional homogeneity.
Voxel-Mirrored Homotopic Connectivity (VMHC)65: The functional connectivity between a pair of geometrically symmetric, inter-hemispheric voxels.
Amplitude of Low-Frequency Fluctuations (ALFF)66: The total power in the low frequency range (0.01–0.1 Hz) of an fMRI image, normalized by the total power across all frequencies measured in that same image.
Cerebral Blood Flow (CBF)67: The rate of delivery of arterial blood to the capillary bed in brain tissue and is quantified in milliliters of blood per 100 g of brain tissue per minute.
We note that each metric was calculated within the group-level surface mask. The CT metric was generated and extracted from the structural outcomes of ciftify in the fsaverage_LR32k space. The LGI metric was produced by the FreeSurfer scripts (https://surfer.nmr.mgh.harvard.edu/fswiki/LGI). The SA, ReHo, ALFF and VMHC metrics were calculated by the CCS scripts from the sMRI and rfMRI outcomes of ciftify. CCS also provided a computational workflow for dMRI data to derive both FA and MD for gray matter and white matter voxels in individual volume spaces. The results were named as FA-WM, MD-WM, FA-GM and MD-GM, respectively. The bbregister command in FreeSurfer was then applied to the native individual maps of CBF, SA, FA and MD to register these individual volume maps onto the fsaverage surface template. The wb_command -metric-resample command in the HCP workbench was used to map the native individual CBF, SA, FA and MD metrics to fs_LR and resample them to the 32k space. The vertex-wise group-level maps across all qualified participants are shown on the fsLR_32k surface as in Fig. 3 (mean) and Fig. 4 (SD).
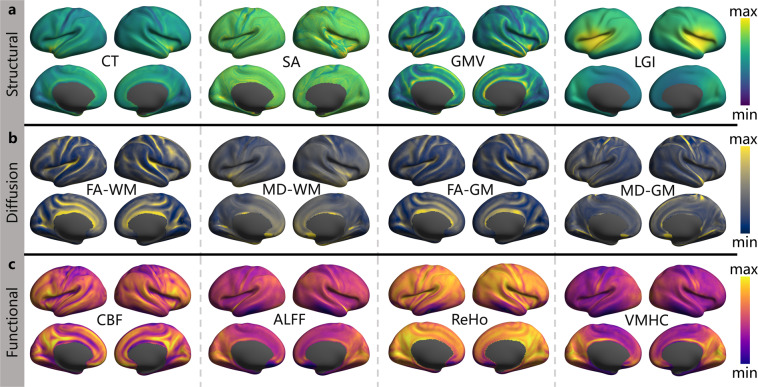
Fig. 3.
The group mean maps of the ISYB multi-modal neuroimaging data on the fsLR_32k cortical surfaces. Given each vertex on the surface, the mean values were calculated across all the 215 participants for the 12 metrics based on the following three MRI modalities: (a) sMRI (CT, SA, GMV, LGI), (b) dMRI (FA-WM, MD-WM, FA-GM, MD-GM), (c) rfMRI and aMRI metrics (CBF, ALFF, ReHo and VMHC).
Fig. 4.
The group variability maps of the ISYB multi-modal neuroimaging data on the fsLR_32k cortical surfaces. Given each vertex on the surface, the standard deviation values were calculated across all the 215 participants for the 12 metrics based on the following three MRI modalities: (a) sMRI (CT, SA, GMV, LGI), (b) dMRI (FA-WM, MD-WM, FA-GM, MD-GM), (c) rfMRI and aMRI metrics (CBF, ALFF, ReHo and VMHC).
Usage Notes
We encourage other labs to use this dataset in publication under the requirement of citing this article and contact us for additional data sharing and cooperation. The user of the ISYB database should acknowledge the contributions of the original authors and research lab, and properly cite this article. We note that the impact of the brain template on the facial anonymization procedure is not negligible. In this section, to evaluate the anonymization of different racial brain template, we used high-resolution sMRI images from the database. The ISYB database contains the 215 Chinese multi-modal MRI images55 and is part of the CHIMGEN consortium (see more details in the consortium paper27). We split the ISYB dataset into two subsets for replicability validation (Discovery sample: ISYB-1, n = 89; Validation sample: ISYB-2, n = 126).
Implementation of analytic framework
As illustrated in Fig. 5, we implemented an analytical framework for evaluation of the template effects on face masking. We noted that the Western brain template (i.e., MNI152 template) was employed as the default brain template in the face masking toolkit. We thus substituted the MNI152 template with the CN200 and Chinese2020 templates to establish the corresponding relationship (i.e., registration) between the two Chinese templates and the individual data space. The corresponding relationship and different ethnic templates were paired and integrated into the face masking process. The three different pipelines were then used to implement the anonymous processing of the MRI data with the different templates. As for the evaluation of the face anonymization performance, we first subtracted the anonymized image from the original image to get the facial information, which was removed from the original face. We then identified the facial areas blurred by the face masking toolkit and created a mask for each blurred surface based on the individual MRI image.
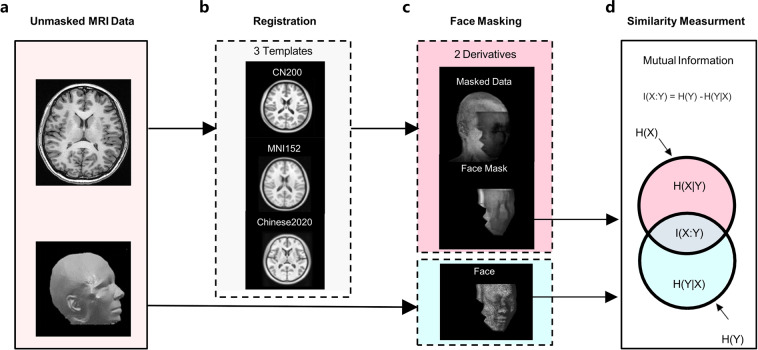
Fig. 5.
The analytic framework of the evaluation of template effects on face anonymization. (a) The face masking toolkit takes raw individual unmasked MRI data with the registration from the unmasked head image to the head template. (b) The head templates include the default MNI152 and other two Chinese (CN200 and Chinese2020) templates. (c) During the face masking, a face mask is generated and used for restoring the face removed by the face masking pipeline. (d) The normalized mutual information between the removed face and the original face is calculated.
Normalized mutual information (NMI) was used to quantify the efficiency of performing the face-masking process with different templates. NMI is widely used to evaluate the similarity between two sets of information. It serves as an important indicator in information theory and can be used to evaluate the spatial similarity or information shared between images. Its value ranges from 0 to 1. A higher NMI value indicates more spatial information shared between a pair of brain images. In the present study, a higher NMI value reveals more facial information picked up by the face-masking pipeline, and thus more efficient in removing private facial information from the original MRI head image. It means that the privacy information of the participant is blurred more efficiently. Therefore, we hypothesize that the face-masking pipelines with a race-appropriate head template would be more effective and recommendable for the anonymization of the facial information.
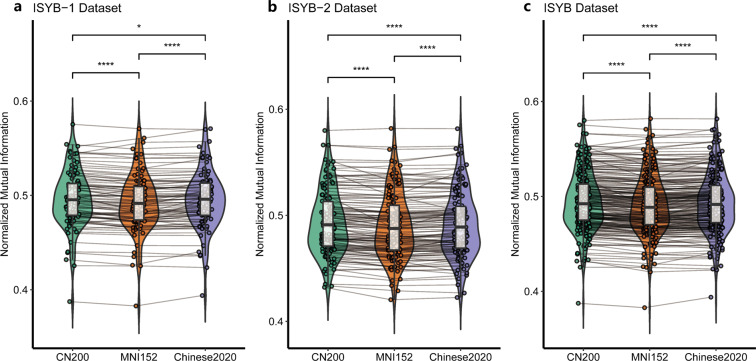
When the original brain images from Chinese (ISYB-1 and ISYB-2) passed through the face masking procedure with three different head templates (CN200, Chinese2020 and MNI152), six sets of facial masked images under the 2 × 3 different conditions were generated. The NMI values between the facial information removed under different conditions and the corresponding original data were calculated for each participant. A two-way analysis of variance with repeated measures (database versus template) was performed on the NMI values with the head template as the within-participant factor for testing the interaction effect between template and database on the face masking pipeline. The statistical model revealed a highly significant template effect (DoF = 4, F = 7.08, p = 1.42 × 10−5). We further carried out a set of post-hoc paired two-sample tests to demonstrate the directions of the effect detected. Multiple comparisons of the estimated marginal means based on the three templates in the repeated measures model were also applied. The details of the results are presented in the Table 1. Specifically, for ISYB dataset, both CN200 and Chinese2020 templates exhibited better performances of face masking than the MNI152 template while the 3.0 T MRI derived CN200 template was better than the 1.5 T MRI derived Chinese2020 template. As illustrated in Fig. 6, such effects were highly replicable across the two data subsets (ISYB-1 versus ISYB-2).
Table 1.
Statistics of the post-hoc tests on the face anonymization with the different templates.
| Dataset | Template_1 | Template_2 | Difference | Standard Error | p-value |
|---|---|---|---|---|---|
| ISYB | CN200 | Chinese2020 | 1.48 × 10−3 | 5.03 × 10−4 | 9.11 × 10−3 |
| ISYB | CN200 | MNI152 | 3.41 × 10−3 | 3.59 × 10−4 | 9.56 × 10−10 |
| ISYB | Chinese2020 | MNI152 | 1.93 × 10−3 | 4.67 × 10−4 | 1.12 × 10−4 |
Fig. 6.
The replicable template effects on face anonymization. Normalized mutual information between the face image from original data and the face image removed by the anonymization are visualized as violin plots with paired lines under the three different template (CN200, MNI152, Chinese2020) conditions. This template effect is replicated across three datasets: (a) ISYB-1, (b) ISYB-2, (c) ISYB. *p<0.05, ****p<0.0001.
Guidance and implication
With the continuous development of neuroimaging, the distinction in the brain structure across different races can become more refined. The comparison of templates of different races provides a basis for more in-depth research on the distinctions of separate races. However, many challenges have also emerged, especially when the private information is easily leaked through images. At the same time, due to the continuous development and maturity of deep learning technology, combined with big data, the entire face can be reconstructed and refabricated using a small portion of facial information. This forces us to pay special attention to protection of subjects’ private information, especially facial information. Masking the face of the data by using the face masking toolkit effectively blur the subjects’ faces, thereby protecting privacy. Meanwhile, it protects the integrity of the brain structure, which makes the structure MRI easier to interface with mainstream brain image analysis software. Follow-up analysis is more convenient without degenerating image quality. Since skull shape differs by racial and ethnic groups in length and width68, systematic biases may be introduced when images of different races are registered to the same template in the face masking toolkit. This kind of registration difference based on data and templates which are not in the same race has been emphasized in preceding articles, which is inevitable due to genetic factors41. This study assumed that using different human brain templates to anonymize facial data would yield different results69. We selected 215 data from Chinese database and three brain templates from two races, and used ethnicity-matched and ethnicity-mismatched templates to blur the facial information of the data, explored the impact of different ethnic templates on data anonymization.
After face masking was performed on Chinese data respectively using brain templates from two races, matching the data according to the ethnic template, quantifying the results according to the index of normalized mutual information, it can be concluded that: (1) There existed a significant difference in the anonymization results when the results based on the two Chinese templates were compared with that based on the Western template. The different ethnicity of the templates lead to the differences in the results when using different ethnic templates to blur face information in structural brain images by the face masking toolbox. Therefore, we believe that such variability is attributed to the race factor. (2) Choosing a template that matches the race of the data will achieve better results in comparison to with a mismatched template when using different templates to anonymize data. In other words, the template selected during anonymization should correspond to the race of the data so that better private information anonymity results can be achieved. In the process of masking the face, the surface mask algorithm proposed by Budin and colleagues was used and performed70. The face area was identified and positioned, and then smoothed by using a blur algorithm to obtain a contaminated face image. However, in the face mask algorithm, the data need to be registered on the specific template, and then the brain tissue position and skull boundary information could be found through the binary image, thereby blurring the part between the brain and skull. The registration effect varies from race to race, resulting in different positions of facial features and causing disparate phenomena based on different templates when using templates of different races as a reference to registration.
The effect of facial anonymization varies by brain templates to a large extent, the results demonstrate that there exist a difference in the effect of facial masking based on two Chinese templates, the difference may attributed to the fact that the two Chinese templates are constructed using data obtained with different field strengths (1.5 T and 3.0 T). What we want to emphasis is that the anonymization effect is determined by the ethnicity of the templates. The brain structure of individuals from different ethnic group is quite different. Although different templates can be used to anonymize structure MRI data, the use of ethnicity-matched templates will lead to the best anonymization effect. This also demonstrated that the accuracy of brain image data processing could be enhanced by using the ethnicity-appropriate templates. Improved facial anonymization, conserving less information, has been demonstrated when the Chinese head template is applied to obscure the faces of data acquired in a Chinese population. The customized pipeline using a native Chinese head template is thus recommended for the anonymization of sharing Chinese neuroimaging data. International data-sharing and the relevant large-scale team work on brain imaging such as building brain charts for the human lifespan (see a recent review21 and progress71) must consider this effect carefully. Its impact on the generalizability of brain-mind association studies10,72 is also an interesting topic in the future.
Supplementary information
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (61976150), the Major Project of National Social Science Foundation of China (20&ZD296), the National Basic Science Data Center “Chinese Data-sharing Warehouse for In-vivo Imaging Brain” (NBSDC-DB-15), the Child Brain-Mind Development Cohort Study in China Brain Initiative (SQ2021AAA010024) and the Start-up Funds for Leading Talents at Beijing Normal University.
Author contributions
X.N. Zuo, H.F. Li: study design and conceptualisation; data acquisition and formal data analysis; manuscript preparation; data curation; management of online repositories. P. Gao, H.M. Dong: data acquisition and formal data analysis; manuscript preparation; data curation; management of online repositories. All authors edited and revised the manuscript.
Code availability
All the scripts and brain templates involved for processing is publicly available as part of the recent CCS updates57 and can be visited at GitHub (https://github.com/zuoxinian/CCS/tree/master/projects/isybdemo).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Peng Gao, Hao-Ming Dong.
Contributor Information
Hai-Fang Li, Email: lihaifang@tyut.edu.cn.
Xi-Nian Zuo, Email: xinian.zuo@bnu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-022-01413-3.
References
- 1.Zuo XN. Mapping the miswired connectome in autism spectrum disorder. Journal of the American Academy of Child & Adolescent Psychiatry. 2020;59:348–349. doi: 10.1016/j.jaac.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Milham MP, et al. Assessment of the impact of shared brain imaging data on the scientific literature. Nature Communications. 2018;9:2818. doi: 10.1038/s41467-018-04976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols TE, et al. Best practices in data analysis and sharing in neuroimaging using MRI. Nature Neuroscience. 2017;20:299–303. doi: 10.1038/nn.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu H, Lei H, Fang X. Big data and the brain: Peeking at the future. Genomics, Proteomics & Bioinformatics. 2019;17:333–336. doi: 10.1016/j.gpb.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SM, Nichols TE. Statistical challenges in “big data” human neuroimaging. Neuron. 2018;97:263–268. doi: 10.1016/j.neuron.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Zuo X-N, et al. Developmental population neuroscience: emerging from ICHBD. Science Bulletin. 2018;63:331–332. doi: 10.1016/j.scib.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Fair DA, Dosenbach NU, Moore AH, Satterthwaite TD, Milham MP. Developmental cognitive neuroscience in the era of networks and big data: Strengths, weaknesses, opportunities, and threats. Annual Review of Developmental Psychology. 2021;3:249–275. doi: 10.1146/annurev-devpsych-121318-085124. [DOI] [Google Scholar]
- 8.Chung J, et al. Statistical connectomics. Annual Review of Statistics and Its Application. 2021;8:463–492. doi: 10.1146/annurev-statistics-042720-023234. [DOI] [Google Scholar]
- 9.Zuo XN, et al. An open science resource for establishing reliability and reproducibility in functional connectomics. Scientific Data. 2014;1:1–13. doi: 10.1038/sdata.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marek S, et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603:654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poldrack RA, Gorgolewski KJ. Making big data open: data sharing in neuroimaging. Nature Neuroscience. 2014;17:1510–1517. doi: 10.1038/nn.3818. [DOI] [PubMed] [Google Scholar]
- 12.Ascoli GA, Maraver P, Nanda S, Polavaram S, Armañanzas R. Win-win data sharing in neuroscience. Nature Methods. 2017;14:112–116. doi: 10.1038/nmeth.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Essen DC, et al. The WU-Minn human connectome project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harms MP, et al. Extending the human connectome project across ages: Imaging protocols for the lifespan development and aging projects. Neuroimage. 2018;183:972–984. doi: 10.1016/j.neuroimage.2018.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casey B, et al. The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Developmental Cognitive Neuroscience. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Littlejohns TJ, et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nature Communications. 2020;11:2624. doi: 10.1038/s41467-020-15948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Martino A, et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry. 2014;19:659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martino AD, et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange ii. Scientific Data. 2017;4:170010. doi: 10.1038/sdata.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biswal BB, et al. Toward discovery science of human brain function. Proceedings of the National Academy of Sciences. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, et al. Chinese Color Nest Project: An accelerated longitudinal brain-mind cohort. Developmental Cognitive Neuroscience. 2021;52:101020. doi: 10.1016/j.dcn.2021.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LZ, Holmes AJ, Zuo XN, Dong Q. Neuroimaging brain growth charts: A road to mental health. Psychoradiology. 2021;1:272–286. doi: 10.1093/psyrad/kkab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan CG, et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proceedings of the National Academy of Sciences. 2019;116:9078–9083. doi: 10.1073/pnas.1900390116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang GY, et al. Sample sizes and population differences in brain template construction. Neuroimage. 2020;206:116318. doi: 10.1016/j.neuroimage.2019.116318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrd JB, Greene AC, Prasad DV, Jiang X, Greene CS. Responsible, practical genomic data sharing that accelerates research. Nature Reviews Genetics. 2020;21:615–629. doi: 10.1038/s41576-020-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schimke, N. & Hale, J. Quickshear defacing for neuroimages. 2nd USENIX Conference on Health Security and Privacy (2011).
- 26.White T, Blok E, Calhoun VD. Data sharing and privacy issues in neuroimaging research: Opportunities, obstacles, challenges, and monsters under the bed. Human Brain Mapping. 2022;43:278–291. doi: 10.1002/hbm.25120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Q, et al. CHIMGEN: a chinese imaging genetics cohort to enhance cross-ethnic and cross-geographic brain research. Molecular Psychiatry. 2020;25:517–529. doi: 10.1038/s41380-019-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteban O, et al. MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLoS One. 2017;12:e0184661. doi: 10.1371/journal.pone.0184661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteban O, et al. Crowdsourced MRI quality metrics and expert quality annotations for training of humans and machines. Scientific Data. 2019;6:30. doi: 10.1038/s41597-019-0035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnotta VA, Friedman L, Birn F. Measurement of signal-to-noise and contrast-to-noise in the fBIRN multicenter imaging study. Journal of Digital Imaging. 2006;19:140–147. doi: 10.1007/s10278-006-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atkinson D, Hill DL, Stoyle PN, Summers PE, Keevil SF. Automatic correction of motion artifacts in magnetic resonance images using an entropy focus criterion. IEEE Transactions on Medical Imaging. 1997;16:903–910. doi: 10.1109/42.650886. [DOI] [PubMed] [Google Scholar]
- 32.Friedman L, et al. Test–retest and between-site reliability in a multicenter fMRI study. Human Brain Mapping. 2008;29:958–972. doi: 10.1002/hbm.20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saad ZS, et al. Correcting brain-wide correlation differences in resting-state FMRI. Brain Connectivity. 2013;3:339–352. doi: 10.1089/brain.2013.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai J, et al. Population differences in brain morphology and microstructure among Chinese, Malay, and Indian neonates. PLoS One. 2012;7:e47816. doi: 10.1371/journal.pone.0047816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JS, et al. Development of Korean standard brain templates. Journal of Korean Medical Science. 2005;20:483–488. doi: 10.3346/jkms.2005.20.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie WZ, Richards JE, Lei D, Lee K, Gong Q. Comparison of the brain development trajectory between Chinese and US children and adolescents. Frontiers in Systems Neuroscience. 2015;8:249. doi: 10.3389/fnsys.2014.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao NP, et al. Population differences in brain morphology: Need for population specific brain template. Psychiatry Research: Neuroimaging. 2017;265:1–8. doi: 10.1016/j.pscychresns.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Tang YC, et al. The construction of a Chinese MRI brain atlas: a morphometric comparison study between chinese and caucasian cohorts. Neuroimage. 2010;51:33–41. doi: 10.1016/j.neuroimage.2010.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie WZ, et al. The construction of MRI brain/head templates for Chinese children from 7 to 16 years of age. Developmental Cognitive Neuroscience. 2015;15:94–105. doi: 10.1016/j.dcn.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang YC, et al. Brain structure differences between Chinese and Caucasian cohorts: A comprehensive morphometry study. Human Brain Mapping. 2018;39:2147–2155. doi: 10.1002/hbm.23994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richards, J. E. & Xie, W. Brains for all the ages: structural neurodevelopment in infants and children from a life-span perspective. In Advances in child development and behavior, 48, 1–52 (Elsevier, 2015). [DOI] [PubMed]
- 43.Zhao TD, et al. Unbiased age-specific structural brain atlases for Chinese pediatric population. Neuroimage. 2019;189:55–70. doi: 10.1016/j.neuroimage.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Dong HM, et al. Charting brain growth in tandem with brain templates at school age. Science Bulletin. 2020;65:1924–1934. doi: 10.1016/j.scib.2020.07.027. [DOI] [PubMed] [Google Scholar]
- 45.Liang PP, et al. Construction of brain atlases based on a multi-center MRI dataset of 2020 Chinese adults. Scientific Reports. 2015;5:1–7. doi: 10.9734/JSRR/2015/14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bischoff GA, et al. A technique for the deidentification of structural brain MR images. Human Brain Mapping. 2007;28:892–903. doi: 10.1002/hbm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milchenko M, Marcus D. Obscuring surface anatomy in volumetric imaging data. Neuroinformatics. 2013;11:65–75. doi: 10.1007/s12021-012-9160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuo XN, Xu T, Milham MP. Harnessing reliability for neuroscience research. Nature Human Behaviour. 2019;3:768–771. doi: 10.1038/s41562-019-0655-x. [DOI] [PubMed] [Google Scholar]
- 49.Xing XX, et al. The anatomy of reliability: a must read for future human brain mapping. Science Bulletin. 2018;63:1606–1607. doi: 10.1016/j.scib.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 50.Marcus DS, et al. Open Access Series of Imaging Studies (OASIS): cross-sectional mri data in young, middle aged, nondemented, and demented older adults. Journal of Cognitive Neuroscience. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 51.Lerch JP, et al. Studying neuroanatomy using MRI. Nature Neuroscience. 2017;20:314–326. doi: 10.1038/nn.4501. [DOI] [PubMed] [Google Scholar]
- 52.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The extensible neuroimaging archive toolkit. Neuroinformatics. 2007;5:11–33. doi: 10.1385/NI:5:1:11. [DOI] [PubMed] [Google Scholar]
- 53.Glasser MF, et al. The human connectome project’s neuroimaging approach. Nature Neuroscience. 2016;19:1175–1187. doi: 10.1038/nn.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorgolewski K, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data. 2016;3:160044. doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao P, Dong HM, Wang YS, Yu CS, Zuo XN. 2021. Imaging Chinese Young Brains (I See Your Brain) [DOI]
- 56.Xu T, Yang Z, Jiang LL, Xing XX, Zuo XN. A Connectome Computation System for discovery science of brain. Science Bulletin. 2015;60:86–95. doi: 10.1007/s11434-014-0698-3. [DOI] [Google Scholar]
- 57.Xing XX, Xu T, Jiang C, Wang YS, Zuo XN. Connectome Computation System: 2015–2021 updates. Science Bulletin. 2022;67:448–451. doi: 10.1016/j.scib.2021.11.021. [DOI] [PubMed] [Google Scholar]
- 58.Zuo XN, et al. Toward reliable characterization of functional homogeneity in the human brain: Preprocessing, scan duration, imaging resolution and computational space. Neuroimage. 2013;65:374–386. doi: 10.1016/j.neuroimage.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pruim RHR, et al. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage. 2015;112:267–277. doi: 10.1016/j.neuroimage.2015.02.064. [DOI] [PubMed] [Google Scholar]
- 60.Pruim RHR, Mennes M, Buitelaar JK, Beckmann CF. Evaluation of ICA-AROMA and alternative strategies for motion artifact removal in resting state fMRI. Neuroimage. 2015;112:278–287. doi: 10.1016/j.neuroimage.2015.02.063. [DOI] [PubMed] [Google Scholar]
- 61.Dickie EW, et al. Ciftify: A framework for surface-based analysis of legacy MR acquisitions. Neuroimage. 2019;197:818–826. doi: 10.1016/j.neuroimage.2019.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panizzon MS, et al. Distinct genetic influences on cortical surface area and cortical thickness. Cerebral Cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lyu I, Kim SH, Girault JB, Gilmore JH, Styner MA. A cortical shape-adaptive approach to local gyrification index. Medical Image Analysis. 2018;48:244–258. doi: 10.1016/j.media.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexander AL, Lee JE, Lazar M, Field AS. Diffusion Tensor Imaging of the Brain. Neurotherapeutics. 2007;4:316–329. doi: 10.1016/j.nurt.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuo XN, et al. Growing together and growing apart: Regional and sex differences in the lifespan developmental trajectories of functional homotopy. Journal of Neuroscience. 2010;30:15034–15043. doi: 10.1523/JNEUROSCI.2612-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuo XN, et al. The oscillating brain: Complex and reliable. Neuroimage. 2010;49:1432–1445. doi: 10.1016/j.neuroimage.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borogovac A, Asllani I. Arterial Spin Labeling (ASL) fMRI: Advantages, theoretical constraints and experimental challenges in neurosciences. International Journal of Biomedical Imaging. 2012;2012:818456. doi: 10.1155/2012/818456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zulkifli, N. S. A. & Kamal, N. F. Does common standard brain template standardize for all brains regardless the different of age, gender and culture? Journal of Life Science and Technologies 150–153 (2013).
- 69.Bhalerao GV, et al. Construction of population-specific Indian MRI brain template: Morphometric comparison with Chinese and Caucasian templates. Asian Journal of Psychiatry. 2018;35:93–100. doi: 10.1016/j.ajp.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Budin F, Zeng D, Ghosh A, Bullitt E. Preventing facial recognition when rendering MR images of the head in three dimensions. Medical Image Analysis. 2008;12:229–239. doi: 10.1016/j.media.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bethlehem RAI, et al. Brain charts for the human lifespan. Nature. 2022;604:525–533. doi: 10.1038/s41586-022-04554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yarkoni T. The generalizability crisis. Behavioral and Brain Sciences. 2022;45:e1. doi: 10.1017/S0140525X20001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Gao P, Dong HM, Wang YS, Yu CS, Zuo XN. 2021. Imaging Chinese Young Brains (I See Your Brain) [DOI]
Supplementary Materials
Data Availability Statement
All the scripts and brain templates involved for processing is publicly available as part of the recent CCS updates57 and can be visited at GitHub (https://github.com/zuoxinian/CCS/tree/master/projects/isybdemo).