Abstract
On Bird Island, South Georgia, albatrosses (n = 140), penguins (n = 100), and fur seals (n = 206) were sampled for Campylobacter jejuni. C. jejuni subsp. jejuni was recovered from three macaroni penguins (Eudyptes chrysolophus). These isolates, the first reported for the subantarctic region, showed low genetic diversity and high similarity to Northern Hemisphere C. jejuni isolates, possibly suggesting recent introduction to the area.
Campylobacter jejuni subsp. jejuni is one of the most common causes of human bacterial enteritis in the world and is commonly found in the intestines of a variety of animals, with birds considered to be a major natural reservoir (4, 17). In domestic birds, the bacteria spread rapidly within flocks by coprophagy and through contaminated air or water (1, 5), but vertical transmission of Campylobacter is considered unlikely (13). The impact of C. jejuni subsp. jejuni on wildlife populations has not yet been clarified.
Antarctica and the subantarctic region are often regarded as virgin, pristine landscapes, unaffected by human activity. Little is known about naturally occurring, and possibly introduced, infectious agents there and their influence on animal population dynamics. Accordingly, we sampled seabirds and fur seals on Bird Island (54°00′S, 38°02′W) in the South Georgian archipelago to investigate the possible occurrence of zoonotic enteropathogens and to determine if certain bacteria could be used as tools for detecting biological pollution in this and other remote areas. Bird Island was an ideal location for our study because of its large colonies of Antarctic fur seals (Arctocephalus gazella) and seabirds, its relatively isolated location, and its comparatively low level of human activity. In this study we successfully isolated C. jejuni subsp. jejuni from 3% of Macaroni penguins (Eudyptes chrysolophus) sampled, with this apparently being the first recorded finding of such isolates for the subantarctic region.
In March 1998, fecal samples from 100 macaroni penguin nestlings, 40 black-browed albatross (Diomedea melanophrys) nestlings, 100 grey-headed albatross (Diomedea chrysostoma) nestlings, and 206 Antarctic fur seal pups were collected on Bird Island. The samples were collected with cotton wool swabs inserted into the cloaca or rectum. Samples were kept in charcoal transport medium (Transwab; BioDisc, Solna, Sweden) at 5 to 10°C and analyzed within 3 weeks of the sampling date. Samples were plated on Campylobacter selective medium (42.5 g/liter of Columbia agar base [Becton Dickinson, Cockeysville, Md.] supplemented with 5% citrated horse blood, 10 mg of vancomycin per liter, 500 IU of polymyxin B per liter, 5 mg of trimethoprim per liter) and incubated for 48 h at 42°C under microaerobic conditions. Isolates showing a gram-negative seagull-like cell morphology under light microscopy, positive reactions in catalase, oxidase and hippurate hydrolysis tests, and an ability to grow at 42°C under microaerobic conditions but not at 37°C under aerobic conditions, were presumptively identified as C. jejuni subsp. jejuni. Samples were stored at −80°C in trypticase soy broth supplemented with 15% glycerol until further species confirmation by phenotypic and genotypic methods. A total of 58 phenotypic characteristics were determined for the three isolates by standardized methods, and the results were subjected to computer-assisted analysis, as described previously (8). The 16S ribosomal DNA (rDNA) (from two isolates) and flaA (from three isolates) genes were amplified by PCR with chemically lysed (2) or boiled cells as the template (Tables 1 and 2). PCR products were purified and ligated into pGEM-T Easy Vector (Scandinavian Diagnostic Services, Falkenberg, Sweden) and subsequently transformed into Escherichia coli DH5α. Plasmids were then isolated (QIAprep Spin Miniprep kit; Qiagen, Hilden, Germany) and used as the templates for DNA sequencing (dye terminator cycle ready reaction; Perkin-Elmer, Calif.; and Thermo Sequenase II dye terminator cycle sequencing premix kit; Amersham Pharmacia Biotech, Buckinghamshire, England) (Tables 1 and 2). Nucleotide sequence analyses were performed by using the University of Wisconsin Genetics Computer Group (Madison, Wis.) sequence analysis software. Macrorestriction profiling by pulsed-field gel electrophoresis (PFGE) of SmaI- and KpnI-digested DNA was performed by a modified version of previously described methods (9, 11). Briefly, cultures were suspended to an optical density of 1.4 at 405 nm, and a 400-μl aliquot of the cell suspension was mixed with an equal amount of 2% low melt preparative grade agarose (Bio-Rad Laboratories, Sundbyberg, Sweden). DNA fragments were separated on an automated PFGE apparatus (Gene Path strain typing system; Bio-Rad). Ramping parameters for SmaI were 5.3 to 32.3 s for 4.9 h, 32.3 to 44.5 s for 7.7 h, and 44.5 to 49.9 s for 7.1 h, and ramping parameters for KpnI were 1.0 to 14.3 s for 4.6 h, 14.3 to 20.4 s for 7.2 h, and 20.4 to 23.0 s for 6.7 h. Restriction profile photographs were analyzed using the GelCompar software version 4.0 (Applied Maths, Kortrijik, Belgium). Similarities between banding patterns were assessed by using the Dice coefficient. Results were clustered by the unweighted pair group method with arithmetic average. A band position tolerance of 1.2% was applied.
TABLE 1.
PCR and sequencing primers for 16S rDNA
| Primera | Sequence | Priming siteb | Purpose |
|---|---|---|---|
| C16SF | 5′ TATGGAGAGTTTGATCCTGGCTCAG 3′ | 268 to 292 | PCR |
| B37 | 5′ TACGGYTACCTTGTTACGA 3′ | 1749 to 1731 | PCR |
| PUC1 | 5′ ACGCCAGGGTTTTCCCAG 3′ | Vector | Seqc |
| B12 | 5′ TGGCGCACGGGTGAGTAA 3′ | 366 to 383 | Seq |
| C16SF2 | 5′ GAGGATGATCAGTCACACTGGAA 3′ | 562 to 584 | Seq |
| C16SF3 | 5′ CAGGATTAGATACCCTGGTAGTC 3′ | 1022 to 1044 | Seq |
| C16SF4 | 5′ TCAGCTCGTGTCGTGAGATGT 3′ | 1308 to 1328 | Seq |
| M13Rev | 5′ CAGGAAACAGCTATGAC 3′ | Vector | Seq |
| C16SR3 | 5′ CGCGTATCTTCGAATTAAACCA 3′ | 1212 to 1191 | Seq |
| C16SR4 | 5′ GGTGATATCTACGGAATTTA 3′ | 950 to 931 | Seq |
| C16SR5 | 5′ GCAATATTCCCTACTGCTGCCT 3′ | 633 to 612 | Seq |
For primers B37 and B12, see the study by Fox et al. (2). Primers PUC1 and M13Rev were from Amersham Pharmacia Biotech. The other primers were our own constructs.
The numbers indicate the relative position on the 16S rDNA sequence of C. jejuni ATCC 43431 (GenBank accession no. Z29326).
Seq, sequencing.
TABLE 2.
PCR and sequencing primers for flaA
| Primer | Sequence | Priming sitea | Purpose |
|---|---|---|---|
| FLA4Fb | 5′ GGATTTCGTATTAACACAAATGGTGC 3′ | 86 to 111 | PCR |
| FLA1728Rb | 5′ CTGTAGTAATCTTAAAACATTTTG 3′ | 1810 to 1787 | PCR |
| FLA242FUc | 5′ CTATGGATGAGCAATTWAAAAT 3′ | 324 to 345′ | Seqf |
| FLA442Fc | 5′ CAAATCGGCGCAAGTTC 3′ | 524 to 540 | Seq |
| FLA845AFc | 5′ AAGATACCACAGGTGTTGAAGC 3′ | 927 to 948 | Seq |
| FLA1290Fmode | 5′ GGATTTTCGGCAGGTTCAGG 3′ | 1310 to 1329 | Seq |
| PUC1d | 5′ ACGCCAGGGTTTTCCCAG 3′ | Vector | Seq |
| FLA442Rc | 5′ GAACTTGCGCCGATTTG 3′ | 540 to 524 | Seq |
| FLA625RUc | 5′ CAAGWCCTGTTCCWACTGAAG 3′ | 725 to 705 | Seq |
| FLA845ARc | 5′ GCTTCAACACCTGTGGTATCTT 3′ | 948 to 927 | Seq |
| FLA1290Rmode | 5′ CCTGAACCTGCCGAAAATCC 3′ | 1329 to 1310 | Seq |
| M13Revd | 5′ CAGGAAACAGCTATGAC 3′ | Vector | Seq |
We obtained three C. jejuni subsp. jejuni isolates, all from Macaroni penguin chicks. We believe this to be the first report of C. jejuni subsp. jejuni in the Antarctic region and the first report of it being part of the microbial flora of a wild penguin species. No differences between the three strains were noted in any of the 58 phenotypic tests used to characterize them. Comparison of the phenotypic profile of the Bird Island isolates with similar data on 37 campylobacterial taxa by probabilistic methods (8) clearly identified the strains as C. jejuni subsp. jejuni, since a score (Willcox probability) exceeding 99.9% for identity to this taxon was obtained. Several results atypical of C. jejuni subsp. jejuni were observed, namely, failure to grow on unsupplemented nutrient agar or potassium permanganate-, cefoperazone-, or sodium deoxycholate nutrient agar-based media and ability to grow on MacConkey agar. However, key reactions in hippurate hydrolysis, hydrogen sulfide production, growth on a minimal medium and alpha-hemolysis tests were all typical of C. jejuni subsp. jejuni (8). The identity of the isolates as C. jejuni subsp. jejuni was validated by comparative analysis of the 16S rRNA gene sequences obtained. These were identical among the Bird Island isolates and differed in one and four bases, respectively, from published sequences derived from the type strain and a serotype reference strain of C. jejuni (GenBank accession no. L04315 and Z29326, respectively).
Two genotyping methods were applied to the penguin strains to investigate their epidemiological relationships. Sequence analysis of the flaA gene has been suggested as a sensitive tool for molecular typing of C. jejuni subsp. jejuni (6). This gene shows high intraspecies variability in Campylobacter and was 100% identical for the three isolates. In a comparison of the flaA sequence of the Bird Island isolates to published sequences, the homology was between 77.6 and 95.9%. The highest percentage of similarity obtained was to the flaA sequence of strain D772 (GenBank accession no. AF050185), isolated in 1983 or 1984 from a retail chicken in Seattle, Washington (R. J. Meinersmann, personal communication).
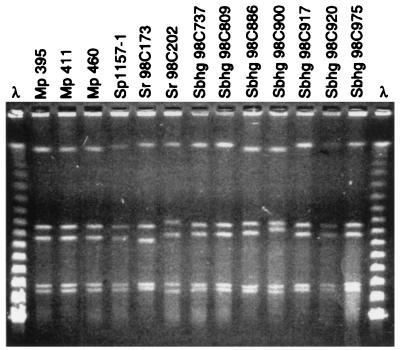
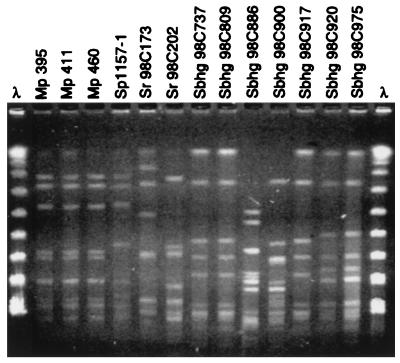
Macrorestriction profiling of whole-cell DNA by PFGE is a powerful method for epidemiological studies of Campylobacter species (3, 11). Two subsequent digestions with SmaI and KpnI, respectively, were performed, as it has been shown that KpnI can distinguish further certain SmaI-defined macrorestriction types (3, 11). A previous study of 43 (8.3%) C. jejuni subsp. jejuni isolates sampled from migratory birds (n = 498) arriving in Sweden and of 12 isolates originating from poultry showed a high variability in macrorestriction profiles (MRPs) (T. Broman, unpublished data). In contrast, SmaI- and KpnI-derived MRPs of the three Bird Island isolates were indistinguishable (Fig. 1 and 2). In accord with previously published criteria (11), these results, combined with those of the other phenotypic and genotypic analyses presented here, strongly indicate that the strains represent a single clone and may be genetically identical.
FIG. 1.
C. jejuni subsp. jejuni strains from different birds and having indistinguishable or near-identical MRPs after digestion by restriction endonuclease SmaI. Sbhg, Swedish black-headed gull; Sr, Swedish rook; Mp, Macaroni penguin; Sp, Swedish poultry. Lanes λ, molecular weight marker (λ ladder). Swedish gulls were sampled in an urban park during a 2-week period.
FIG. 2.
Endonuclease KpnI MRPs of the C. jejuni subsp. jejuni isolates listed in Fig. 1. Sbhg, Swedish black-headed gull; Sr, Swedish rook; Mp, Macaroni penguin; Sp, Swedish poultry. Lanes λ, molecular weight marker (λ ladder).
Should we consider C. jejuni subsp. jejuni endemic or introduced to Bird Island? Despite the comparatively remote location of Bird Island, a number of possible vectors, or routes of infection, exist. Seabirds are known to cover large distances in their search for food and could encounter infectious organisms found in areas with higher levels of human activity. Further, an increasing number of humans visit Antarctica and the subantarctic region, for professional or recreational reasons, and such human activity could result in the introduction of exotic microorganisms to the area. Some of our results suggest that C. jejuni subsp. jejuni may have been transmitted to the area relatively recently. If C. jejuni subsp. jejuni was indigenous to the area, it would be reasonable to expect other bird species on Bird Island to be carriers as well, especially since this bacterium is known to have a broad host range (14). It is also noteworthy that the same clone was isolated from each of the three birds. This could be a reflection of the existence of a well-adapted strain that has out-competed all others, of this particular strain having a higher ability to survive prolonged transportation, or of contamination of samples. The latter is most unlikely, due to the susceptibility of C. jejuni subsp. jejuni to excessive amounts of oxygen, which reduces its ability to survive in the environment. Also, at the time of culturing the samples, no other bird samples were handled in the laboratory.
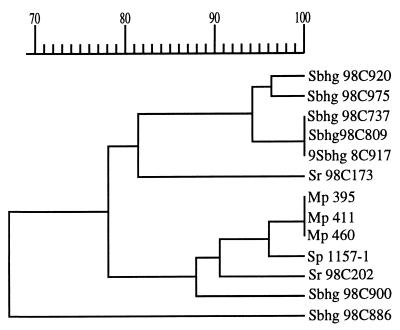
The similarity of the isolates could also be the result of a common source of infection for the three carrier penguins. In many respects a Macaroni penguin colony could be considered analogous to a domestic poultry flock. In both cases, large numbers of individuals are congregated together in a relatively small area. Macaroni chicks leave the nest at an early age and roam around in groups, and the birds therefore are at increased risk of encountering and spreading infectious organisms compared to more sedentary or solitary species. Poultry frequently peck the floor or ground looking for food, while penguins have the habit of pecking at each other and picking up objects like stones from the ground. Considering the low number of C. jejuni subsp. jejuni bacteria needed to establish an infection (12), the natural behavior of both chickens and Macaroni penguins could facilitate the rapid spread of this microorganism through a flock or colony. Consequently, when a single C. jejuni subsp. jejuni strain is introduced to a previously uninfected domestic poultry flock, individual birds may be rapidly colonized (1), carrying the same clone in their feces. Genetic diversity in Campylobacter strains can arise by several different mechanisms and may be detected by a variety of genotypic methods (10, 16). These phenomena may account for the results of a recent study of C. jejuni subsp. jejuni in poultry, which indicated that different genotypes could be found within the same flock or the same batch of poultry products from a manufacturer (15, 16). Similar effects could be expected with penguins. In view of the above-mentioned studies, we cannot exclude the possibility that C. jejuni subsp. jejuni was recently introduced to the area. In this respect, it is noteworthy that the dendrogram (Fig. 3) based on combined SmaI- and KpnI-based PFGE patterns (Fig. 1 and 2) indicates that the Bird Island isolates resemble those found among many Swedish bird isolates (69 to 95% similarity). Moreover, for one poultry isolate investigated (Sp 1157-1), the SmaI MRP is indistinguishable from those of the Bird Island isolates, and the corresponding KpnI MRP shows a higher resemblance to those of the Bird Island isolates than to any others. These similarities are reflected in the dendrogram (Fig. 3). Nevertheless, further investigations of the natural microbiological flora of wildlife in the subantarctic region are needed before we can determine if the presence there of C. jejuni subsp. jejuni is a result of microbial pollution.
FIG. 3.
Dendrogram showing relationship of combined SmaI and KpnI MRPs from C. jejuni subsp. jejuni of different bird origin. Sbhg, Swedish black-headed gull; Sr, Swedish rook; Mp, Macaroni penguin; Sp, Swedish poultry.
Acknowledgments
We gratefully acknowledge the support given to this project by the late Peter Prince. We also thank Simon Berrow and staff at the British Antarctic Survey Base on Bird Island for assistance in collecting samples and Ian Collinge and Kath Nicholson (BAS) for arranging the transport of materials. Swedish poultry samples were kindly provided by the Department of Food Hygiene, Faculty of Veterinary Medicine, Uppsala, Sweden. Furthermore, we thank Penny Jordan for excellent technical assistance, and John Croxall, Peter Vandamme, and Paul Haemig for valuable comments on the manuscript.
This work was financially supported by the Center for Environmental Research, the Medical Faculty of Umeå University, the Swedish Council for Forestry and Agricultural Research (23.0161), the Swedish Society of Medicine, and the Swedish Medical Research Council (07922).
REFERENCES
- 1.Berndtson E. Campylobacter in broiler chickens. Ph.D. thesis. Uppsala, Sweden: Swedish University of Agricultural Sciences; 1996. [Google Scholar]
- 2.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson J R, Fitzgerald C, Owen R J. Comparison of PFGE, ribotyping and phage-typing in the epidemiological analysis of Campylobacter jejuni serotype HS2 infections. Epidemiol Infect. 1995;115:215–225. doi: 10.1017/s0950268800058349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapperud G, Rosef O. Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Appl Environ Microbiol. 1983;45:375–380. doi: 10.1128/aem.45.2.375-380.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapperud G, Skjerve E, Vik L, Hauge K, Lysaker A, Aalmen I, Ostroff S M, Potter M. Epidemiological investigation of risk factors for Campylobacter colonization in Norwegian broiler flocks. Epidemiol Infect. 1993;111:245–255. doi: 10.1017/s0950268800056958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meinersmann R J, Helsel L O, Fields P I, Hiett K L. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J Clin Microbiol. 1997;35:2810–2814. doi: 10.1128/jcm.35.11.2810-2814.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachamkin I, Bohachick K, Patton C M. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J Clin Microbiol. 1993;31:1531–1536. doi: 10.1128/jcm.31.6.1531-1536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.On S L W, Holmes B, Sackin M J. A probability matrix for the identification of campylobacters, helicobacters, and allied taxa. J Appl Bacteriol. 1996;81:425–432. doi: 10.1111/j.1365-2672.1996.tb03529.x. [DOI] [PubMed] [Google Scholar]
- 9.On S L W, Vandamme P. Identification and epidemiological typing of Campylobacter hyointestinalis subspecies by phenotypic and genotypic methods and description of novel subgroups. Syst Appl Microbiol. 1997;20:238–247. [Google Scholar]
- 10.On S L W. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol Lett. 1998;165:341–346. doi: 10.1111/j.1574-6968.1998.tb13167.x. [DOI] [PubMed] [Google Scholar]
- 11.On S L W, Nielsen E M, Engberg J, Madsen M. Validity of Sma-defined genotypes of Campylobacter jejuni examined by SalI, KpnI and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol Infect. 1998;120:231–237. doi: 10.1017/s0950268898008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz-Palacios G M, Escamilla E, Torres N. Experimental Campylobacter diarrhea in chickens. Infect Immun. 1981;34:250–255. doi: 10.1128/iai.34.1.250-255.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shane S M, Lee A, Sorrell T C. Campylobacter jejuni in broilers: the role of vertical transmission. J Hyg Camb. 1986;96:153–159. doi: 10.1017/s002217240006592x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skirrow M B. Diseases due to Campylobacter, Helicobacter and related bacteria. J Comp Pathol. 1994;111:113–149. doi: 10.1016/s0021-9975(05)80046-5. [DOI] [PubMed] [Google Scholar]
- 15.Thomas L M, Long K A, Good R T, Panaccio M, Widders P R. Genotypic diversity among Campylobacter jejuni isolates in a commercial broiler flock. Appl Environ Microbiol. 1997;63:1874–1877. doi: 10.1128/aem.63.5.1874-1877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wassenaar T M, Geilhausen B, Newell D G. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yogasundram K, Shane S M, Harrington K S. Prevalence of Campylobacter jejuni in selected domestic and wild birds in Louisiana. Avian Dis. 1989;33:664–667. [PubMed] [Google Scholar]