Summary
Gene functions can be assessed in mouse embryonic stem (ES) cells and in mutant mice derived from mutant ES cells. Here, we describe an approach for efficient isolation of the ES clones carrying deletion mutations at the target genes by CRISPR-Cas9. Two sgRNAs against a target gene are co-expressed with puromycin-resistant gene in ES cells through co-transfection followed by transient puromycin selection. Deletion mutations are identified by PCR from individual ES clones that are picked from puromycin-selected ES cells.
Subject areas: Genetics, Molecular Biology, CRISPR, Stem Cells
Graphical abstract

Highlights
-
•
Express two different sgRNAs against a target gene in ES cells by co-transfection
-
•
Deletion mutation generated with two sgRNA target sites
-
•
Enrichment of transfected ES cells by puromycin selection for 2 days
-
•
Screening for candidate deletion mutant ES clones by PCR
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Gene functions can be assessed in mouse embryonic stem (ES) cells and in mutant mice derived from mutant ES cells. Here, we describe an approach for efficient isolation of the ES clones carrying deletion mutations at the target genes by CRISPR-Cas9. Two sgRNAs against a target gene are co-expressed with puromycin-resistant gene in ES cells through co-transfection followed by transient puromycin selection. Deletion mutations are identified by PCR from individual ES clones that are picked from puromycin-selected ES cells.
Before you begin
CRITICAL: For this protocol, it is essential to culture ES cells on the puromycin-resistant feeder cells during selection of the transfected ES cells with puromycin. This significantly results in higher proportion of mutant ES cells when individual ES cell colonies are picked and cultured for identification of the candidate mutant ES clones. We take the following steps to prepare feeder cells that are resistant to puromycin.
-
1.
We derive mouse embryonic fibroblast (MEF) cells that are resistant to four drugs from the mice expressing these four drug resistance genes that were originally obtained from The Jackson Laboratory (Tucker et al., 1997). We have named these derived MEF cells as DR4 MEF cells in this protocol.
-
2.
We use irradiation to make puromycin-resistant feeder cells from DR4 MEF cells.
We have named these feeder cells as DR4 feeder cells in the protocol.
CRITICAL: For transfection, we prefer to use freshly plated ES cells on the DR4 feeder cells seeded on a small 3.5-cm dish plate or on a well of 6-well plate. Good quality of DNA samples should be used in order to achieve optimal transfection efficiency. If necessary, purify the DNA samples with the PCR purification columns and elute the DNA samples with sterile PBS solution first before transfection into ES cells.
Note: We have successfully utilized this protocol to obtain deletion mutations in three different mouse ES cell lines (D1911, D1906 and TC1) in our lab, with D1911 ES cells used for most experiments (Deng et al., 1995; Lau et al., 2016). This protocol should be applicable to other available mouse ES cell lines. It may be also suitable for generation of deletion mutations in human ES cells with some necessary changes in the protocol because human ES cell culture condition is different from the mouse ES cell culture condition. In addition, this approach might be tested in feeder-free or serum-free ES culture experiments in the future to find out if it will work equally well.
Institutional permissions
The animal protocol for this study was approved by the Institutional Animal Care and Use Committee (IACUC) of ShanghaiTech University in China. We have followed it carefully in our animal experiments.
Preparation of feeder cells for ES cell culture
Timing: 9 days
The feeder cells were made from both DR4 MEF cells and SNL cells (McMahon and Bradley, 1990; Tucker et al., 1997). The feeder cells made from DR4 MEF cells were used for ES cell transfection followed by puromycin selection, whereas the feeder cells made from SNL cells had been employed for all other ES cell culture experiments in this protocol.
The dosage of irradiation is determined for the MEF cells to produce feeder cells. Then MEF cells are expanded in culture before being irradiated to make large quantities of feeder cells for ES cell culture experiments in the future. The feeder cells are aliquoted in freezing vials and then stored in liquid nitrogen for long-term use.
-
3.Determine the irradiation conditions for MEF cells to make feeder cells.
-
a.Test a few different dosage conditions for irradiation of the MEF cells.
-
b.The irradiated MEF cells are plated on a dish plate and then split 1:5 when they are attached to the plates next day.
-
c.Examine the irradiated MEF cells for growth arrest and viability on the new plates for a week or longer. Then determine the optimal irradiation condition for making feeder cells out of the MEF cells.
-
a.
CRITICAL: The irradiated MEF cells are arrested in cell growth, but remain alive in culture for more than one week. Use this irradiation condition for the expanded MEF cells to make feeder cells in the following steps.
Note: To make the DR4 feeder cells, subject the DR4 MEF cells to irradiation with a dosage of 3750 rads. We use the dosage of 7500 rads to make the SNL feeder cells by irradiation.
-
4.Grow MEF cells.
-
a.Thawing of frozen MEF cells.Thaw approximately 2 × 106 MEF cells from one freezing vial and then plate them onto two 10-cm dishes in a total volume of 10 mL of feeder cell media per plate with about 1 × 106 MEF cells on each plate.
-
b.Expansion of MEF cells.
-
i.About 3 days later when they become relatively confluent, the MEF cells on each plate are passaged onto five 10-cm dish plates to obtain ten 10-cm dish plates of MEF cells in total at this step.
-
ii.It may take another 3 days for the MEF cells to become confluent again. Then the MEF cells on each plate are passaged onto five 10-cm dish plates again to obtain a total of fifty 10-cm dish plates of MEF cells after two successive passages.
-
i.
-
a.
-
5.Irradiation of MEF cells to make feeder cell.
-
a.Harvest expanded MEF cells.Harvest all MEF cells from fifty 10-cm dish plates. Combine the MEF cells after centrifuge and resuspension of the cell pellets. Then resuspend the combined MEF cells in two 50-mL conical tubes before irradiation in the next step below.
-
b.Irradiation of MEF cells.After the dosage is determined for irradiation to make feeder cells in the testing step above, the harvested MEF cells in two 50-mL conical tubes are subject to this irradiation condition.
-
a.
-
6.Making aliquots of the feeder cells in freezing vials for long-term storage.Note: The irradiated MEF cells can be used as the feeder cells for the ES cell culture right now. Since a lot of feeder cells are obtained from the MEF cells grown on fifty 10-cm dish plates after irradiation, they are usually sufficient for many ES cell culture experiments. The feeder cells in freezing medium are usually stored in liquid nitrogen and they stay alive for many years. The frozen feeder cells may be thawed later when they are needed for the ES cell culture experiments in the future.
-
a.Resuspend the irradiated MEF cells by pipetting 15 times with a 15-mL serological transfer pipette and then perform serial dilutions to calculate the total number of the irradiated MEF cells, i.e., feeder cells, obtained from the experiment.
-
b.Centrifuge two 50-mL conical tubes with feeder cells at 1,000 g for 10 min. Remove feeder cell medium from two 50-mL conical tubes after centrifuge.
-
c.Resuspend the feeder cells in the right volume of the feeder cell freezing medium. About 5 × 106 feeder cells are resuspended in 1 mL of freezing feeder cell medium and stored in a freezing vial in liquid nitrogen for later use.
 Pause point: This step completes the production of feeder cells. They may be used for ES cell culture immediately. Alternatively, they are stored in freezing vials in liquid nitrogen until they are needed. Then the frozen feeder cells are thawed and seeded on the plates the day before ES cells are grown on the feeder cells.
Pause point: This step completes the production of feeder cells. They may be used for ES cell culture immediately. Alternatively, they are stored in freezing vials in liquid nitrogen until they are needed. Then the frozen feeder cells are thawed and seeded on the plates the day before ES cells are grown on the feeder cells.
-
a.
Preparation of the constructs for expressing sgRNAs against the gene of the interest
Timing: 3 days
The CRISPR-Cas9 genome engineering system has been widely used to generate mutations in the endogenous loci since its discovery (Cong et al., 2013; Doudna and Charpentier, 2014; Hsu et al., 2014; Jinek et al., 2012; Mali et al., 2013). We had used this CRISPR-Cas9 method to generate mutant ES clones for Zfp57 and Zfp445, two genes involved in the maintenance of genomic imprinting (Jiang et al., 2021; Li et al., 2008; Takahashi et al., 2019). We had also successfully applied this approach to other genes in the lab.
Note:Zfp445 is used as an example for the study (Figure 1A). Two primers (Zfp445-sgRNA1-top and Zfp445-sgRNA1-bottom) are annealed to make the double-strand DNA fragment for cloning into pX330 to express one target sgRNA of Zfp445 after transfection into ES cells, whereas another two primers (Zfp445-sgRNA2-top and Zfp445-sgRNA2-bottom) are annealed before being cloned into pX330 to express the other target sgRNA of Zfp445 upon ES cell transfection (see key resources table below) (Pyzocha et al., 2014; Ran et al., 2013). These two sgRNA constructs will produce two sgRNA molecules targeting two different regions of Zfp445 when they are co-transfected into ES cells so that deletion mutation may be generated for Zfp445 upon CRISPR-mediated excision targeted by these two sgRNAs (Figure 1).
CRITICAL: In general, we selected the sgRNA target sites out of the ones with the best scores based on the website for designing sgRNA primers (http://crispr.tefor.net) (Concordet and Haeussler, 2018). (Troubleshooting 1) For good targeting efficiency and relatively easy screening of the deletion mutations by PCR, we recommend to use two sgRNA target sites that are 100–1,000 bp apart on the genomic sequence, which was the case for generation of deletion mutations for Zfp445 (Figure 1B). In order to generate null deletion mutations, at least one exon or part of the exonic sequence is located between two sgRNA target sites.
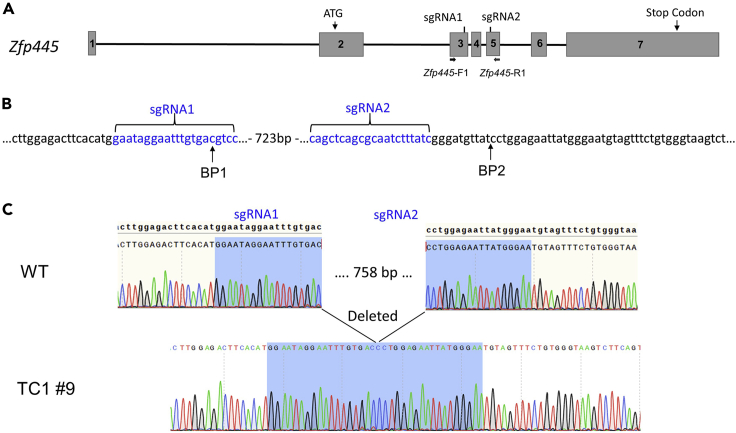
Figure 1.
Zfp445 is used as an example for CRISPR-mediated deletion mutation
(A) A schematic diagram is shown for Zfp445 and its two sgRNA target sites as well as the primers used for screening of deletion mutations of Zfp57. Mouse Zfp445 gene contains 7 exons, with the start codon and stop codon located in Exon 2 and Exon 7, respectively. Two sgRNA target sites (sgRNA1 and sgRNA2) residing in Exon 3 and Exon 5 will lead to excision of Exon 4 and part of Exon 3 and Exon 5 upon CRISPR-mediated deletion. The forward primer Zfp445-F1 is located 5′ to sgRNA1 on Exon 3, whereas the reverse primer Zfp445-R1 is located 3′ to sgRNA2 on Exon 5. The forward primer Zfp445-F2 is located on Exon 4.
(B) Two primers Zfp445-F1 and Zfp445-R1 were used for PCR-based screening of Zfp445 deletion mutations after CRISPR-mediated excision. Two sgRNA target sites (sgRNA1 and sgRNA2) are 765 bp apart in the genomic sequence. The wild-type PCR product is 1039 bp after PCR amplification with Zfp445-F1 and Zfp445-R1. The expected PCR product from the deletion mutations is around 274 bp accordingly.
Two oligos for each sgRNA are phosphorylated and annealed in the same time. The annealed sgRNA DNA fragment is cloned into the pX330 vector. Then the plasmid containing the right sgRNA DNA fragment is identified and confirmed by sequencing. The purified plasmid DNA sample will be used for transfection into ES cells afterward.
-
7.Phosphorylation and annealing of sgRNA oligos.
-
a.Two oligos for the respective top and bottom strands of the DNA fragment expressing each sgRNA targeting the gene of interest are designed based on the web-based tool (http://crispr.tefor.net).
-
b.Add 100 pmol of each oligo to the buffer for T4 DNA ligase in a total volume of 10 μL, together with 0.5 μL of T4 polynucleotide kinase (PNK) (Table 1). Mix it by pipetting for 15 times.
-
c.Incubate the oligo mixture at 37°C for 30 min, and then at 95°C for 5 min. Cool it gradually to 20°C–25°C.
-
a.
Note: Two primers, Zfp445-sgRNA1-top and Zfp445-sgRNA1-bottom, are added in one mixture, whereas Zfp445-sgRNA2-top and Zfp445-sgRNA2-bottom are added in the other mixture. ddH2O, double distilled water.
-
8.Ligation of the annealed sgRNA DNA fragment into the pX330 vector.
-
a.Add 2 μL of the oligo mixture containing the annealed sgRNA DNA fragment to the digestion buffer that also contains 100 ng of pX330, 1 mM of DTT, 1 mM of ATP, 1 μL of BpiI and 0.5 μL of T4 DNA ligase (Table 2). Mix it by pipetting for 15 times.
-
b.Incubate the digestion and ligation mixture at 37°C for 5 min, and then at 23°C for 5 min.
-
c.Repeat 5 more times for two incubation periods in step 4b at 37°C for 5 min and at 23°C for 5 min. After 6 cycles of repeated digestion and ligation, the mixture is ready for transformation into the bacterial competent cells in the next step.
-
a.
-
9.Identification and purification of the plasmid containing the sgRNA DNA fragment.
-
a.Use 2 μL of the above ligation mixture for transformation into T1 or other bacterial competent cells.
-
b.Plate the transformed bacteria on LB agar plates containing 50 μg/mL carbenicillin at 37°C for 16–20 h until large bacterial colonies appear on the LB agar plates.
-
c.Pick single bacterial colonies to inoculate 3 mL of LB liquid medium containing 50 μg/mL carbenicillin for each bacterial colony. Grow the bacterial culture at 37°C for 10–16 h.
-
d.Purify the plasmid DNA sample from the bacterial culture and then send it for sequencing to identify and confirm the construct containing the right sgRNA DNA fragment with the sequencing primer U6-Forward.
-
e.After identification of the right construct, grow the bacterial culture for this construct in LB medium containing 50 μg/mL carbenicillin at 37°C for 10–16 h.
-
f.Purify the DNA sample from 100 mL of bacterial culture with AxyPrep Endo-Free Plasmid Midiprep Kit and the manufacturer’s suggested protocol.
-
a.
Pause point: This completes the preparation of sgRNA plasmid constructs. The purified DNA samples are ready for transfection into the ES cells. They are usually stored at −20°C until they are needed for the experiments.
Table 1.
The mixture for phosphorylation and annealing of sgRNA oligos
| Top strand for the sgRNA primer | 100 pmol |
| Bottom strand for the sgRNA primer | 100 pmol |
| 10× T4 DNA ligase buffer | 1 μL |
| T4 polynucleotide kinase (PNK) (10 U/ μL) | 0.5 μL |
| ddH2O | Add ddH2O to a total volume of 10 μL |
Table 2.
The mixture for cloning of sgRNA DNA fragment into pX330
| Annealed sgRNA mixture from Table 1 | 2 μL |
| pX330 | 100 ng |
| 10× BpiI Digestion Buffer | 1 μL |
| 10 mM DTT | 1 μL |
| 10 mM ATP | 1 μL |
| BpiI | 1 μL |
| T4 DNA ligase (350 U/ μL) | 0.5 μL |
| ddH2O | Add ddH2O to a total volume of 10 μL |
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| T1 competent cells | Shanghai Weidi Biotechnology Co, Ltd | DL1015S |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal bovine serum (FBS) | Sigma, USA | F2442 |
| Pennicillin/Streptomycin solution | Beyotime, China | C0222 |
| Trypsin-EDTA solution | Sigma, USA | T4049 |
| Leukemia inhibitory factor (LIF) or ESGRO | Millipore, USA | ESG1107 |
| T4 DNA ligase | Takara, Japan | 2011A |
| T4 polynucleotide kinase (PNK) (10 U/ μL) | Takara, Japan | 2021S |
| Bpil | Thermo Scientific, USA | FD1014 |
| Gelatin | Sinopharm Chemical Reagent (SCR), China | 9000-70-8 |
| Bacto-yeast extract | Sangon, China | A515245-0500 |
| Bacto-tryptone | Sangon, China | A505247-0500 |
| Agar | Macklin, China | A800730 |
| Lipofectamine 2000 | Invitrogen, USA | 11668019 |
| DMEM | Invitrogen, USA | C11995500CP |
| Non-essential amino acid (NEAA) solution (100×) | HyClone, USA | SH30238.01 |
| Dithiothreitol (DTT) | Fisher Biotec, USA | BP172-25 |
| Dimethyl sulfoxide (DMSO) | Amethyst, China | 966629 |
| Carbenicillin | YEASEN, China | 60202ES08 |
| β-mercaptoethanol | Sigma, USA | M3148 |
| 84 disinfection solution (with sodium hypochlorite) | Aitefu, China | 92651097 |
| Sodium Chloride (NaCl) | Sangon, China | A501218-0001 |
| PBS (20×) | Sangon, China | B548117-0500 |
| Proteinase K | Abcone, China | P78893 |
| Tris | Beyotime Biotechnology, China | ST761 |
| SDS | Sigma, USA | L4390 |
| Agarose | Abcone, China | A88490 |
| GeneGreen nucleic acid dye | TIANGEN, China | RT210 |
| EDTA | Amethyst, China | 976151-500G |
| EcoRV | NEB, USA | R0195S |
| HindIII | NEB, USA | R0104S |
| Critical commercial assays | ||
| T7E1 assay | Vazyme, China | EN303-01 |
| Experimental models: Mouse wild-type ES cell lines | ||
| D1911 ES cell line (129/DBA hybrid background) | Li Lab, ShanghaiTech | (Lau et al., 2016) |
| D1906 ES cell line (129/DBA hybrid background) | Li Lab, ShanghaiTech | (Lau et al., 2016) |
| TC1 ES cell line (129 background) | Leder Lab, Harvard | (Deng et al., 1995) |
| Oligonucleotides | ||
| CACCGAATAGGAATTTGTGACGTCC | GENEWIZ | Zfp445-sgRNA1-top |
| AAACGGACGTCACAAATTCCTATTC | GENEWIZ | Zfp445-sgRNA1-bottom |
| CACCGAGCTCAGCGCAATCTTTATC | GENEWIZ | Zfp445-sgRNA2-top |
| AAACGATAAAGATTGCGCTGAGCT | GENEWIZ | Zfp445-sgRNA2-bottom |
| AGTGCGTCCTTCGTTACCTG | GENEWIZ | Zfp445-F1 |
| GTGAAGGTAGCTGGGGATAC | GENEWIZ | Zfp445-R1 |
| GAGGGCCTATTTCCCATGATTCCT | GENEWIZ | U6-Forward |
| GGACCTACAAGCAAGCAGAC | GENEWIZ | Zfp445-F2 |
| GTCACCGAGCTGCAAGAACT | GENEWIZ | Puro-F |
| GCTCGTAGAAGGGGAGGTTG | GENEWIZ | Puro-R |
| GGAAACAGCTATGACCATG | GENEWIZ | M13 Reverse Primer |
| Recombinant DNA | ||
| pX330 | Addgene, USA | 42230 |
| PGL3-U6-sgRNA-PGK-puromycin | Youbio, China | VT8203 |
| pBluescript | GenBank | X52331.1 |
| Other | ||
| AxyPrep DNA Gel Extraction Kit | Axygen of Corning | 35717KE1 |
| AxyPrep Endo-Free Plasmid Midiprep Kit | Axygen of Corning | 09318KA2 |
| Advantage GC2 PCR Kit | Takara, Japan | 639119 |
| TA cloning with the pUCm-T vector | Sangon, China | B522213 |
Materials and equipment
CRITICAL: The same fetal bovine serum (FBS) is used in our ES and feeder cell culture experiments in this protocol. Fetal bovine serum (FBS) needs to be incubated at 55°C for 30 min first before use. Disinfection solution is added to bacterial culture and cell culture before disposal. The bacterial LB plates are grown at 37°C in the incubators, whereas liquid bacterial culture samples are grown at 37°C in 500-mL flasks in the shakers. All ES cell experiments are performed under sterile condition inside a biosafety hood. MEF cells, feeder cells and ES cells are cultured at 37°C with 5% CO2 in cell culture incubators.
LB liquid medium
| Reagent | Amount |
|---|---|
| Bacto-tryptone | 10 g |
| Bacto-yeast extract | 5 g |
| NaCl | 10 g |
| ddH2O | Add total volume to 1 liter |
Note: Adjust the pH to 7 with 2 M NaOH when it is needed. After it is dissolved, the LB liquid medium is autoclaved and stored at 4°C for up to 6 months. For preparation of LB agar plates, add 15 g of agar to 1 liter of LB liquid medium and then autoclave it before pouring the petri dish plates. Carbenicillin is added right before use of LB medium.
Feeder cell medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM with glutamine | 1× | 500 mL |
| Fetal bovine serum | Roughly 10% | 50 mL |
| Penicillin/Streptomycin | About 1% | 6 mL |
| Total | n/a | 556 mL |
Note: DMEM contains high glucose and glutamine. The feeder cell medium is sterilized with a 500-mL filter unit of 0.2 μm in the pore size. It is stored at 4°C and it is recommended to use within 6 months. The medium needs to be warmed to 37°C before use. n/a, not applicable.
Feeder cell freezing medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM with glutamine | 1× | 400 mL |
| Fetal bovine serum | Roughly 25% | 100 mL |
| Penicillin/Streptomycin | About 1% | 6 mL |
| DMSO | 10% | 56.5 mL |
| Total | n/a | 562.5 mL |
Note: DMEM contains high glucose and glutamine. Add DMSO to the DMEM medium mixture first before filtration. The feeder cell freezing medium is sterilized with a 500-mL filter unit of 0.2 μm in the pore size. It is stored at 4°C and it is recommended to use within 6 months. The medium does not need to be warmed before use. n/a, not applicable.
ES cell growth medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM with glutamine | 1× | 500 mL |
| Fetal bovine serum | Roughly 15% | 75 mL |
| Non-essential amino acid | About 1% | 6 mL |
| Penicillin/Streptomycin | About 1% | 6 mL |
| β-mercaptoethanol | <0.001% | 4 μL |
| Leukemia inhibitory factor (LIF) | <0.01% | 50 μL |
| Total | n/a | 556 mL |
Note: Leukemia inhibitory factor (LIF) is not required when the SNL feeder cells are used for ES cell culture as they express LIF already (McMahon and Bradley, 1990; Takikawa et al., 2013). It is recommended, but it may not be essential to include LIF in the medium for ES cell culture on the DR4 feeder cells. DMEM contains high glucose and glutamine. The ES cell growth medium is sterilized with a 500-mL filter unit of 0.2 μm in the pore size. The ES cell growth medium is stored at 4°C for up to 6 months although it is better to use the LIF-containing ES cell medium within a month since LIF may not be stable after a few months in storage. It needs to be warmed up in a water bath at 37°C for about 15 min before use. n/a, not applicable.
ES cell freezing medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM with glutamine | 1× | 400 mL |
| Fetal bovine serum | Roughly 25% | 100 mL |
| Penicillin/Streptomycin | About 1% | 6 mL |
| Non-essential amino acid | About 1% | 6 mL |
| β-mercaptoethanol | <0.001% | 4 μL |
| DMSO | 10% | 57 mL |
| Total | n/a | 569 mL |
Note: DMEM contains high glucose and glutamine, with sodium pyruvate. Add DMSO to the DMEM medium mixture first before filtration. The ES cell freezing medium is sterilized with a 500-mL filter unit of 0.2 μm in the pore size. It is stored at 4°C and it is still good to use for up to half year. The freezing medium can be used directly without the need to be warmed up. n/a, not applicable.
Lysis buffer for genomic DNA isolation from the ES cell culture
| Reagent | Final concentration | Amount |
|---|---|---|
| ddH2O | n/a | 89 mL |
| Tris buffer (pH = 7.6) | 20 mM | 2 mL of 1 M solution |
| NaCl | 100 mM | 2 mL of 5 M solution |
| EDTA | 10 mM | 2 mL of 0.5 M solution |
| SDS | 0.5% | 5 mL of 10% solution |
| Total | n/a | 100 mL |
Note: The lysis buffer is stored on the bench shelf at 20°C–25°C and it is still good to use after a few years. Add Proteinase K to the lysis buffer right before genomic DNA preparation to the final concentration 100 μg/mL of Proteinase K. n/a, not applicable.
TE buffer (pH=8)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris buffer (pH = 8) | 10 mM | 10 mL of 1 M solution |
| EDTA | 1 mM | 2 mL of 0.5 M solution |
| ddH2O | n/a | 988 mL |
| Total | n/a | 1000 mL |
Note: Autoclave the TE buffer for 20 min at 121°C. Store it at 20°C–25°C and it is still good to use after a few years. n/a, not applicable.
Step-by-step method details
Transfection of ES cells
Timing: About 8 h
Two sgRNA constructs targeting Zfp445 are co-transfected into ES cells by Lipofectamine, together with the plasmid expressing the puromycin resistance gene. This results in simultaneous expression of two sgRNAs targeting two different regions of Zfp445 in ES cells. Deletion mutations of Zfp445 may occur in the co-transfected ES cells expressing these two sgRNAs.
-
1.Prepare Lipofectamine mixture in the DMEM medium.
-
a.Add 150 μL of DMEM without serum to a labeled 1.5-mL Eppendorf tube for transfection.
-
b.Then add 6 μL of Lipofectamine 2000 (or Lipofectamine 3000) reagent to the DMEM medium in the tube because either Lipofectamine could work well. Mix by tapping the tubes with the fingers a few times.
-
a.
Note: Leave the tube containing the transfection reagent DMEM mixture at 20°C–25°C while you start mixing the DNA mixture for transfection.
-
2.Prepare DNA mixture in the DMEM medium.
-
a.Mix two CRISPR constructs at 1.6 μg each with 0.8 μg of a plasmid containing the puromycin resistance gene in a clean autoclaved 1.5-mL Eppendorf tube. Mix well by pipetting 15 times because we think it will be easier to mix first and co-transfection efficiencies may be better with the premixed DNA sample. (troubleshooting 2).
-
b.Add 4 μg of the DNA mixture to another labeled tube containing 150 μL of DMEM medium without serum. Pipet 15 times to make the diluted DNA mixture.
-
a.
CRITICAL: The DNA samples containing two sgRNA constructs targeting Zfp445 are mixed first before DNA is mixed with the transfection reagent to make the transfection mixture. This ensures that almost all transfected ES cells contain a portion of all DNA samples in the transfection mixture. We also use less DNA containing the puromycin resistance gene so that most likely the puromycin-resistant ES colonies have undergone CRISPR-Cas9 editing events. The plasmid PGL3-U6-sgRNA-PGK-puromycin or its derivatives may be used for expression of the puromycin resistance gene in the co-transfection experiments, without expressing another sgRNA. Other constructs expressing puromycin resistance gene product can be also used in the co-transfection experiments in place of PGL3-U6-sgRNA-PGK-puromycin.
-
3.Prepare transfection mixture.
-
a.Add the diluted DNA mixture to the diluted Lipofectamine mixture in the DMEM medium.
-
b.Mix it by tapping the tube a few times with the fingers to make the transfection mixture. You may mix it again a couple minutes later.
-
c.Keep the transfection mixture at 20°C–25°C for 5 min.
-
a.
-
4.Transfection of ES cells.
-
a.Remove the feeder cell medium from the DR4 feeder cells and plate 1 mL of freshly digested ES cells of about 1 million ES cells on the DR4 feeder cells for transfection.
-
b.Add the transfection mixture dropwise to the freshly plated ES cells on the DR4 feeder cells. Distribute the transfection mixture evenly on the ES cells by moving the cell culture plate sideways a few times.
-
c.Transfer the ES cell plate with the transfection mixture to the cell culture incubator at 37°C with 5% of CO2.
-
a.
Note: The ES cells are grown to become relatively confluent before being harvested by trypsin digestion (Figure 2A). Then about one million ES cells are plated onto the DR4 feeder cells seeded on a well of 6-well plate or a small 3.5-cm dish plate right before addition of the transfection mixture.
-
5.
Post-transfection addition of ES cell growth medium.
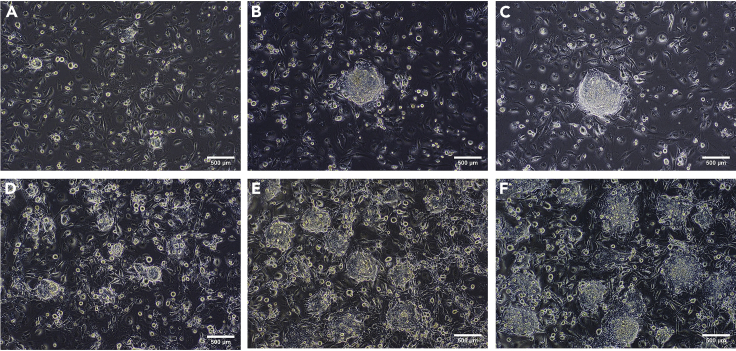
Figure 2.
The images of ES cell colonies are shown after transfection of ES cells followed by puromycin selection
The images were taken under an inverted microscope with a scale bar of 500 μm.
(A) D1911 ES cells were relatively confluent right before being harvested for ES cell transfection experiments.
(B) D1911 ES cell colonies are shown one day after transfection.
(C) D1911 ES cells were dying quickly after one day in the ES cell culture medium with puromycin.
(D) Most of D1911 ES cells died after two days in the ES cell culture medium with puromycin.
(E) D1911 ES cells were recovering quickly after one day in the ES cell growth medium without puromycin.
(F) D1911 ES cells became relatively confluent again after two days in the ES cell growth medium without puromycin.
About 6 h later after addition of the transfection mixture onto the ES cells, add more ES cell growth medium to increase the total volume to about 3–4 mL of ES cell growth medium per well. Then move the transfected ES cell plate back to the cell culture incubator at 37°C with 5% of CO2.
Enrichment of transfected ES cells
Timing: About 5 to 6 days
Puromycin is added to the ES cell culture medium for 2 days to select for ES cells expressing the puromycin resistant gene. These puromycin-resistant ES cells will probably express two sgRNAs targeting two different regions of Zfp445 in the same time. As a result, the ES cell culture is enriched with the co-transfected ES cells after puromycin selection.
To enrich the transfected ES cells, we use puromycin drug selection to kill most of un-transfected ES cells.
-
6.
We usually start drug selection next day about 24 h later after transfection (Figure 2B). For puromycin selection, 3–4 mL of fresh ES cell growth medium with 1 μg/mL puromycin is added to the ES cells after removal of the old growth medium one day later after the transfection experiment.
-
7.
Remove the old medium from the ES cell culture one day after puromycin selection, which is also two days after transfection. Add 3 mL of fresh ES cell growth medium with 1 μg/mL puromycin to the ES cells again (Figure 2C).
Note: Since ES cells die quickly in the medium containing puromycin, usually only 2 days of puromycin selection is required to enrich the transfected ES cells so that most untransfected ES cells are dead and removed from the ES cell culture during daily change of the ES cell medium (Figure 2D).
-
8.
Remove the old medium containing puromycin from the ES cell culture after two days of puromycin selection, and add fresh ES cell growth medium to the transfected ES cells.
-
9.
Change the medium for ES cell culture next day. Change the ES cell medium daily until the ES cell culture becomes confluent.
Note: Usually ES cell colonies will grow back quickly and become easily visible under microscope one day after removal of puromycin (Figure 2E). Change the medium for the ES cell culture daily until the ES cell colonies are large enough (Figure 2F). Typically 2–3 mL of fresh ES cell growth medium is needed for each well when ES cells are relatively sparsely populated. 4–5 mL of fresh ES cell growth medium is added to the transfected ES cells if the ES cells become more confluent.
Pause point: This completes the puromycin selection and enrichment of transfected ES cells. We may go to step 10 directly for serial dilution and plating of ES cells in order to pick individual ES colonies for screening and identification of candidate mutant ES clones. Alternatively, we may test the mutation rate of the transfected ES cells by the optional assay described immediately below. Then we can estimate how many ES cell colonies need to be individually picked for identification of the mutant ES clones before we proceed with the serial dilution and plating of ES cells in step 10.
Optional: Testing targeting efficiency before plating and picking single ES clones
Timing: About 10 days
We may test the targeting efficiency of Zfp445 in the ES cell culture first after co- transfection of two sgRNA constructs followed by puromycin selection. Then only ES cell culture with good targeting efficiency will be plated for picking ES cell colonies individually later. This will ensure that mutant ES clones can be relatively easily identified after certain number of singly picked ES clones are screened for the mutations in the Zfp445 gene.
Note: T7 endonuclease I (T7E1) assay is often used to assess the mutation frequency mediated by CRISPR-Cas9 (Li et al., 2018). However, we found the mutation frequencies were quite high after the enrichment of the transfected ES cells through puromycin selection. It is not necessary to use T7E1 assay to test the mutation frequency first before plating and picking of individual ES clones. However, the deletion mutation efficiency may be relatively low when two sgRNA target sites are far apart in the genome (e.g., much longer than 1 kb apart). Then it may be better to test the mutation efficiencies of the transfected ES cells with T7E1 assay first so that the required number of single ES colonies may be picked in order to find the candidate ES clones carrying the deletion mutations.
To test mutation rate in the transfected ES cell culture by T7E1 assay, it is necessary to grow a well of ES cells from the puromycin-treated transfected ES cells. Then isolate genomic DNA samples for the T7E1 assay as described below.
-
a.
Coat the new wells of a 6-well plate or a small 3.5-cm dish plate with 2 mL of 0.1% autoclaved gelatin in ddH2O the day before use.
-
b.Once the ES cells are relatively confluent, you may harvest the ES cells by trypsin digestion.
-
i.Add 2 mL of fresh ES cell medium to the well of gelatin-coated 6-well plate or a small gelatin-coated 3.5-cm dish plate after removal of the gelatin solution. Then harvest ES cells from the confluent ES cell culture that have regrown after removal of puromycin and add about 0.5 mL of the harvested ES cells to a well of gelatin-coated 6-well plate or gelatin-coated 3.5-cm dish plate containing 2 mL of ES cell medium.
-
ii.Spin down the rest of transfected ES cells in a 15-mL conical tube by centrifuge for 5–10 min at 1,000 g. Aspirate off the top medium from the 15-mL conical tube. Resuspend the cell pellet in 2 mL of freezing medium for ES cells and split the ES cells in two 2-mL freezing vials at 1 mL each. Store the freezing vials containing the resuspended transfected ES cells in freezing medium at −80°C in a cryo-freezer or in a liquid nitrogen tank.
-
i.
-
c.
Allow the plated ES cells on gelatin-coated plates to grow to relatively confluent without the need to change the medium at the interval. Then harvest the ES cells for genomic DNA preparation according to the standard protocol as described below in step 14.
-
d.
Perform the T7E1 assay for the genomic DNA sample according to the manufacturer’s suggested protocol.
-
e.
If the mutation frequency is high enough so that roughly more than 5% of the target region is mutated based on the T7E1 assay, we will plate the feeder cells derived from the SNL MEF cells on 6-well plates first the day before one frozen vial of transfected ES cells is thawed. Thaw the frozen ES cell sample according to the standard procedure and plate the thawed ES cells onto the SNL feeder cells in 2–3 mL of fresh ES cell growth medium.
-
f.
Change the medium 2 days later and then change the medium for the growing ES cells daily with the increasing volume of ES cell growth medium until about 5 mL of fresh ES cell growth medium is used for relatively confluent ES cells.
Plating and picking of single ES cell colonies
Timing: About 5 days
The ES cells harvested from the ES cell culture after puromycin selection are serially diluted, and then plated on the feeder cells so that well separated single ES cell colonies appear a few days later. These single ES cell colonies are picked individually and transferred to grow on the wells of 24-well plates seeded with feeder cells.
-
10.Picking single ES colonies grown from serially diluted ES cells.
 CRITICAL: The ES cells need to be serially diluted and plated onto the SNL feeder cells at relatively low densities so that well separated ES cell colonies can be picked individually after they grow up. SNL feeder cells are recommended for the ES cell culture after transfection since they express LIF and it is not necessary to add LIF in the ES cell growth medium. This is advantageous because LIF is quite expensive.
CRITICAL: The ES cells need to be serially diluted and plated onto the SNL feeder cells at relatively low densities so that well separated ES cell colonies can be picked individually after they grow up. SNL feeder cells are recommended for the ES cell culture after transfection since they express LIF and it is not necessary to add LIF in the ES cell growth medium. This is advantageous because LIF is quite expensive.-
a.Plating serially diluted ES cells.Note: Seed SNL feeder cells on a 10-cm dish plate the day before the ES cells are harvested and serially diluted for plating single ES cell colonies.
-
i.Harvest the ES cells from the well containing relatively confluent ES cells by trypsin digestion based on the standard cell culture procedure. Count the total number of cells for the harvested ES cell sample.
-
ii.Perform the serial dilutions of the harvested ES cells at 1:10 dilution for each dilution until the diluted ES cell density is low enough (about 100–200 ES cells/mL) to allow plating of ES cells with a 5-mL transfer pipet.
-
iii.Then plate the resuspended ES cells onto the SNL feeder cells on the 10-cm dish plate at the density of about 500 ES cells per 10-cm dish plate (Figure 3A). The rest of the harvested ES cells may be frozen again in 1 or 2 freezing vials as backup.
-
i.
-
b.Growing ES cell culture for picking individual ES colonies later.
-
i.Start changing the ES cell medium 2 days later with increasing fresh ES cell medium, from 10 mL for the first change of medium up to 20 mL a few days later.
- ii.
-
i.
-
c.Picking well separated single ES cell colonies.Note: Seed SNL feeder cells on the 24-well tissue culture plates the day before the ES cell colonies are large enough for picking individually.
-
i.Remove the medium from the 10-cm dish plate with ES cell colonies by aspiration.
-
ii.Circle the large ES colonies on the bottom of the plate with a marker pen quickly.
-
iii.Carefully wash the plate with 10 mL of autoclaved PBS solution. Aspirate off the PBS solution. Add 10 mL of fresh autoclaved PBS solution to the plate marked with ES cell colonies.
-
iv.Place the plate under the microscope inside the hood. Then pick a relatively big well-separated undifferentiated ES cell colony with good morphology by circling around it with a sterile tip (200-ul filter tip) to break the feeder cell layer, suck off the whole ES cell colony and transfer it to a well of 96-well plate.
-
i.
-
d.Dissociation and plating of individually picked ES cell colonies.Note: It is recommended to pick 24 well-separated ES cell colonies for each transfection. We usually start trypsin digestion after we have picked 12 ES cell colonies.
-
i.Mix trypsin solution with autoclaved PBS solution at the ratio of 1:1 by volume. Add 1 drop (about 50–100 μL) of mixed trypsin/PBS solution to the wells of the 96-well plate containing the picked ES cell colonies. Leave the 96-well plate at 20°C–25°C for about 5 min. Then stop trypsin digestion by addition of two drops of ES cell growth medium to each well.
-
ii.Dissociate the individually picked ES cell colonies by pipetting 15 times. To save time, we usually use multi-channel pipetman to dissociate ES cell colonies.
-
iii.After dissociation, transfer the resuspended ES cells for each ES cell colony to a well of 24-well plates containing 1 mL of fresh ES cell growth medium per well, which have been seeded with SNL feeder cells the day before.
-
i.
-
a.
Figure 3.
Single ES cell colonies were plated and picked for isolation of candidate deletion mutation ES clones
The images of the ES cell colonies were taken under an inverted microscope with a scale bar of 500 μm.
(A) Transfected D1911 ES cells after enrichment by puromycin selection were serially diluted and plated onto the 10-cm dish plate seeded with the SNL feeder cells.
(B) Well separated ES cell colonies appeared a few days after being plated onto the 10-cm dish plate seeded with the SNL feeder cells.
(C) Single ES cell colonies were large enough for being picked individually.
(D) Single ES cell colonies were plated onto the wells of 24-well plate seeded with the SNL feeder cells after being picked individually and dissociated by trypsin digestion.
(E) ES cell colonies were rapidly growing on the SNL feeder cells in the wells of 24-well plate.
(F) ES cells were ready for being harvested and frozen for temporary storage before PCR-based screening with the genomic DNA samples derived from a portion of the ES cells for each singly picked ES cell colony.
Growth and freezing of singly picked ES clones
Timing: About 5 days
These singly picked ES cell colonies are frozen after the cell culture of these ES clones has become confluent on the wells of 24-well plates with feeder cells. The frozen ES cell samples are stored at −80°C until the candidate ES clones are identified by PCR screening. These stored ES cell samples can then be amplified later when the confirmed ES clones are needed for other experiments.
The individually picked ES clones will be grown to become relatively confluent before they are harvested and frozen for temporary storage. In the same time, genomic DNA samples will be isolated from these ES clones for PCR screening to identify the candidate ES clones in the following steps.
Note: Normally it is not necessary to change the medium next day.
-
11.
Change ES cell medium two days later with 1 mL of fresh ES cell growth medium per well (Figure 3D).
-
12.
Change the medium daily until the ES cells become relatively confluent in the wells of the 24-well plate (Figures 3E and 3F).
-
13.Freezing individually picked ES clones after they grow up.Note: The ES clones will be temporarily frozen afterward while the candidate ES clones are being screened by PCR and confirmed by sequencing.
-
a.Harvest ES cell culture by trypsin digestion for freezing.
-
i.To harvest ES clones, remove the ES cell medium from the wells and wash once with 1 mL of PBS per well for the 24-well plate.
-
ii.Add 4 drops (approximately 200–400 μL) of 1:1 mixed trypsin/PBS solution to each well of 24-well plates. Leave the plate at 20°C–25°C for about 5 min until the ES cell colonies are dissociated from the plates. Then pipet 15 times with a 1-mL pipet to resuspend the ES cells for each ES cell colonies that are confluent.
-
i.
-
b.Freeze and store the ES cell samples from the ES cell culture of individually picked ES cell colonies.
-
i.Add 1 mL of ES cell freezing medium to each well of the 24-well plate. Use 1-mL pipetman to resuspend ES cells in each well by pipetting for 15 times.
-
ii.Transfer most of the resuspended ES cells in ES cell freezing medium to a labeled 2-mL freezing vial. Store these freezing vials containing the ES cells in ES cell freezing medium in a freezer of −80°C or in a liquid nitrogen tank for temporary storage until the candidate ES clones have been identified.
-
i.
-
a.
Identification of candidate ES clones
Timing: About 5 days
Genomic DNA samples are isolated from the cell culture of the picked ES clones. Then these genomic DNA samples are screened for candidate mutant ES clones carrying deletion mutations of Zfp445 by PCR with two primers flanking two sgRNA target sites.
Note: Genomic DNA samples isolated from the ES clones will be subjected to PCR-based screening with two primers spanning two CRISPR target sites in order to identify the candidate ES clones carrying the deletion mutations of the target gene. The mutant PCR product is sequenced to find out the exact mutation caused by CRISPR-Cas9.
-
14.Grow cell culture for genomic DNA preparation of the originally singly picked ES clones.
-
a.For genomic DNA sample isolation of these ES clones, 1 mL of ES cell growth medium is added to the remaining resuspended ES cells in each well of the 24-well plate of step 13.
-
b.The remaining ES cells in the well will grow to become relatively confluent in a few days without the need to change the medium during this interval.
-
a.
-
15.Prepare genomic DNA samples for originally individually picked ES clones.Note: We harvest ES cells for genomic DNA sample preparation by using the standard procedure for genomic DNA preparation from the adherent cells (Thomas et al., 1992). The detailed experimental procedure is provided below for genomic DNA sample preparation that we have employed in our own lab.
-
a.Aspirate off the medium from the confluent ES cell culture.
-
b.Add 200 μL of the lysis buffer containing 100 μg/mL of Proteinase K to each well of the 24-well plate with the cultured ES cells and then the lysed cell sample is transferred to a clean Eppendorf tube.
-
c.Incubate the Eppendorf tubes containing ES cell lysate samples in the water bath at 37°C for 8–12 h.
-
d.Add 100 μL of saturated 6 M NaCl solution to each Eppendorf tube. Close the lid of each tube tightly.
-
e.Transfer the tubes into a storage paper box and shake the box vigorously for 200 times to mix the cell lysate. Leave these tubes on ice for 10 min.
-
f.Centrifuge these tubes at 3,500 g for 10 min.
-
g.Transfer 200 μL of the supernatant to a clean tube containing 400 μL of ethanol. Mix the solution by inverting the closed Eppendorf tubes a few times until DNA precipitate appears.
-
h.Move DNA in the tube with a pipet tip to a clean tube.
-
i.Rinse DNA once with 1 mL of 70% ethanol.
-
j.Dry the DNA in air for 15 min.
-
k.Add 100 μL of TE buffer (pH=8) to the tube containing DNA and leave the tube at 25°C for 12 h to dissolve the genomic DNA sample.
-
l.This genomic DNA solution can be used for PCR or other experiments. Store the genomic DNA samples at 4°C.
-
a.
-
16.Identification of the candidate mutant ES clones carrying deletion mutations.
-
a.Screen for candidate ES clones with deletion mutations by PCR.
 CRITICAL: To identify the candidate ES clones, generally we use PCR-based approaches to amplify the target regions from the purified genomic DNA samples. Routinely we use two constructs expressing two different sgRNAs targeting two different regions of the target gene so that a relatively large piece of genomic region including part of the exons will be removed from the target gene. The ES clones containing the desired deletion in the gene of interest will be identified by PCR amplification of the genomic DNA samples derived from these ES clones followed by regular DNA gel electrophoresis. (troubleshooting 3).
CRITICAL: To identify the candidate ES clones, generally we use PCR-based approaches to amplify the target regions from the purified genomic DNA samples. Routinely we use two constructs expressing two different sgRNAs targeting two different regions of the target gene so that a relatively large piece of genomic region including part of the exons will be removed from the target gene. The ES clones containing the desired deletion in the gene of interest will be identified by PCR amplification of the genomic DNA samples derived from these ES clones followed by regular DNA gel electrophoresis. (troubleshooting 3).-
i.Prepare the PCR reaction master mix with Advantage GC2 PCR Kit according to the following format:PCR reaction master mix
Reagent Amount DNA template 1 μg of genomic DNA Taq DNA Polymerase 0.5 μL Primer 1 0.5 μL of 20 μM Primer 2 0.5 μL of 20 μM PCR Buffer 5 μL of 5× buffer dNTPs 1 μL of 10 mM ddH2O 16.5 μL Total volume 25 μL -
ii.Use the PCR cycling condition below for amplification of the target gene region from the genomic DNA samples of these ES clones.PCR cycling conditions
Steps Temperature Time Cycles Initial Denaturation 95°C 120 s 1 Denaturation 95°C 30 s 35 cycles Annealing 58°C 30 s Extension 72°C 1 min Final extension 72°C 5 min 1 Hold 12°C forever
-
i.
-
b.Determine the deletion mutations by sequencing.Then the amplified PCR product containing the deletion will be sequenced to find out if it is indeed derived from the deleted allele of the candidate ES clone.
-
i.The PCR product of the right deletion may be separated on the agarose gel and purified with AxyPrep DNA Gel Extraction Kit by using the manufacturer’s suggested protocol.
-
ii.To find out the precise deleted sequence of the target gene in the candidate ES clones, the purified DNA fragment derived from the deletion mutation is cloned into the pUCm-T vector by using the TA cloning with the pUC-m vector, or into a common bacterial vector such as pBluescript.
-
iii.Then the plasmid containing the PCR product is sequenced with the M13 Reverse Primer to find out the exact deleted region of the target gene.Note: The PCR product derived from the target gene region containing the deletion mutation may be directly cloned into the pUCm-T vector first with the TA cloning strategy. The PCR product can be also cloned into the EcoRV site of pBluescript through blunt-end ligation. Alternatively, a unique restriction enzyme site (e.g., HindIII) may be introduced at both ends of the PCR product by another round of PCR with two primers containing the restriction enzyme recognition sequence. Then the new PCR product is cut with the restriction enzyme and ligated into the same restriction site present in pBluescript.
-
i.
-
a.
Alternative reagents: Other similar TA cloning systems may be used in place of the pUCm-T vector system for TA cloning for cloning of the PCR product. These include the pGEM-T vector from Promega, USA (Cat. # A3610) and the TOPO TA cloning system from Invitrogen of Thermo Fisher Scientific, USA (Cat. # 451641).
Expansion and storage of identified ES clones
Timing: About 4 to 5 days
The identified candidate mutant ES clones are thawed from the stored ES cell samples at −80°C and plated on the feeder cells seeded on 6-well plates. Then the mutant ES clones are expanded in cell culture on 6-well plates and may be further expanded on 10-cm dish plates if more frozen ES cell samples are needed for long-term storage of the confirmed mutant ES clones.
Note: Plate SNL feeder cells on the 6-well tissue culture plates, with 2-mL of feeder cell medium per well, the day before the candidate ES cell clones are thawed and cultured for expansion of the ES cells on the SNL feeder cells.
-
17.
After they have been confirmed by PCR and sequencing, the candidate ES clones may be thawed from the freezing vials containing the frozen ES cells that have been stored temporarily at −80°C or in a liquid nitrogen tank in step 13.
-
18.
Remove the freezing medium from the thawed ES cells by washing with 10 mL of ES cell growth medium in a 15-mL conical tube followed by centrifuge at 1,000 g for 5 min.
-
19.
Aspirate off the feeder cell medium from the 6-well plate, and then add the thawed ES cells for each candidate ES clone onto the feeder cells in 2 mL of ES cell growth medium.
Note: Normally we do not need to change the medium next day.
-
20.
Change ES cell medium daily with the increasing fresh ES cell growth medium from 2 mL per well initially to 5 mL per well a few days later as the ES cells grow. When they become relatively confluent, the ES cells in the wells are ready for being harvested and put away for long-term storage.
-
21.
If necessary, the candidate ES clones can be further expanded on the SNL feeder cells on the 10-cm dish plate so that more freezing vials containing the frozen ES cells may be obtained for the desired ES clones for long-term storage in liquid nitrogen.
Optional: Determining homozygous or heterozygous mutant ES clones
Timing: About 2 days
The identified candidate mutant ES clones are either homozygous or heterozygous for Zfp445. To find out if they carry one deleted allele or two deleted alleles of Zfp445, the genomic DNA samples of these mutant ES clones may be subjected to PCR with a third primer located in the deleted region besides two primers flanking two sgRNA sites. Alternatively, western blot analysis or immunostaining may be carried out to test if there is no wild-type protein in the mutant ES clones.
-
22.Test if the mutant ES clones are homozygous or heterozygous.
-
a.PCR screening to identify wild-type or deletion mutant allele of Zfp445 in the ES clones.If homozygous mutant ES clones are needed for the experiments, the candidate ES clones carrying deletion mutation of the target gene may be subjected to another PCR-based screening with three PCR primers so that we can determine if they are homozygous and heterozygous mutant ES clones, with two primers spanning two CRISPR target sites and one primer located in-between. Then the PCR product is examined after agarose gel electrophoresis to find out if there is PCR product derived from the wild-type allele of the target gene.Note: Two PCR primers (Primer 1 and Primer 2 of the Table below for PCR reaction master mix) were used for amplification of small PCR product derived from the deleted allele for initial PCR-based screening of mutant ES clones carrying deletion mutations. Then we use three PCR primers (Primer 1, Primer 2 and Primer 3 of the Table below for PCR reaction master mix) to amplify both the wild-type and mutant alleles for the target gene to identify the homozygous mutant ES clones carrying deletion mutations on both alleles. If Zfp445 is used as an example, Primer 1 and Primer 2 are Zfp445-F1 and Zfp445-R1 listed in the key resources table (Figure 1). Primer 3 is another new primer Zfp445-F2 located in the deleted region of Zfp445 (Figure 1A).
-
i.Prepare PCR reaction master mix with Advantage GC2 PCR Kit according to the following format:PCR reaction master mix
Reagent Amount DNA template 1 μg of genomic DNA Taq DNA Polymerase 0.5 μL Primer 1 0.5 μL of 20 μM Primer 2 0.5 μL of 20 μM Primer 3 (optional) 0.5 μL of 20 μM PCR Buffer 5 μL of 5× buffer dNTPs 1 μL of 10 mM ddH2O 16.5 μL Total volume 25 μL -
ii.Perform PCR to identify wild-type or deletion mutant allele as illustrated below.PCR cycling conditions
Steps Temperature Time Cycles Initial Denaturation 95°C 120 s 1 Denaturation 95°C 30 s 35 cycles Annealing 58°C 30 s Extension 72°C 1 min Final extension 72°C 5 min 1 Hold 12°C forever
-
i.
-
b.Test if wild-type protein product is absent in the mutant ES clones.
 CRITICAL: Good antibodies are available for either western blot analysis or immunostaining of wild-type protein in the ES clones.
CRITICAL: Good antibodies are available for either western blot analysis or immunostaining of wild-type protein in the ES clones.-
i.In case there is good antibody available, the protein lysate samples will be prepared from these identified mutant ES clones and subjected to western blot analysis to test if there is wild-type protein in the mutant ES clones. Then we will be able to tell if they are indeed homozygous or heterozygous mutant ES clones.
-
ii.Immunostaining may be also performed to determine if the mutant ES clones lack any wild-type protein.Note: Some antibodies may be good for western blot analysis, whereas others may be suitable for immunostaining. Some may be good for both analyses. Antibodies may be generated to confirm the mutant ES clones. These antibodies may be useful for other experiments later such as ChIP analysis of the target gene product in the wild-type ES cells compared with the mutant ES clones. It can be also employed in characterization of the mutant mice derived from the mutant ES clones.
-
i.
-
a.
Expected outcomes
We have obtained desired deletion mutations for multiple genes with three different mouse ES cell lines tested in our lab. For example, two sgRNA constructs were generated in the pX330 vector to express two different sgRNA molecules (sgRNA1 and sgRNA2) targeting two different regions of Zfp445 (Figure 1A). Deletion mutations could be induced in the Zfp445 gene when they were co-transfected into ES cells.
Usually we pick 24 single ES clones for each transfection following this protocol. We used two primers of Zfp445 (Zfp445-F1 and Zfp445-R1) spanning two sgRNA target sites of Zfp445 for PCR-based screening of candidate ES clones carrying deletion mutations of Zfp445 (Figure 1B). The PCR primers may be placed at least 50 bp away from the sgRNA target sites so that most deletion mutations may be captured by PCR. Indeed, 7 out of 24 ES clones appeared to contain deletion mutations in the Zfp445 gene when the D1911 ES cells were co-transfected with two sgRNA constructs targeting two different sites of Zfp445 (Figure 4). Similarly, 8 out of 24 ES clones carried deletion mutations of Zfp445 when the TC1 ES cells were co-transfected with these two sgRNA constructs targeting Zfp445 (Figure 5). The PCR product of all 15 candidate ES clones with potential deletion mutations was subjected to sequencing (Figure 6). Based on sequencing results, these ES clones indeed carried expected deletion mutations of Zfp445, which is exemplified by sequencing of the mutant ES clone TC1 #9 in Lane 9 of Figure 5. Since two sgRNAs targeted two different exons of Zfp445, there were two breakpoints for Zfp445 deletion mutant ES clones (Figures 6A and 6B). For the mutant ES clone TC1 #9, one breakpoint (BP1) was at the sgRNA1 recognition site and the other breakpoint (BP2) was just outside the sgRNA2 recognition site (Figure 6B). The deleted sequence was 758 bp according to the sequencing result of the amplified PCR product from the deleted allele in the mutant ES clone TC1 #9 (Figure 6C).
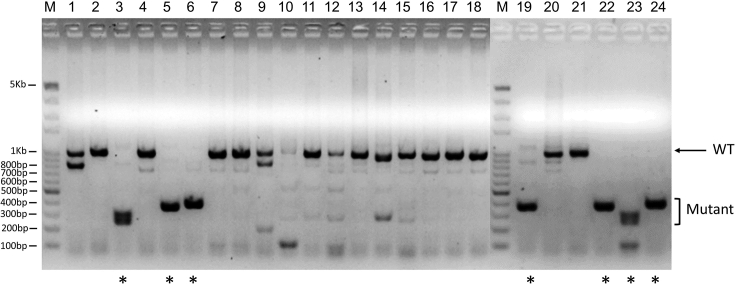
Figure 4.
Candidate deletion mutant ES clones were identified by PCR for individually picked ES cell colonies after transfection of D1911 ES cells with the constructs targeting Zfp445
The genomic DNA samples derived from the cells of the 24-well plate with singly picked ES cell colonies were subjected to PCR with two primers of Zfp445 (Zfp445-F1 and Zfp445-R1) spanning two sgRNA target sites that were approximately 765 bp apart. The expected wild-type (WT) PCR product for Zfp445 with these two PCR primers is 1039 bp in length. M, DNA ladder with molecular weight markers on the agarose gel. ∗, candidate mutant ES clones carrying the deletion mutations of Zfp445. 7 out of 24 ES clones appeared to contain deletion mutations in the Zfp445 gene, with smaller PCR product derived from the mutant allele (Mutant).
Figure 5.
Candidate deletion mutant ES clones were identified by PCR for individually picked ES cell colonies after transfection of TC1 ES cells with two CRISPR constructs targeting Zfp445
The genomic DNA samples derived from 24 singly picked ES cell colonies were screened by PCR with two primers (Zfp445-F1 and Zfp445-R1) spanning two sgRNA target sites that were 765 bp apart. The expected wild-type (WT) PCR product for Zfp445 is 1039 bp in length. M, DNA ladder with molecular weight markers on the agarose gel. ∗, candidate mutant ES clones carrying the deletion mutations of Zfp445. 8 out of 24 ES clones carried deletion mutations of Zfp445 that gave rise to smaller PCR product derived from the mutant allele (Mutant).
Figure 6.
The deletion mutation in Zfp445 mutant ES clones was confirmed by sequencing of the mutant PCR product
The smaller PCR product derived from the candidate deletion mutant ES clones was sent for sequencing and further confirmed by bacterial colony sequencing after it was cloned into the pBluescript vector.
(A) A schematic diagram is shown for Zfp445 and the primers used for screening of deletion mutations of Zfp445 generated with two sgRNA target sites (sgRNA1 and sgRNA2). Zfp445-F1 and Zfp445-R1 were used for PCR-based screening for Zfp445 deletion mutations.
(B) Two sgRNAs were targeted to two different regions of Zfp445 that were separated by 723 bp. Two breakpoints (BP1 and BP2) were generated by CRISPR-mediated deletion with these two sgRNAs. For the mutant ES clone TC1#9, one breakpoint (BP1) was at the sgRNA1 recognition site, whereas the other breakpoint (BP2) was just outside the sgRNA2 recognition site.
(C) Direct sequencing of the PCR product confirmed that the smaller PCR product in the ES clone (TC1 #9 in Lane 9 of Figure 5) carried an expected deletion of Zfp445, with the deleted sequence of 758 bp including the sgRNA2 recognition site. WT, wild-type ES clone. The sequences of two blue shaded regions that were separated by 758 bp in the wild-type allele of Zfp445 were next to each other in the deletion mutation allele of Zfp445 indicated by one merged shaded region in blue.
A few candidate ES clones carrying deletion mutations may also have visible but faint PCR product derived from the wild-type allele of Zfp445, indicating that these may be heterozygous for Zfp445. The PCR product is less for the wild-type allele because PCR amplification efficiency is usually much higher for the smaller PCR product carrying the deletion mutations. For the candidate mutant ES clones with a very faint wild-type Zfp445 PCR product, it could mean that there were residual feeder cells in the ES cell culture even though the ES cell culture is grown on gelatin-coated plates for one generation to get rid of most feeder cells before it is harvested for genomic DNA preparation. For the extra bands in the middle between the PCR products of wild-type allele and deletion mutant allele on the agarose gel, they could be some non-specific PCR product or more likely the annealed hybrid DNA containing the wild-type and deletion mutant PCR product that were present in a few candidate mutant ES cell samples. Since all 15 candidate mutant ES clones were confirmed to carry deletion mutations of Zfp445 based on sequencing of the mutant PCR product, we think the deletion efficiency for Zfp445 in D1911 and TC1 ES cells should still be considered relatively high, with 29.2% (7 out of 24) and 33.3% (8 out of 24), respectively.
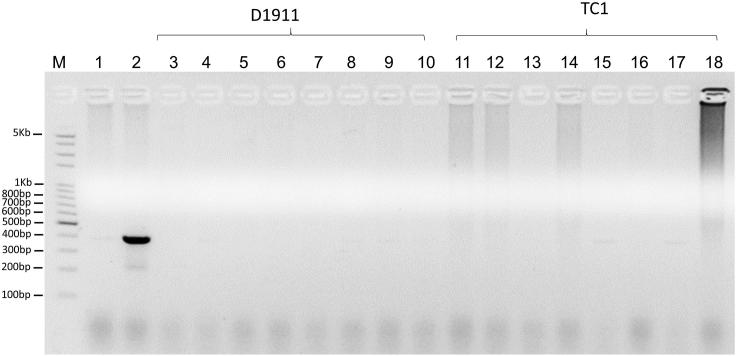
To test if these mutant ES clones may carry stably integrated puromycin resistance gene, genomic DNA samples derived from these deletion mutant ES clones were subjected to PCR amplification with two primers in the puromycin resistance gene (Puro-F and Puro-R). As a positive control, an expected PCR product of 382 bp was amplified from the genomic DNA sample of the parental D1911 ES clone plus 0.5 ng of PGL3-U6-sgRNA-PGK-puromycin (Lane 2 of Figure 7). But there was no amplified product from the genomic DNA samples of parental D1911 ES clone (Lane 1 of Figure 7), the ES clone that did not contain deletion mutation (Lane 6 of Figure 7) and all 15 deletion mutant ES clones (Lane 3–5 and Lane 7–18 of Figure 7). Therefore, these mutant ES clones did not appear to contain stably integrated puromycin resistance gene after two days of puromycin selection.
Figure 7.
Zfp445 mutant ES clones did not appear to carry integrated puromycin resistance gene
The genomic DNA samples were prepared from 15 Zfp445 mutant ES clones identified through transfections into D1911 (7 candidate mutant clones in Figure 5 and 8 candidate mutant clones in Figure 6) as well as from the parental D1911 ES clone and an ES clone that did not contain deletion mutation (Lane 10 of Figure 4). Approximately 1 μg of genomic DNA was used for each PCR amplification of puromycin resistance gene with the primers Puro-F and Puro-R for an expected PCR product of 382 bp. M, DNA ladder with molecular weight markers on the agarose gel. Lane 1, D1911 parental ES cells. Lane 2, 1 μg of genomic DNA of D1911 ES cells plus 0.5 ng of PGL3-U6-sgRNA-PGK-puromycin. Lane 3, mutant ES clone in Lane 3 of Figure 4. Lane 4, mutant ES clone in Lane 5 of Figure 4. Lane 5, mutant ES clone in Lane 6 of Figure 4. Lane 6, ES clone in Lane 10 of Figure 4. Lane 7, mutant ES clone in Lane 19 of Figure 4. Lane 8, mutant ES clone in Lane 22 of Figure 4. Lane 9, mutant ES clone in Lane 23 of Figure 4. Lane 10, mutant ES clone in Lane 24 of Figure 4. Lane 11, mutant ES clone in Lane 2 of Figure 5. Lane 12, mutant ES clone in Lane 9 of Figure 5. Lane 13, mutant ES clone in Lane 11 of Figure 5. Lane 14, mutant ES clone in Lane 13 of Figure 5. Lane 15, mutant ES clone in Lane 19 of Figure 5. Lane 16, mutant ES clone in Lane 20 of Figure 5. Lane 17, mutant ES clone in Lane 21 of Figure 5. Lane 18, mutant ES clone in Lane 22 of Figure 5.
Limitations
So far, we have successfully obtained deletion mutations in all target genes that we tested in our experiments. The only limitation to this approach is that there may not be two suitable sgRNA target sites for the target gene within a reasonable distance. The sgRNA target sites can be located in either introns or exons. To create a null deletion mutation, it is ideal to remove an exon or at least part of the exonic sequence after CRISPR-mediated excision of the target gene. For PCR-based screening of deletion mutations, it is easier to distinguish the PCR product of the deletion mutation from that of the wild-type allele on the agarose if the deletion is more than 100 bp in size. As long as these considerations can be met, deletion mutations may be isolated with good efficiencies for the target genes. The deletion mutation frequencies may decrease significantly if two sgRNA sites of the target gene are too far apart in the genomic sequence (e.g., much longer than 1 kb in distance). However, we think it is still possible to obtain the desired deletion mutations of the target gene with this approach just by picking more ES clones for screening.
Another potential problem is the off-target effect by CRISPR-Cas9. We employ the popular web-based tool for CRISPR sgRNA primer design that has been extensively utilized in the community. We usually choose the primers with lowest possibilities for off-target. Therefore, we believe the potential off-target sites for each set of sgRNA primers used in this manuscript should be comparable to other published studies. To reduce or even eliminate the off-target effect in the mutant mice derived from these mutant ES clones, the mice carrying the deletion mutations of the target genes are recommended to be backcrossed with the wild-type mice a few times before the mutant mice are used for phenotypic analyses. In this way, most of the off-target mutations will be removed from the backcrossed mice carrying the deletion mutations of the target genes.
Troubleshooting
Problem 1
One potential problem is that one or two sgRNA target sites may not have worked as expected after co-transfection of two sgRNA constructs.
Potential solution
Find new good sgRNA target sites and then design new sgRNA primers in order to obtain better targeting and deletion efficiencies. The sgRNA constructs can be tested in a few different combinations to find out which two sgRNA constructs may work the best based on the T7E1 assay mentioned in step 9.
Problem 2
One potential problem for the failure to obtain the desired deletion mutations is low co-transfection efficiency of ES cells by two sgRNA constructs.
Potential solution
Good mixing of DNA samples is important for high co-transfection efficiency. Try to pipet more times to mix DNA samples more thoroughly before adding the DNA mixture to the Lipofectamine mixture. This ensures that most transfected ES cells will likely contain both sgRNA constructs. Since DNA purity and its concentration are also important for the transfection efficiency, it is possible that new good DNA preparation may be required in order to improve the transfection and deletion efficiencies.
Another potential solution is to use 0.4 μg or even 0.2 μg of the plasmid expressing puromycin resistance gene product in the DNA mixture instead of 0.8 μg mentioned above in step 2. In this way, there may be increased probabilities for the transfected ES cells to carry both sgRNA constructs after puromycin selection.
Problem 3
It is possible that PCR screening may have failed to pick up the deletion mutations.
Potential solution
A solution to this potential problem is to include a positive control carrying deletion mutation of Zfp445 and a negative control with just wild-type ES genomic DNA for PCR screening. A deletion mutation can be created in a plasmid carrying Zfp445 DNA if an endogenous deletion mutant allele is not available. Then find out if PCR primers have worked well as expected. After PCR primers are confirmed to be good for screening of deletion mutations, then redo the PCR screening for ES clones again to find out if there is any candidate mutant ES clone carrying deletion mutation of Zfp445.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiajun Li, E-mail: lixj1@shanghaitech.edu.cn.
Materials availability
D1911 and D1906 ES cell lines used in this study are available upon request from the lead contact, Xiajun Li, E-mail: lixj1@shanghaitech.edu.cn. TC1 ES cell line is the property of Harvard University (USA) and the material transfer agreement may need to be reached with Harvard University first before it is available.
Acknowledgments
The work in the authors’ laboratories has been supported by grants from the Science and Technology Commission of Shanghai Municipality and the Ministry of Science and Technology of the People’s Republic of China (grant #2018YFC1005004). We appreciate the assistance received in the facility of Protein Center of Shanghai Zhangjiang Laboratory for irradiation of MEF cells. We also would like to thank the Molecular and Cell Biology Core Facility (MCBCF) and Multi-Omics Core Facility (MOCF) at the School of Life Science and Technology in ShanghaiTech University for providing technical support. The graphical abstract was created with the template and some assets provided by STAR Protocols.
Author contributions
X.L. designed the experiments and wrote the manuscript. Y.L., Q.C., C.S., Z.X., and S.Y. performed the experiments to develop the full protocol, under the guidance of X.L. Y.L., Z.X., and X.L. analyzed data.
Declaration of interests
X.L. and Y.L. have filed a patent application for this approach.
Data and code availability
The article includes all necessary information for the experiments and expected outcome.
References
- Concordet J.P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Zhang P., Wade Harper J., Elledge S.J., Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Shi J., Zhao J., Wang Q., Cong D., Chen F., Zhang Y., Liu Y., Zhao J., Chen Q., et al. ZFP57 dictates allelic expression switch of target imprinted genes. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2005377118. e2005377118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H.T., Liu L., Ray C., Bell F.T., Li X. Derivation of hybrid ES cell lines from two different strains of mice. Stem Cell Res. 2016;16:252–255. doi: 10.1016/j.scr.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Zeng C., Dong Y. Design and assessment of engineered CRISPR-Cpf1 and its use for genome editing. Nat. Protoc. 2018;13:899–914. doi: 10.1038/nprot.2018.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., Ferguson-Smith A.C. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A.P., Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Pyzocha N.K., Ran F.A., Hsu P.D., Zhang F. RNA-guided genome editing of mammalian cells. Methods Mol. Biol. 2014;1114:269–277. doi: 10.1007/978-1-62703-761-7_17. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Coluccio A., Thorball C.W., Planet E., Shi H., Offner S., Turelli P., Imbeault M., Ferguson-Smith A.C., Trono D. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 2019;33:49–54. doi: 10.1101/gad.320069.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa S., Ray C., Wang X., Shamis Y., Wu T.Y., Li X. Genomic imprinting is variably lost during reprogramming of mouse iPS cells. Stem Cell Res. 2013;11:861–873. doi: 10.1016/j.scr.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K.R., Deng C., Capecchi M.R. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol. Cell Biol. 1992;12:2919–2923. doi: 10.1128/mcb.12.7.2919-2923.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K.L., Wang Y., Dausman J., Jaenisch R. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 1997;25:3745–3746. doi: 10.1093/nar/25.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The article includes all necessary information for the experiments and expected outcome.