Abstract
A new SimPlate heterotrophic plate count (HPC) method (IDEXX Laboratories, Westbrook, Maine) was compared with the pour plate method at 35°C for 48 h. Six laboratories tested a total of 632 water samples. The SimPlate HPC method was found to be equivalent to the pour plate method by regression analysis (r = 0.95; y = 0.99X + 0.06).
Water utilities are required to maintain a detectable disinfection residual in water distribution systems or measure for heterotrophic plate count (HPC) bacteria (6). The standard HPC pour plate method is an approved U.S. Environmental Protection Agency USEPA method (5) for reporting HPC in lieu of testing for residual disinfectant concentration or for testing when residual disinfectant levels are less than 0.2 mg/liter in finished waters (4). This method, as well as other HPC methods, such as membrane filtration or spread plating, may be also used to collect data for internal purposes (nonreporting). All the methods (1) for testing of heterotrophic bacteria require time-consuming preparation of media and can be difficult to read. Recently, the SimPlate total plate count method for determining the most probable number (MPN) of microorganisms in food was developed by IDEXX Laboratories, Westbrook, Maine (3), and approved by the Association of Official Analytical Chemists (AOAC) International Research Institute (2). The formulation was modified to allow for the detection of heterotrophic bacteria in water. The test known as SimPlate for HPC medium contains substrates that are hydrolyzed by microbial enzymes to release 4-methylumbelliferone, which fluoresces blue under a long-wavelength (365-nm) 6-W UV light after incubation for 48 h at 35°C. The bacteria are detected as fluorescent wells on the SimPlate. The bacterial density of a water sample is determined by determining the number of positive wells and by using the MPN table provided for SimPlate. This format will allow a MPN/milliliter value up to 738 without any dilution. The objective of this study was to compare the performance of SimPlate and the HPC pour plate method (1) for the enumeration of heterotrophic bacteria. Six laboratories in different regions of the United States participated in this study.
Between May and June 1997, naturally occurring heterotrophic bacterium samples were collected in sterile vessels at each site. The samples consisted of chlorinated drinking waters (neutralized with sodium thiosulfate) (1), well waters, untreated natural (raw) waters (lakes and streams), and secondary chlorinated sewage effluents (neutralized with sodium thiosulfate). Since chlorinated drinking waters have relatively few or no heterotrophic bacteria, the sites prepared composites of raw waters and/or secondary effluent with neutralized chlorinated drinking water. The composite samples were prepared in ratios of 1:1, 1:2, and 2:1 to allow as broad a range of bacterial counts as possible to be represented in the study. Each site was requested to test 100 samples for the evaluation. Approximately 40% of the samples were to be from finished waters (defined in this study as chlorinated drinking water and well water), 30% were to be from raw waters, and 30% were to be from composites of finished and raw waters.
No dilution was required for finished waters and was labeled as 100; dilutions of 10−1 and 10−2 and/or 10−3 were required for raw waters and composites. All samples were tested in duplicate.
SimPlate for HPC is available as a unit-dose (1 test) or as a multi-test (10 tests) medium (used for this study) from the manufacturer. This kit consists of 10 foil-wrapped, sterile polystyrene vessels with dehydrated medium and four plastic sleeves holding 25 sterile SimPlates each. The foil wrap enclosing the vessel was removed, and the vessel was opened aseptically to hydrate the medium with 100 ml of sterile diluent (sterile deionized water or sterile buffered water). SimPlates to be used were removed from the sterile plastic sleeve, and the remaining SimPlates were stored in the sleeve sealed with tape. A blank was performed with each run using sterile diluent to verify SimPlate sterility.
One milliliter of the water sample was placed on the center-landing pad of the SimPlate by using a sterile pipette. Nine milliliters of hydrated medium was then placed on top of the sample, by using a sterile pipette, to achieve a total volume of 10 ± 0.2 ml. The mixture of the sample and the medium was distributed in all wells by gently swirling and tilting the plate. Air bubbles were observed in some of the wells, although IDEXX Laboratories indicated that this would not have any effect on the test (3). This was not evaluated during the study. Approximately half of this mixture was removed by pouring off all of the excess through the indented spout on the base of the SimPlate. The SimPlates were inverted to prevent condensation on the covers and incubated at 35 ± 0.5°C for 48 ± 2 h. The number of wells in each SimPlate exhibiting a blue fluorescence when exposed to a long-wavelength (365 nm), 6-W UV light was recorded as positive results. The MPN/milliliter was obtained by using the IDEXX MPN table provided, and where applicable, this value was multiplied by the dilution factor to obtain the corrected MPN/milliliter.
The pour plate count agar was prepared as described in Standard Methods (1). One milliliter of the sample was placed on the center of a sterile petri dish (100-mm diameter) by using a sterile pipette. Sterile, molten (44 to 46°C) plate count agar (pH 7.0; Difco, Detroit, Mich.) was added and mixed with the sample by swirling the plate. The samples were allowed to cool at room temperature until solidified and then were inverted and incubated at 35 ± 0.5°C for 48 ± 2 h. Colonies formed in or on the plate count medium within 48 ± 2 h were counted as described in Standard Methods (1), and the results were reported as CFU/milliliter. Where applicable, this value was multiplied by the dilution factor to obtain the corrected CFU/milliliter.
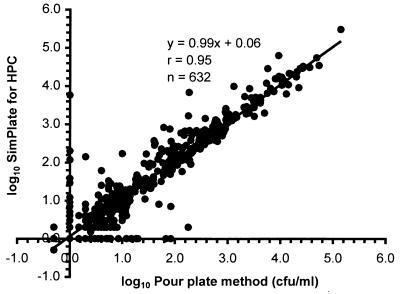
The MPN/milliliter of the samples for SimPlate was compared to the CFU/milliliter from the pour plate method. All of the data were first converted to log10 values. For the samples yielding zero counts, a value of 1 MPN/ml or 1 CFU/ml was used to enable inclusion on scatter plots showing log10 MPN/milliliter for SimPlate against log10 CFU/milliliter for the HPC pour plate method. Linear regression analysis (correlation coefficient [r], y intercept, and slope) was used to compare the two methods. Table 1 indicates the number of samples tested at each site along with the linear regression analysis comparing results of the two methods. Figure 1 represents the combined linear regression graph for all six sites.
TABLE 1.
SimPlate HPC versus HPC pour plate field trial data
| Site | na | r | Slope | y intercept |
|---|---|---|---|---|
| Maine | 136 | 0.96 | 0.96 | 0.09 |
| Massachusetts | 95 | 0.92 | 1.08 | −0.03 |
| California | 157 | 0.98 | 1.04 | −0.15 |
| Georgia | 100 | 0.98 | 1.04 | −0.04 |
| Texas | 114 | 0.86 | 0.99 | 0.22 |
| Indiana | 30 | 0.95 | 0.87 | 0.47 |
| Combined | 632 | 0.95 | 0.99 | 0.06 |
Samples composed of approximately 50% finished and 50% raw and composite water samples.
FIG. 1.
SimPlate combined field test sites (comparison of 632 samples, SimPlate versus pour plate).
A total of 632 samples consisting of 320 finished waters (50.6%), 222 raw waters (35.1%), and 90 composites (14.2%) were evaluated by the six sites. Twenty-four additional samples were eliminated from the analysis because the value from one or both methods was greater than the upper limit. A strong positive correlation was found between both methods (r = 0.95), with a slope of 0.99 and a y intercept of 0.06 for all sites. The slope and correlation were not different from 1, and the y intercept was not different from zero. All of this strongly suggests that SimPlate for HPC produced results equivalent to those of the standard HPC method, indicating suitability of the SimPlate as an alternative test method for HPC in water. SimPlate is easy to use and does not require preparation of media or sterilization. Counting of positive fluorescent wells is an easy process, does not require a colony counter, and takes less time than counting colonies on the standard HPC plate.
Acknowledgments
We thank Gil Dichter of IDEXX Laboratories for technical assistance and for the materials to perform this study. We also acknowledge the following who participated in the evaluation: Shirley Bellock, Clinical Labs of San Bernardino #2; Ceclia O'Byran, Raxa Patel, William Wilson, and Richard Dufour, Indiana State Department of Health; and a Massachusetts Water Utility (who requested anonymity).
REFERENCES
- 1.American Public Health Association. Section 9215A and B. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 2.AOAC Research Institute. The referee, inside lab management, P35-36. Washington, D.C.: Association of Official Analytical Chemists; 1997. [Google Scholar]
- 3.Beuchat L R, Copeland F, Curiale M S, Danisavich T, Gangar V, King B W, Lawlis T W, Likin R O, Okwusoa J, Smith C F, Townsend D E. Comparison of the SimPlate™ total plate count method with Petrifilm, Redigel, and conventional pour plate methods for enumerating aerobic microorganisms in foods. J Food Prot. 1998;61:14–18. doi: 10.4315/0362-028x-61.1.14. [DOI] [PubMed] [Google Scholar]
- 4.Code of Federal Regulations. 1994. CFR40 Part 141.74 (a) 3.
- 5.Code of Federal Regulations. 1994. CFR40 Part 141.73.
- 6.Federal Register. 1989. 54:27486–27568.