Vitamin B12 shapes gut microbiome and supports mitochondrial metabolism of ileal epithelial cells to synergistically regulate epithelial oxygenation, controlling aerobic Salmonella infection. This mechanistic interplay highlights the significance of this crucial micronutrient in maintaining intestinal homeostasis against pathogen infections.
Abstract
Deprivation of vitamin B12 (VB12) is linked to various diseases, but the underlying mechanisms in disease progression are poorly understood. Using multiomic approaches, we elucidated the responses of ileal epithelial cells (iECs) and gut microbiome to VB12 dietary restriction. Here, VB12 deficiency impaired the transcriptional and metabolic programming of iECs and reduced epithelial mitochondrial respiration and carnitine shuttling during intestinal Salmonella Typhimurium (STm) infection. Fecal microbial and untargeted metabolomic profiling identified marked changes related to VB12 deficiency, including reductions of metabolites potentially activating mitochondrial β-oxidation in iECs and short-chain fatty acids (SCFAs). Depletion of SCFA-producing microbes by streptomycin treatment decreased the VB12-dependent STm protection. Moreover, compromised mitochondrial function of iECs correlated with declined cell capability to utilize oxygen, leading to uncontrolled oxygen-dependent STm expansion in VB12-deficient mice. Our findings uncovered previously unrecognized mechanisms through which VB12 coordinates ileal epithelial mitochondrial homeostasis and gut microbiota to regulate epithelial oxygenation, resulting in the control of aerobic STm infection.
Introduction
Micronutrients control cellular molecular dynamics, microbial structure, and the associated metabolism to calibrate intestinal homeostasis and human health (Nicholson et al., 2012). One of these micronutrients is vitamin B12 (VB12), which participates in one-carbon metabolism (Selhub, 2002), drives DNA synthesis, and supports cellular energy (O’Leary and Samman, 2010). VB12 is synthesized by a subset of bacteria and archaea in nature (Sokolovskaya et al., 2020), and its biosynthesis is tightly regulated by bacterial structured noncoding RNAs, known as riboswitches (Li et al., 2020). This vitamin consists of a central cobalt atom in a corrin ring attached to the lower axial ligand 5,6-dimethylbenzimidazole (Elias-Arnanz, 2020), pivotally important for binding VB12 to the intrinsic factor (IF). The formed IF–VB12 complex is recognized by cubilin and megalin, which facilitate receptor-mediated endocytosis by ileal epithelial cells (iECs; Andersen et al., 2010). Upon lysosomal degradation of IF, VB12 serves as a cofactor for two enzymes: the cytoplasmic methionine synthase (MS) and the mitochondrial methylmalonyl-CoA mutase (MCM), directing the methylation of homocysteine to methionine and the conversion of methylmalonyl-CoA to succinyl-CoA, respectively (O’Leary and Samman, 2010).
Clinically, VB12 deficiency is primarily the result of dietary restriction (e.g., malnutrition) or loss of the IF (Stabler, 2013). VB12 deficiency may result in pernicious anemia and neuropsychiatric complications, which can often be reversed by early diagnosis and prompt treatment (Toh et al., 1997). Additionally, VB12 deficiency reduces MS enzymatic activity, leading to uncontrolled homocysteine, a risk factor for cardiovascular diseases and ischemic stroke (Yahn et al., 2021), and impaired biosynthesis of S-adenosylmethionine, the universal methyl donor critical for DNA methylation and genomic stability (Fenech, 2001). Further, VB12 deficiency may induce micronuclei formation and chromosomal abnormalities (Fenech et al., 1998) and contribute to DNA hypomethylation that initiates significant epigenomic changes in the epithelium, potentially leading to colorectal cancer (Goelz et al., 1985). Moreover, when VB12 is transported across the placenta to the fetus, it influences pre- and postnatal health by preventing megaloblastic anemia, DNA damage, and osteoporosis (Roman-Garcia et al., 2014; Stabler and Allen, 2004), and its deficiency highly contributes to adverse pregnancy and the neurodevelopmental morbidity of neonates (Molloy et al., 2008).
Although the clinical manifestations of VB12 deficiency have been extensively studied, the VB12-dependent molecular mechanisms regulating genomic and metabolic homeostasis of iECs to resist disease progression still require rigid elucidation. Further, VB12, as an enzymatic cofactor, is required for bacterial metabolic functions (Sokolovskaya et al., 2020), and its deficiency may be involved in affecting the gut microbial ecology (Degnan et al., 2014). Yet conflicting results hint at VB12’s role in gut microbiota regulation, especially under inflammatory conditions. Some studies showed the correlation between VB12 status and gut microbiota (Franco-Lopez et al., 2020), while other studies indicated that VB12 deficiency demonstrates no impact on microbial composition in healthy animals, but instead impairs the fecal microbiota in experimental dextran sulfate sodium–induced colitis (Lurz et al., 2020). This is in stark contrast to earlier findings demonstrating that dietary active VB12 exhibits proinflammatory effects (Zhu et al., 2019) and that VB12 deficiency protects against dextran sulfate sodium–induced colitis in mice (Benight et al., 2011).
Additionally, a recent clinical study revealed that there are no fecal microbial changes in infants with VB12 deficiency (Boran et al., 2020). Interestingly, supplementing acne patients with VB12 affects the transcriptome of skin microbiota, inducing the generation of inflammatory porphyrins that contribute to cutaneous disease pathogenesis (Kang et al., 2015). Thus, comprehensive mechanistic insights are still necessary to gain further insights into VB12’s properties.
Here, we elucidated VB12-dependent regulation of the genomic fitness of iECs and gut microbiome and associated metabolic responses during steady-state and Salmonella Typhimurium (STm) infection. Our results correlatively suggest that VB12 oral supplementation reprograms the transcriptional and metabolic machinery of iECs and supports microbiota composition and activity, which may cooperatively regulate the mitochondrial metabolisms of iECs to restrict intestinal oxygen-dependent STm infection.
Results
VB12 deficiency impairs the transcriptional and metabolic homeostasis of iECs
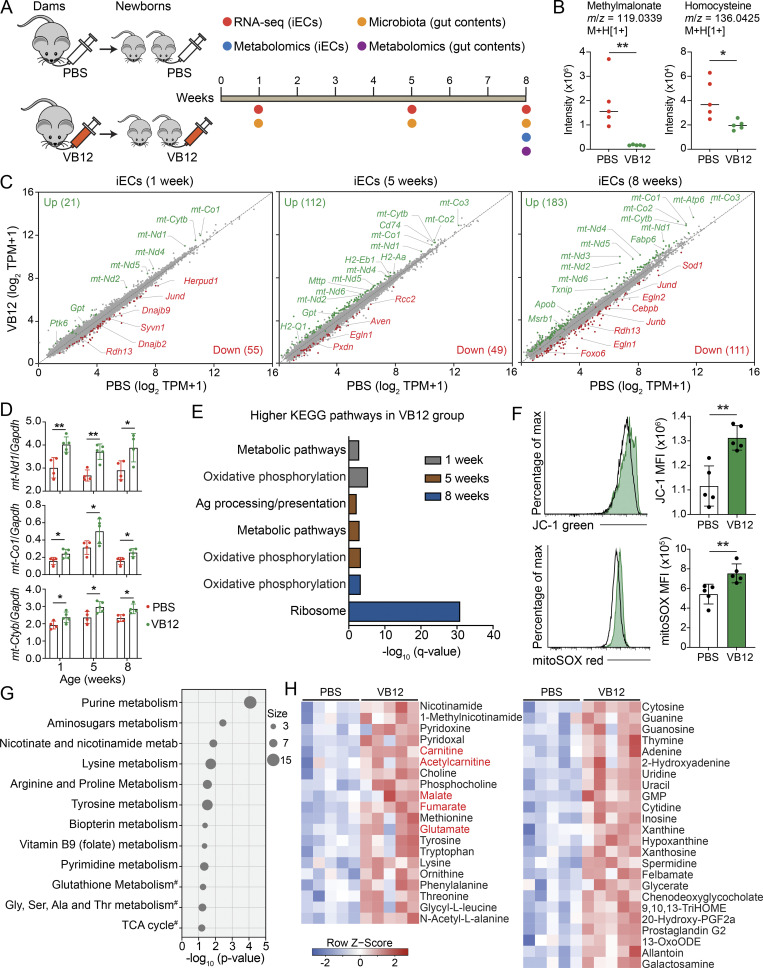
iECs absorb VB12 via the cubilin-dependent classic endocytosis pathway (Andersen et al., 2010). Thus, iECs serving as the entry site for VB12 may be functionally impacted. To interrogate this notion, groups of pregnant C57BL/6 mice kept on a VB12-deficient diet were gavaged with VB12 or PBS for 21 d. After birth, newborn mice of the respective dams were continuously fed a VB12-deficient diet and gavaged with VB12 or PBS. These mice were then euthanized at 1, 5, and 8 wk of age to FACS-sort iECs for transcriptomic analysis (Fig. 1 A). As expected, VB12 administration significantly reduced the levels of serum methylmalonate and homocysteine (Fig. 1 B), two critical markers that have been widely used for evaluating VB12 deficiency owing to their high sensitivity of measurement (Oh and Brown, 2003). Additionally, VB12 deficiency caused growth retardation in mice after 8 wk (Fig. S1 A). RNA sequencing (RNA-seq) analysis demonstrated that compared with VB12 deficiency, VB12 supplementation resulted in transcriptional modifications of iECs, whereupon the numbers of differentially expressed genes (DEGs) steadily increased over time (Fig. 1 C). Notably, a set of mitochondrial DNA (mtDNA) genes (e.g., mt-Nd1, mt-Co1, and mt-Cytb) were significantly enriched in VB12-supplemented mice (Fig. 1, C and D). VB12 deficiency also activated genes associated with stress responses, including Herpud1, Syvn1, Dnajb2, and Dnajb9 (1 wk); Egln1, Pxdn, and Aven (5 wk); and Sod1, FoxO6, Egln1, and Egln2 (8 wk; Fig. 1 C). Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses demonstrated oxidative phosphorylation as the major pathway that was augmented in the VB12-gavaged group (Fig. 1 E), and no significant pathway suppression was observed in those mice.
Figure 1.
VB12 deficiency impacts the transcriptional and metabolic activities of iECs. (A) Experimental scheme. Groups of pregnant C57BL/6 dams fed a VB12-deficient diet were gavaged with VB12 or PBS twice a week until the delivery of their newborns. Newborn mice were continuously kept on the same VB12-deficient diet and gavaged with VB12 or PBS twice a week for 8 wk. Mice were euthanized at ages of 1, 5, and 8 wk to FACS-sort iECs for RNA-seq and to collect gut contents for 16S rRNA sequencing. Metabolomic analyses were performed for gut contents and iECs of 8-wk-old mice. (B) Liquid chromatography–mass spectrometry analysis of serum methylmalonate and homocysteine in VB12- and PBS-gavaged mice at the age of 8 wk (n = 5/group). The m/z and adduct ion were labeled for each metabolite. (C) RNA-seq results depicted as scatterplots showing the DEGs in iECs between VB12- and PBS-gavaged mice at the age of 1, 5, and 8 wk (n = 3–5/group). The number of upregulated (green) and downregulated (red) DEGs in the VB12 group is shown in parentheses. (D) qPCR validation of representative mitochondrial genes (n = 4–5/group). (E) KEGG pathways of DEGs enriched in VB12 versus PBS group. (F) Mitochondrial membrane potential (JC-1) and mROS (mitoSOX) of iECs isolated from VB12- or PBS-gavaged mice at 8 wk (n = 5/group). (G and H) Metabolic pathways (G) and differentially enriched metabolites (H) in iECs isolated from mice gavaged with VB12 or PBS for 8 wk (n = 5/group). #, interesting pathways with P values >0.05. Metabolites involved in carnitine shuttle and TCA cycle are highlighted in red. FACS data are representative of three independent experiments (F). Bar graphs show mean ± SD. *, P < 0.05; **, P < 0.01; two-tailed unpaired t test (B, D, and F).
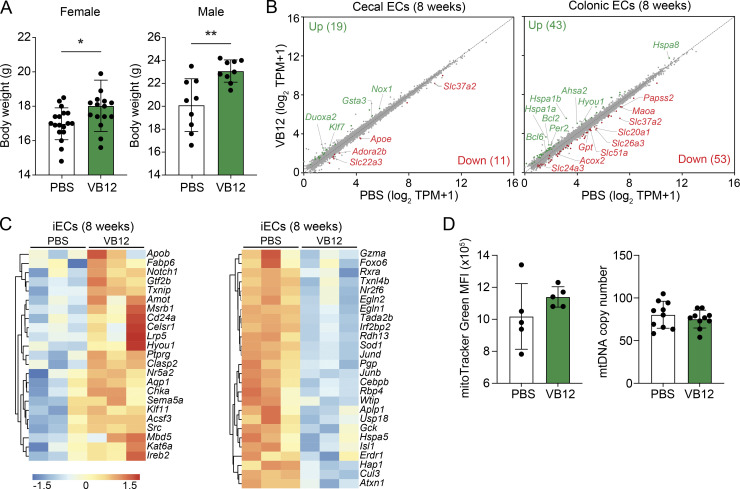
Figure S1.
VB12 deficiency differentially impacts the transcriptomes of gut ECs. (A) Body weight analysis of VB12-deficient and VB12-replete female (n = 15–18/group) and male (n = 9/group) mice. (B) Scatterplots of DEGs in cecal ECs (n = 4/group) and colonic ECs (n = 4/group) isolated from mice gavaged with VB12 versus PBS for 8 wk. The number of upregulated (green) and downregulated (red) DEGs in the VB12 group is shown in parentheses. (C) Heatmap of representative DEGs enriched in iECs of VB12- or PBS-gavaged mice. (D) Mitochondrial mass (n = 5/group) and mtDNA copies (n = 9–10/group) in iECs isolated from mice gavaged with VB12 or PBS for 8 wk. FACS results are representative of three independent experiments, and qPCR results are from one experiment. Bar graphs show mean ± SD; *, P < 0.05; **, P < 0.01; two-tailed unpaired t test (A).
Given that iECs were steadily regulated in VB12-gavaged mice within 8 wk, we then spatially analyzed the transcriptomes of cecal and colonic epithelial cells (ECs) and compared them with iECs from 8-wk-old mice. Compared with cecal and colonic ECs (Fig. S1 B), iECs were transcriptionally modified by VB12 at a greater magnitude (Fig. 1 C). These iECs were enriched with genes related to hypoxia (Cd24a, Src, Aqp1, Hyou1, and Notch1), EC migration (Clasp2, Amot, Ptprg, and Sema5a), antioxidation (Msrb1), and lipid metabolic process (e.g., Apob and Fabp6; Fig. S1 C). Intriguingly, the mitochondrially encoded genes were elevated solely in iECs derived from VB12-gavaged mice (Fig. 1, C and D), which also had increased mitochondrial membrane potential and reactive oxygen species (mROS; Fig. 1 F), but not mitochondrial mass or mtDNA copies (Fig. S1 D), suggesting that VB12 transcriptionally supports the mitochondrial respiration of iECs.
Untargeted metabolomic analysis of iECs derived from VB12-deficient compared with VB12-supplemented mice at 8 wk of age (scheme in Fig. 1 A) demonstrated differential pathways associated with nucleotide, amino acid, and vitamin metabolisms (Fig. 1 G). Mitochondrial TCA cycle and antioxidant glutathione metabolism were also likely impaired by VB12 deficiency. Interestingly, VB12 deficiency led to metabolite reductions in iECs, including amino acids (e.g., methionine and lysine) and nucleobases (e.g., cytosine and uridine; Fig. 1 H). Additionally, iECs of VB12-depleted mice exhibited lower levels of B vitamins and cofactors, such as nicotinamide, 1-methylnicotinamide, pyridoxine, and choline, which mitigate cellular oxidative stress. Long-chain fatty acids (LCFAs; e.g., 9,10,13-TriHOME and 13-OxoODE), carnitine and acetylcarnitine of carnitine shuttle, which are both required for mitochondrial LCFA β-oxidation, were also decreased owing to VB12 deficiency. Moreover, declined levels of malate, fumarate, and glutamate in iECs devoid of VB12 (Fig. 1 H) may be attributed to the suppressed TCA cycle that fuels mitochondrial electron transport chain (ETC). Thus, these results suggest that VB12 deficiency impairs mitochondrial metabolism and promotes oxidative stress in iECs.
VB12 deficiency reduces the mitochondrial respiration and β-oxidation of iECs during STm infection
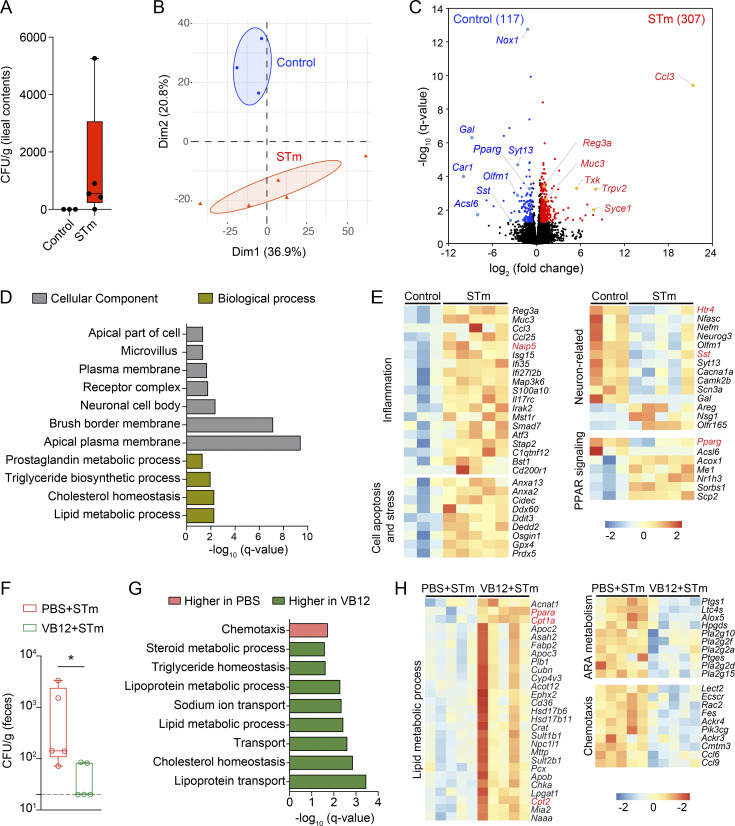
STm invades intestinal ECs (Ginocchio et al., 1992) and induces transcriptional changes 2 d after infection (Haber et al., 2017). To investigate the iEC response to STm in VB12-deficient mice, we analyzed the transcriptomes of iECs isolated from VB12-deficient mice infected with STm for 2 d and compared them with uninfected controls (Fig. S2 A). Here, STm infection modified the transcriptomes of iECs (Fig. S2, B and C) and impaired lipid metabolic pathways and the cellular component of neuronal cell body (Fig. S2 D). Further, a panel of genes associated with inflammation (e.g., Ccl3, Ccl25, Reg3a, Isg15, and Naip5), cell stress, and death (e.g., Cidec, Ddit3, Dedd2, and Osgin1) were increased in iECs of STm-infected mice, whereupon neuron-related transcripts, including serotonin receptor Htr4 and neuropeptide somatostatin (Sst), both of which are important for controlling gut motility, were decreased (Fig. S2 E). Interestingly, the expression of peroxisome proliferator-activated receptor (PPAR) γ critically involved in mitochondrial β-oxidation was also suppressed in iECs by STm.
Figure S2.
VB12 deficiency impacts iEC response to STm. (A) STm colonization in the ileal contents of VB12-deficient mice 2 d after infection (n = 3–5/group). (B–E) PCA plot (B), volcano plot (C), significant DAVID Gene Ontology (GO) pathways (D), and representative DEGs (E) in iECs isolated from VB12-deficient mice infected with STm versus uninfected controls. (F) STm colonization in the feces of VB12- and PBS-gavaged mice (n = 5/group) 2 d after infection. (G) GO pathways enriched in VB12- or PBS-gavaged mice infected with STm. (H) Heatmap of DEGs associated with indicated pathways. ARA, arachidonic acid. Genes of interest are highlighted in red in heatmaps. STm colonization data are from one experiment (A) or representative of three independent experiments (F). *, P < 0.05; Mann–Whitney U test (F).
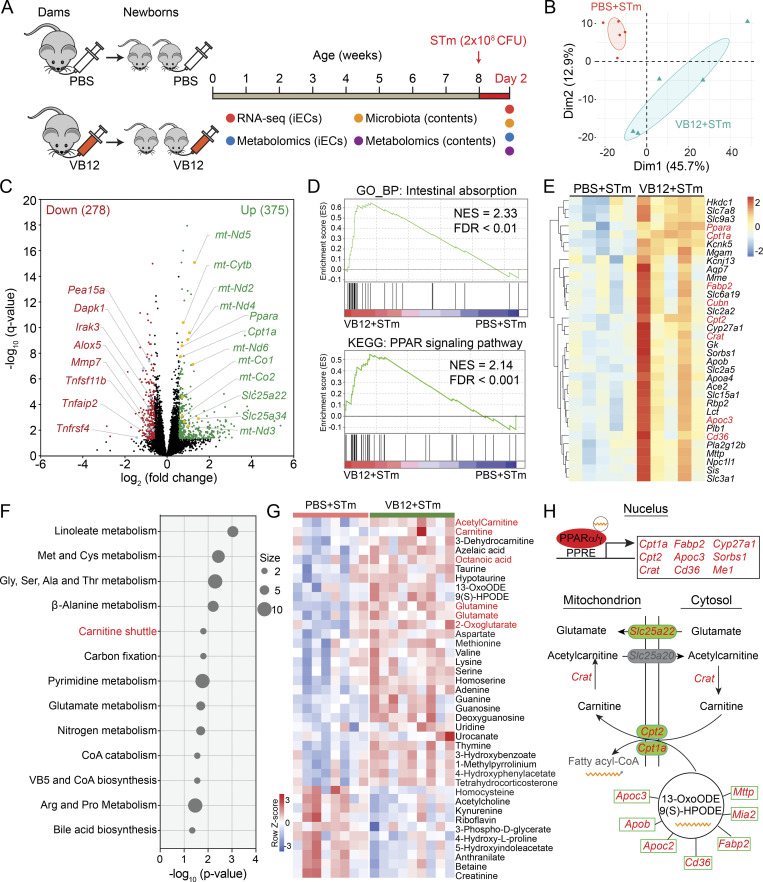
To elaborate on the VB12-dependent iEC regulation in response to STm infection, we analyzed the transcriptomes and metabolomes of iECs isolated from mice gavaged with VB12 or PBS for 8 wk and subsequently infected with STm for 2 d (Figs. 2 A and S2 F). Results demonstrated that VB12 significantly modified the transcriptomes of iECs during STm infection (Fig. 2 B), resulting in 375 upregulated and 278 downregulated DEGs in iECs derived from mice received VB12 compared with those with VB12 deficiency (Fig. 2 C). Mitochondrially encoded genes (e.g., mt-Nd5, mt-Cytb, mt-Nd2, and mt-Nd4), along with nuclear-encoded mitochondrial transporters (Slc25a22 and Slc25a34), were notably activated in iECs through VB12 supplementation (Fig. 2 C). These cells were primarily defined by pathways related to intestinal absorption, PPAR signaling pathway (Fig. 2 D), lipoprotein transport, and lipid metabolic process (Fig. S2 G). Accordingly, iECs of VB12-gavaged mice were enriched with transcripts of the VB12 receptor (Cubn), the fatty acid uptake receptor (Cd36), and chylomicron components (Mttp, ApoB, ApoC2, and ApoC3; Fig. 2 E), which are critical for fatty acid absorption and systemic distribution.
Figure 2.
VB12 deficiency reduces the mitochondrial respiration and fatty acid β-oxidation of iECs during STm infection. (A) Experimental scheme. Groups of pregnant C57BL/6 dams fed a VB12-deficient diet were gavaged with VB12 or PBS twice a week until the delivery of their newborns. Newborn mice were gavaged continuously with VB12 or PBS twice a week for 8 wk and then orally infected with STm. 2 d later, iECs and intestinal contents were processed for transcriptomic, metabolomic, and microbiota analyses. (B) PCA plot of iEC transcriptomes of VB12- and PBS-gavaged mice (n = 5/group). (C) Volcano plot of DEGs between iECs of VB12- and PBS-gavaged mice. The number of upregulated (green) and downregulated (red) DEGs in the VB12 group is shown in parentheses. (D) Representative GSEA plots showing “intestinal absorption” and “PPAR signaling pathway” were enriched in iECs of VB12-supplemented mice. NES, normalized enrichment score. (E) Heatmap of DEGs for intestinal absorption and PPAR signaling pathways. Genes related to PPAR signaling and the VB12 receptor are highlighted in red. (F) Metabolic pathway analysis of metabolites; the intensity significantly differed in the two groups of iECs (n = 8–9/group). (G) Heatmap of differentially enriched metabolites. Metabolites of carnitine shuttle and TCA cycle and the cellular PPAR agonist octanoic acid are highlighted in red. (H) Summary gene and metabolic networks. Upon binding to fatty acids, PPARα/γ activated genes associated with lipid metabolic process and carnitine shuttle that facilitated the translocation of fatty acids into mitochondria. Additionally, the mitochondrial glutamate carrier (Slc25a22) and glutamate were consistently enriched in iECs of VB12-gavaged mice. The significantly enriched genes in VB12 group are highlighted in red and metabolites in black. Genes and metabolites in gray are not significant. PPRE, PPAR response element. Statistical significance was determined using the DESeq2 package (C and E) or two-tailed unpaired t test (G).
The PPAR signaling pathway, which can be activated by free fatty acids, regulates the transcription of genes involved in peroxisomal and mitochondrial β-oxidation (Minnich et al., 2001). Here, VB12 supplementation enhanced the expression of Ppara and its target genes in iECs, particularly carnitine palmitoyltransferase 1A (Cpt1a), carnitine palmitoyltransferase 2 (Cpt2), and carnitine O-acetyltransferase (Crat; Fig. 2 E), all of which are essential components of the carnitine shuttle. The expression of PPARγ-activated genes, including Cd36 and the mitochondrial enzyme Cyp27a1, was also enhanced in iECs of VB12-gavaged mice. In contrast, VB12 deficiency exacerbated inflammatory responses during intestinal STm infection, as demonstrated by enriched transcripts associated with inflammation (e.g., Tnfsf11b, Tnfrsf4, and Tnafaip2), chemotaxis (e.g., Ccl6, Ccl9, Ackr3, and Ackr4), and arachidonic acid and prostaglandin metabolism (e.g., Alox5, Ptges, Ptgs1, and Hpgds), as well as genes implicated in cell death and apoptosis (e.g., Dapk1, Pea15a, and Irak3; Figs. 2 C and S2 H).
Importantly, cellular metabolomic analysis demonstrated that linoleate metabolism and carnitine shuttle were the top altered pathways associated with VB12 deficiency (Fig. 2 F). Consistent with the steady-state condition, iECs isolated from STm-infected mice were also characterized by differential pathways related to amino acid and nucleotide metabolism (Fig. 2 F). Interestingly, VB12 deficiency reduced the mitochondrial glutaminolysis (glutamine, glutamate, 2-oxoglutarate) supplying the TCA cycle and decreased the activity of carnitine shuttling (carnitine and acetylcarnitine) crucial for fatty acid β-oxidation. Furthermore, iECs of VB12-deprived mice had diminished levels of fatty acids (octanoic acid, 13-OxoODE, and 9[S]-HPODE), taurine, and hypotaurine (Fig. 2 G). These iECs also had decreased aspartate necessary for cellular proliferation (Birsoy et al., 2015) and substrates for nucleotide metabolism (e.g., adenine, guanine, uridine, and thymine). During STm infection, VB12 deficiency reduced azelaic acid and proline, exerting antioxidant properties, and enriched kynurenine, associated with toxic inflammation. Unsurprisingly, VB12 deficiency elevated homocysteine and decreased methionine levels in iECs (Fig. 2 G). These results indicate that VB12 deficiency reduces the capacity of iECs for mitochondrial respiration and fatty acid β-oxidation, potentially contributing to intestinal pathogen expansion.
VB12 deficiency impacts the development of gut microbiota
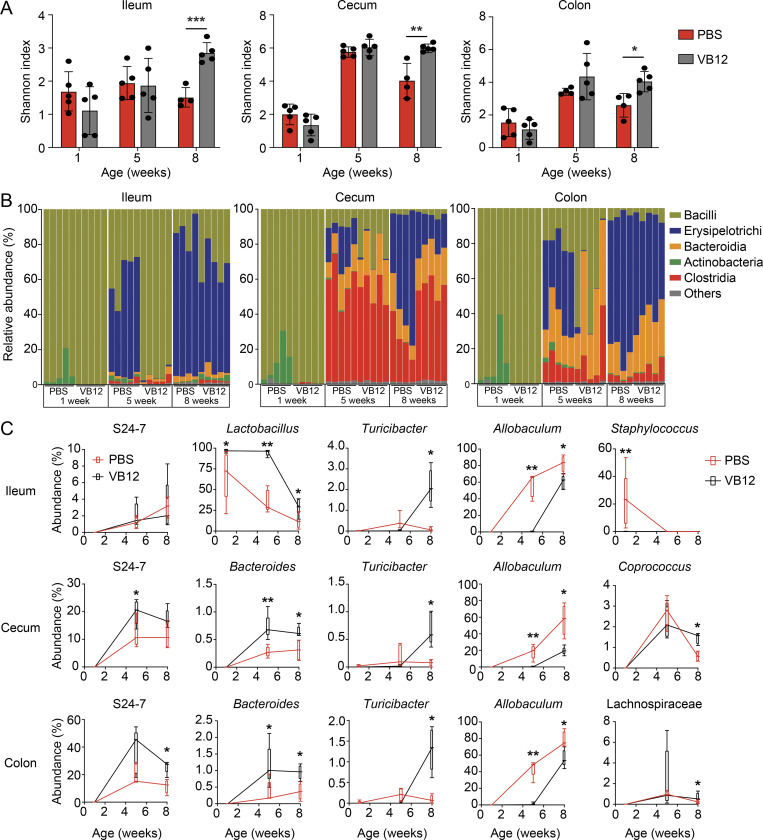
Having shown that VB12 supplementation affects the transcriptomes and metabolome of iECs, we posited that VB12 may also support the host gut microbiota to potentially fortify the regulation of molecular machinery in iECs (Kelly et al., 2015). Additionally, although VB12 is absorbed at the ileal region, residual dietary VB12, as a cofactor, may also reach the cecum and colon to potentially influence microbial growth in these regions (Kelly et al., 2019). Thus, we first analyzed the ileal, cecal, and colonic microbial compositions of mice gavaged with VB12 or PBS for 1, 5, and 8 wk at steady state (scheme in Fig. 1 A). Data demonstrated that VB12 deficiency hindered the formation of a diverse bacterial community, with a significant reduction in the Shannon index in the ileal, cecal, and colonic contents of mice within 8 wk (Fig. 3 A). At the bacterial class level, VB12 deficiency resulted in the expansion of actinobacteria in 1-wk-old mice and highly enriched Erysipelotrichi in all three intestinal regions after 5 wk, while the relative abundance of Bacteroidia and Clostridia was reduced in the cecum and colon of 8-wk-old mice (Fig. 3 B).
Figure 3.
VB12 deficiency impairs gut microbiota development. 16S rRNA sequencing of ileal, cecal, and colonic contents collected from mice gavaged with VB12 or PBS for 1, 5, and 8 wk (n = 4–5/group). (A) Shannon index measuring bacterial diversity. (B and C) Bacterial class abundance (B) and major differential bacterial genera (C) of the ileum, cecum, and colon samples collected from the indicated mouse groups. Bar graphs show mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; two-tailed unpaired t test (A) or Mann–Whitney U test (C).
Further analysis revealed key bacterial genera associated with VB12 status. When comparing the two groups of mice, slight differences were noticed in 1-wk-old mice; however, significant changes were observed in the bacterial genera of 8-wk-old mice. VB12 deficiency generally reduced the relative abundance of Turicibacter, which has been reported to regulate host lipid metabolism (Fung et al., 2019), and increased the levels of Allobaculum that accounted for 60–80% of total bacteria in 8-wk-old mice (Fig. 3 C). Additionally, gut region–specific microbial changes were identified. In the ileum, the abundance of Lactobacillus was reduced in VB12-deficient mice of all ages (Fig. 3 C). In cecal and colonic regions, members of Bacteroidales (family S24-7 and genus Bacteroides) were present at higher levels than those in the ileum, but their levels were reduced by ∼50% (Fig. 3 C), suggesting that VB12 deficiency influences gut microbiome development.
VB12 deficiency reduces gut bacterial diversity and associated metabolic activity during STm infection
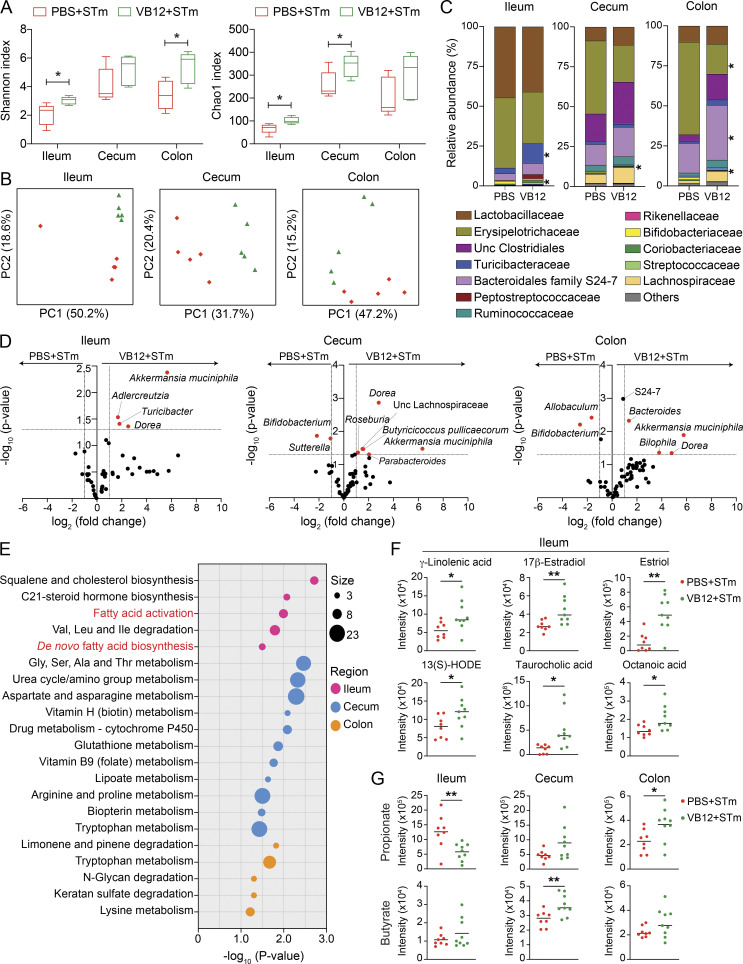
Next, we analyzed the gut microbiota and associated metabolites in mice gavaged with VB12 or PBS for 8 wk and subsequently infected with STm (scheme in Fig. 2 A). Results demonstrated that α diversity, as measured by Shannon and Chao1 indexes (Fig. 4 A), was decreased in the ileal, cecal, and colonic contents of mice receiving PBS compared with VB12. Unweighted UniFrac principal coordinates analysis (PCoA) revealed distinct bacterial community structures in each of the regions (Fig. 4 B). Compositional analysis further demonstrated significant differences in the ileum, cecum, and colon at family and species levels (Fig. 4, C and D). Akkermansia muciniphila, a mucin-degrading bacterium that reduces microbial translocation and inflammation (Derrien et al., 2017), as well as Dorea, which metabolizes sialic acids commonly found as end products of mucin degradation (Schirmer et al., 2016), were both observed at significantly lower levels in all the regions of VB12-deficient mice compared with VB12-gavaged mice (Fig. 4 D). As observed in the steady state (Fig. 3 C), Turicibacter (Turicibacteraceae) was significantly decreased in the ileum of VB12-deficient mice during STm infection (Fig. 4, C and D) and accounted for 3.5% of total bacteria compared with 12.3% in VB12-replete mice. Members of the Lachnospiraceae family (e.g., Butyricicoccus pullicaecorum and Roseburia) producing SCFAs (e.g., butyrate) were reduced in the cecum of VB12-deficient mice. In the colon, these mice also showed a significantly lower abundance of family S24-7 and Bacteroides, along with increased levels of Allobaculum (Erysipelotrichaceae). In contrast, Bifidobacterium, possessing the capability to synthesize VB12, showed decreased relative abundance in the cecal and colonic contents of VB12-supplemented mice (Fig. 4 D).
Figure 4.
VB12 deficiency reduces microbiota-associated fatty acids during STm infection. Ileal, cecal, and colonic contents were collected from VB12- and PBS-gavaged mice (8 wk old) infected with STm for 2 d to analyze microbiota composition (n = 5/group) and associated metabolites (n = 8–9/group). (A) Shannon and Chao1 indexes measuring bacterial diversity. (B) Unweighted UniFrac PCoA plots showing the bacterial community structures. (C) Top bacterial (family) abundance of the ileal, cecal, and colonic contents. (D) Volcano plots of normalized reads showing significantly different taxa. Dashed lines indicate the fold-change of 2 and P value of 0.05. (E) Significant metabolic pathways of the ileal, cecal, and colonic luminal samples collected from VB12- and PBS-gavaged mice infected with STm. The overlapped size indicates the number of significant metabolic features mapped to corresponding pathways. (F) Scatterplots for putatively annotated metabolites, known to activate PPAR signaling pathway. (G) Scatterplots for propionate and butyrate in the ileal, cecal, and colonic contents of VB12- and PBS-gavaged mice. *, P < 0.05; **, P < 0.01; two-tailed unpaired t test (A, C, F, and G).
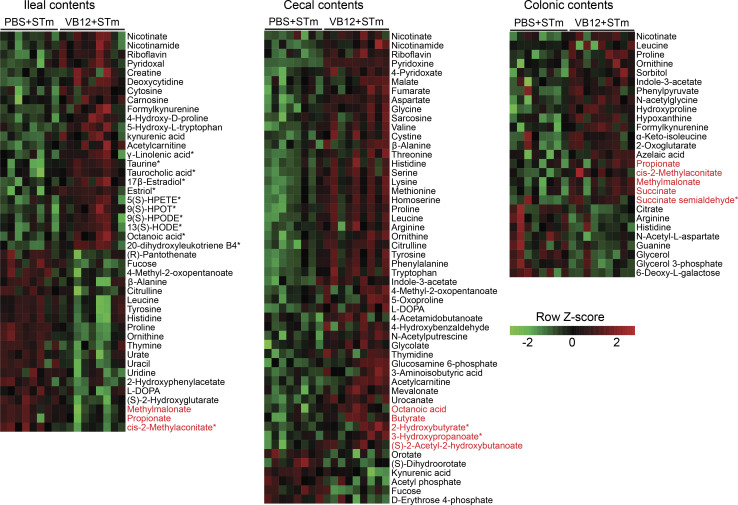
Interestingly, fecal metabolomic analysis demonstrated differential metabolite enrichment in different luminal regions that was associated with microbial composition. Here, vitamins and cofactors (e.g., nicotinate, riboflavin, and pyridoxine) supporting the growth and diversity of gut microbes (Visconti et al., 2019) were decreased in the intestinal contents of VB12-deficient mice compared with VB12-gavaged mice during STm infection (Fig. S3). Further, spatially distinct metabolic profiles were revealed between the two groups of mice. In the ileum, VB12 deficiency suppressed fatty acid activation and biosynthetic pathways (Fig. 4 E) and accordingly decreased luminal levels of fatty acids serving as PPARα/γ agonists. This includes putatively annotated γ-linolenic acid (Evans et al., 2010; Forman et al., 1997), 13(S)-HODE (Cimen et al., 2011; Kammerer et al., 2011), 17β-estradiol (Campbell et al., 2003; Rodriguez-Cuenca et al., 2007), and octanoic acid (Intrasuksri et al., 1998; Liberato et al., 2012; Fig. 4 F), possibly potentiating the PPAR signaling pathway in iECs, as shown by RNA-seq (Fig. 2 D). Instead, amino acid and glycan metabolisms were the major differentially activated pathways in the colon and particularly in the cecum (Fig. 4 E), wherein most amino acids (e.g., aspartate, methionine, proline, and arginine) were reduced in mice kept on the VB12-deficient diet (Fig. S3). These mice also had significantly lower levels of metabolites related to the TCA cycle and cellular respiration (e.g., malate, fumarate, succinate, 2-oxoglutarate, and ubiquinone).
Figure S3.
VB12 deficiency affects microbiota-associated metabolism during STm infection. Heatmap showing the differentially enriched metabolites in the ileal, cecal, and colonic contents collected from VB12- and PBS-gavaged mice infected with STm (n = 8–9/group). Metabolite identification was performed through reference to metabolite library, curated by running each standard. Additional metabolites were identified based on their enrichment in significant metabolic pathways and the m/z match and are labeled with asterisks (*). Metabolites highlighted in red are associated with propionate or butyrate metabolic pathways. Statistical significance was determined by two-tailed unpaired t test.
Members of Bacteroidetes mainly produce propionate, while Firmicutes and particularly Clostridia mostly synthesize butyrate in the gut (Louis and Flint, 2017; Parada Venegas et al., 2019). Here, we observed significantly lower levels of cecal butyrate and colonic propionate in VB12-deficient mice than in those supplemented with VB12 (Fig. 4 G), which was associated with the reduced relative abundance of cecal Clostridia (e.g., Butyricicoccus pullicaecorum and Roseburia) and colonic Bacteroidia (e.g., S24-7 and Bacteroides; Fig. 4 D). Furthermore, metabolites (e.g., 3-hydroxypropanoate, 2-hydroxylbutyrate, and succinate semialdehyde) involved in propionate and butyrate metabolism were also decreased in the cecal and colonic contents of VB12-deficient mice (Fig. S3). In contrast, propionate and its metabolic intermediates (cis-2-methylaconitate and methylmalonate) were increased in the ileal samples of VB12-deficient mice. These results indicate that VB12 administration modifies microbiota function to potentially fine-tune intestinal epithelial signaling in response to STm infection.
Host susceptibility to STm infection is augmented by VB12 deficiency
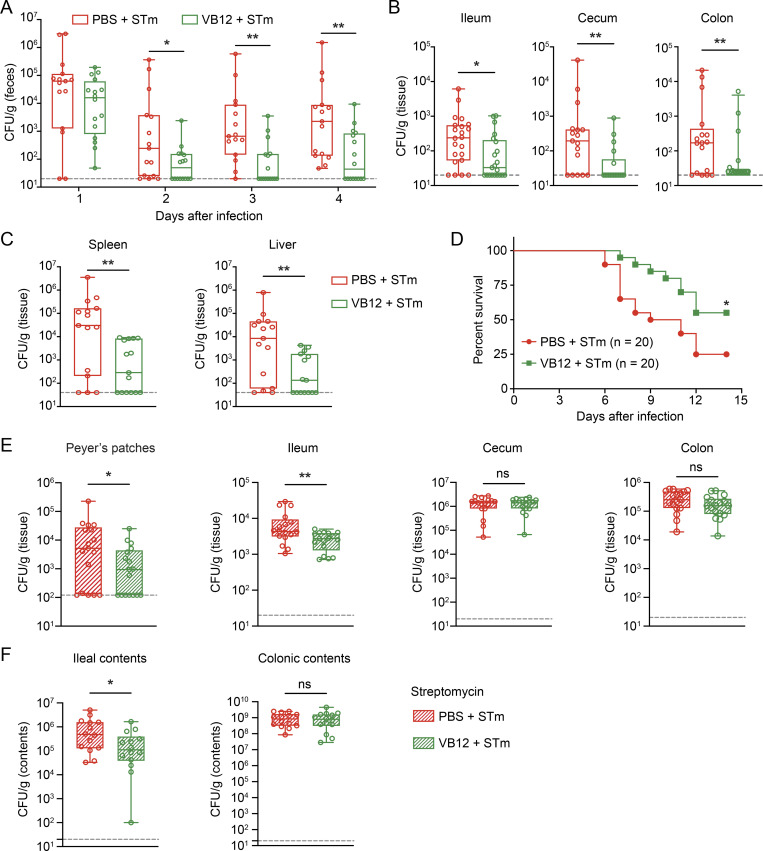
To investigate whether VB12-dependent regulation of iECs and microbiota contributes to host protection against STm infection, we monitored mice gavaged with VB12 or PBS that were subsequently infected with STm at different time points. Data demonstrated that VB12-gavaged mice had reduced STm burdens in the feces as early as 2 d after infection, and these differences were further increased over time (Fig. 5 A). Notably, VB12 supplementation significantly restrained STm tissue invasion (Fig. 5 B) and limited STm dissemination into spleen and liver (Fig. 5 C), resulting in the prolonged survival of mice receiving VB12 compared with those deficient in VB12 (Fig. 5 D).
Figure 5.
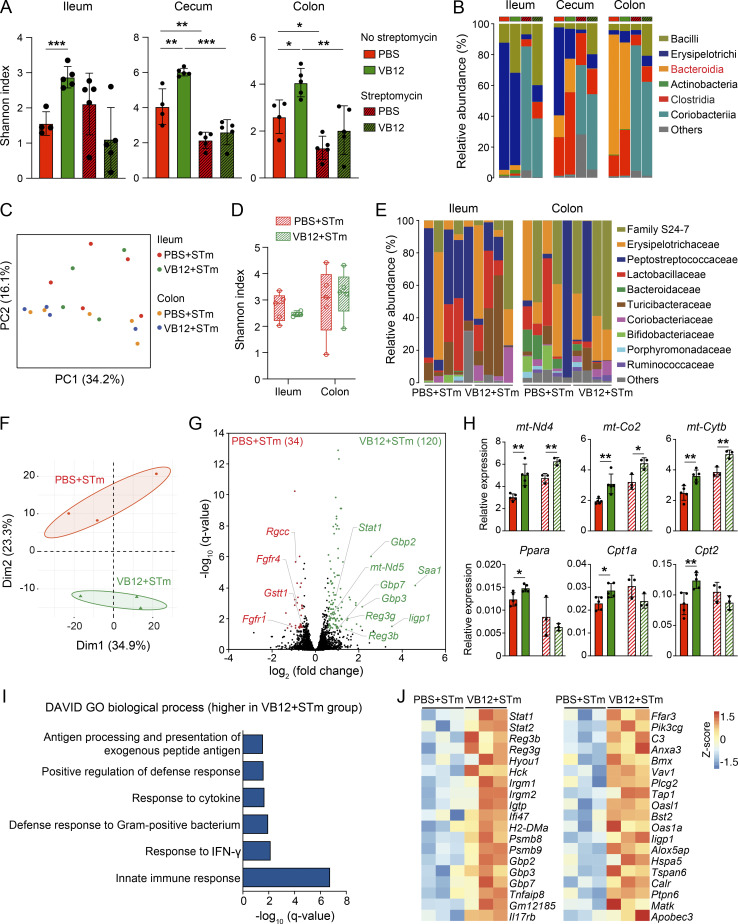
VB12 deficiency renders host susceptible to STm infection. (A) CFU burdens in the feces of VB12- and PBS-gavaged mice (n = 15–16/group) infected with STm. (B) CFU burdens in the ileal, cecal, and colonic tissues dissected from VB12- and PBS-gavaged mice (n = 21–22/group) 2 d after STm infection. (C) CFU burdens in the spleen and liver on day 4 (n = 15–16/group). (D) Survival of VB12- and PBS-gavaged mice (n = 20/group) during STm infection. (E) Tissue CFU burdens of VB12- and PBS-gavaged mice (n = 17/group) treated with streptomycin and then infected with STm for 2 d. (F) CFU burdens in ileal and colonic contents of indicated mice (n = 14/group) treated with streptomycin. All mice were continuously gavaged with VB12 or PBS for 8 wk before STm infection. Data are from one experiment (D) or combined from three experiments (A–C, E, and F). *, P < 0.05; **, P < 0.01; Mann–Whitney U test (A–C, E, and F) or log-rank test (D).
To dissect the roles of functional iECs and microbiota in controlling STm infection, mice were treated with streptomycin to abate gut microbiota, and STm infection was examined in antibiotic-treated mice. Analyzing the ileal, cecal, and colonic microbial compositions of VB12- and PBS-gavaged mice before infection demonstrated that VB12-associated microbial regulation was not seen in streptomycin-treated mice (Fig. S4 A). Notably, Bacteroidia were eliminated at all anatomical sites, and Clostridia were reduced in the cecal contents (Fig. S4 B). Consistently, no significant changes in microbiota composition were observed in mice that were subsequently infected with STm for 2 d (Fig. S4, C–E).
Figure S4.
Streptomycin treatment abates the VB12-dependent microbial regulation without disrupting iEC-mitochondrial gene activation. (A and B) Shannon index (A) and bacterial class abundance (B) in the ileal, cecal, and colonic contents collected from VB12- and PBS-gavaged mice (8 wk old) treated with streptomycin for 24 h (n = 5/group), compared with those derived from untreated mice (n = 4–5/group). (C–E) PCoA plot (C), Shannon index (D), and bacterial family abundances (E) in the ileal and colonic contents of VB12- and PBS-gavaged mice (8 wk old) that were treated with streptomycin and infected with STm for 2 d (n = 5/group). (F) PCA plot of the transcriptomes of iECs isolated from VB12- and PBS-gavaged mice (8 wk old) treated with streptomycin and infected with STm for 2 d (n = 3/group). (G) Volcano plot of DEGs obtained from RNA-seq results. The number of DEGs enriched in VB12 or PBS group is shown in parentheses. (H) qPCR analysis of genes involved in mitochondrial respiration (mt-Nd1, mt-Co2, mt-Cytb) and PPAR signaling (Ppara, Cpt1a, Cpt2) in iECs of VB12- and PBS-gavaged mice that were infected with STm with (pattern fill) or without (solid fill) streptomycin pretreatment. (I) Significant Gene Ontology (GO) pathways in VB12-gavaged mice treated with streptomycin and infected with STm. (J) Heatmap showing DEGs associated with significant GO terms. Bar graphs show mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; two-tailed unpaired t test (A and H).
Interestingly, RNA-seq analysis demonstrated that iECs of VB12-gavaged mice treated with streptomycin and infected with STm still displayed a different transcriptome from that of VB12-deficient mice (Fig. S4 F). The upregulation of mitochondrially encoded genes (e.g., mt-Nd5, mt-Co2, and mt-Cytb) was largely retained in mice gavaged with VB12 (Fig. S4, G and H). However, activation of fatty acid oxidation–related genes (e.g., Ppara, Cpt1a, and Cpt2) and pathways (e.g., PPAR signaling) was absent when ileal microbiota were perturbed by streptomycin treatment (Fig. S4, H–J), potentially highlighting the relevance of VB12 in regulating microbiota-derived signals such as PPAR agonists (Fig. 4 F) to activate mitochondrial fatty acid β-oxidation in iECs.
In comparing VB12- and PBS-gavaged mice treated with streptomycin and infected with STm, a significant reduction of STm CFUs was observed in the ileal tissue and luminal contents, rather than the cecal or colonic samples of the VB12-gavaged group (Fig. 5, E and F). This indicates that sustained iEC-mitochondrial function, along with bacterially associated propionate and butyrate biosynthesis in the cecal and colonic regions, may be requisites to resist intestinal STm infection.
VB12 deficiency reduces host capacity to control oxygen-dependent STm growth
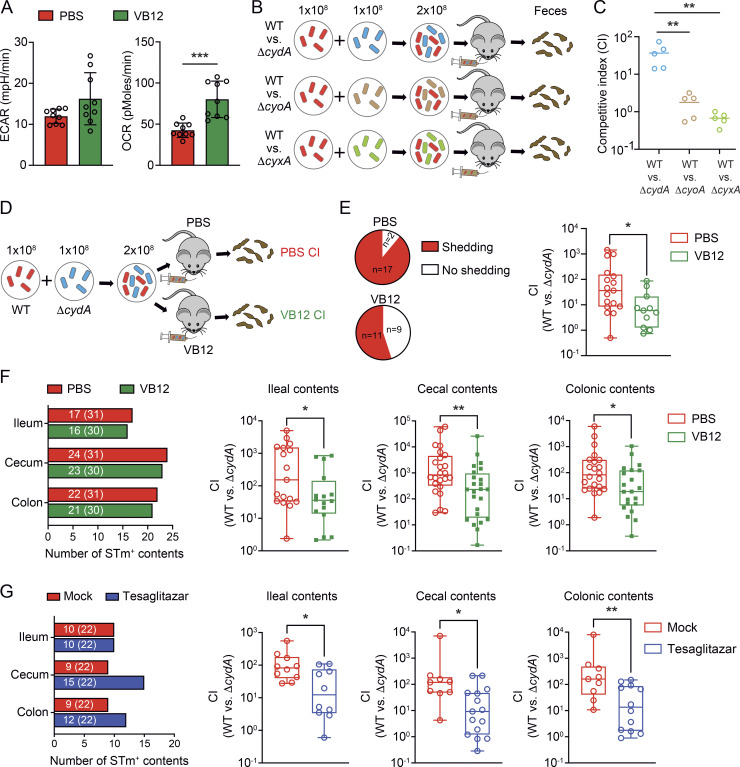
Having shown that VB12 deficiency reduced mitochondrial respiration and β-oxidation in iECs, the two major oxygen-consuming processes inducing epithelial hypoxia in mice (Byndloss et al., 2017; Cevallos et al., 2019), we tested the capability of iECs to consume oxygen by Seahorse analysis. iECs isolated from VB12-deficient mice had significantly reduced basal oxygen consumption rates (OCRs), a measurement of mitochondrial OCR, compared with those derived from VB12-administered mice (Fig. 6 A). Thus, we rationally postulated that VB12 deficiency may exacerbate STm infection at the ileal level by failing to control ileal epithelial oxygenation. To address this notion, we investigated whether STm aerobic respiration (Litvak et al., 2019; Rivera-Chavez et al., 2016), mediated by cytochrome bd oxidase (encoded by cydAB genes), cytochrome bo3 oxidase (encoded by cyoAB genes), or cytochrome bd-II oxidase (encoded by cyxAB genes), conferred a fitness advantage in the gut of VB12-deficient mice (Fig. 6 B). Interestingly, infection of VB12-deficient mice with a 1:1 mixture of the STm WT and a cytochrome mutant (ΔcydA, ΔcyoA, or ΔcyxA) revealed that there was a striking fitness advantage to STm expressing cydA, in contrast to the equal fecal recovery of WT and other mutants (ΔcyoA or ΔcyxA; Fig. 6 C). Subsequently, VB12-deficient and -replete mice were infected with equal amounts of WT and ΔcydA mutant (Fig. 6 D), whereupon VB12 supplementation reduced the fecal competitive index (CI) of the two strains by approximately an order of magnitude (Fig. 6 E). Further, the cydA-conferred growth advantage was significantly reduced in not only the ileum, but also the cecum and colon, through VB12 administration (Fig. 6 F), suggesting the VB12-dependent control of oxygen-conferred STm growth.
Figure 6.
VB12 deficiency reduces the host capacity to control intestinal oxygen-dependent STm growth. (A) Seahorse analysis showing that iECs of VB12-deficient mice had a defect in consuming oxygen compared with those derived from VB12-gavaged mice 2 d after STm infection (n = 9/group). ECAR, extracellular acidification rate. (B) Experimental scheme. The STm WT and mutants lacking an aerobic respiration gene (ΔcydA, ΔcyoA, or ΔcyxA) were equally mixed to infect mice kept on the VB12-deficient diet (1 × 108 CFU/strain/mouse, n = 5/group). The CI was determined based on the relative recovery of the fecal CFU of WT versus the respective mutants. (C) The CI of WT versus indicated mutants in the feces was determined 4 d after infection. (D) Experimental scheme. VB12- and PBS-treated mice were infected with a 1:1 mixture of WT and ΔcydA, and the CI was determined to examine oxygen-dependent STm growth. (E) Pie charts showing the number of VB12- and PBS-gavaged mice (n = 19–20/group) shedding STm 4 d after infection and bar graph showing CI in the feces of the shedders. Nonshedders were excluded from CI calculation. (F) Ileal, cecal, and colonic contents were collected from VB12- and PBS-gavaged mice (n = 31/group) infected with a 1:1 mixture of WT and ΔcydA for 4 d. The number of contents in which CFU was detected is depicted in the left panel (the total number of samples is shown in parentheses). The CI was calculated based on CFU recovered from the shedders. (G) VB12-deficient mice (n = 22/group) were inoculated with a 1:1 mixture of WT and ΔcydA strains and received PPARα/γ dual agonist tesaglitazar or were mock-treated. The left bar graph shows the number of contents containing STm (the total number of samples is in parentheses). The CI was determined in the STm-positive ileal, cecal, and colonic contents 4 d after infection. All mice were continuously gavaged with VB12 or PBS for 8 wk before STm infection. Data are from one experiment (C) or combined from two (A), three (G), or four (E and F) experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; two-tailed unpaired t test (A) or Mann–Whitney U test (C and E–G).
Next, we investigated whether reduced PPAR signaling in VB12-deficient mice could account for the elevated oxygen-dependent STm growth. Thus, VB12-deficient mice were gavaged with PPARα/γ dual agonist tesaglitazar, and the CI of WT and ΔcydA mutant was evaluated. Results showed that tesaglitazar treatment significantly reduced the oxygen-conferred STm expansion in the hosts (Fig. 6 G), suggesting that PPAR signaling in VB12-deficient mice serves as a critical target that can be activated by micronutrients such as VB12 to limit aerobic STm growth.
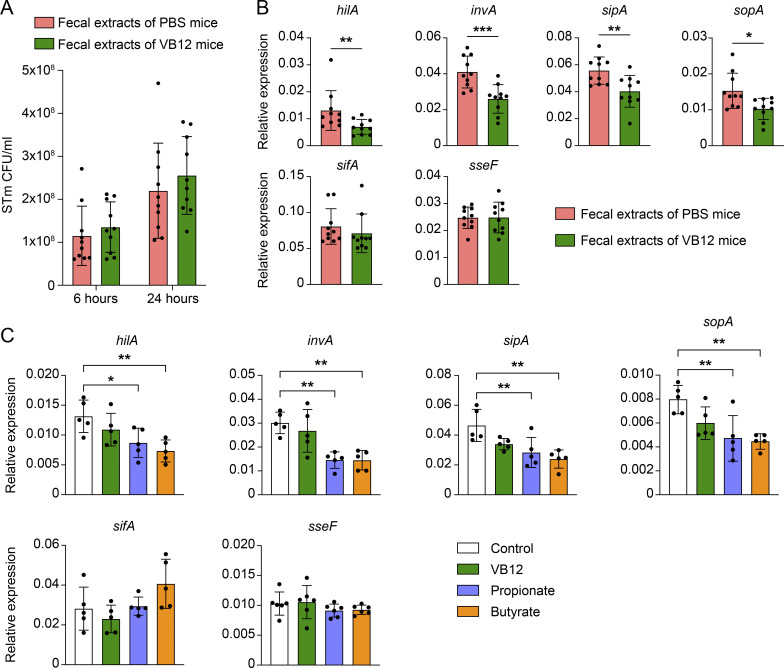
To gain further insights into VB12-dependent STm protection, we investigated whether microbiota-associated products could directly impact STm growth and virulence. Although water extracts of fecal samples derived from VB12-deficient mice exerted no impact on anaerobic STm growth (Fig. S5 A), these fecal water extracts significantly increased the expression of STm pathogenicity island 1 (SPI-1) genes (e.g., invA, sipA, and sopA) critical for invading ECs (Fig. S5 B). Interestingly, fecal water extracts did not impact the expression of SPI-2 genes (sifA and sseF) required for STm replication and survival in phagocytic cells (Fig. S5 B). This could be due to the diminished levels of SCFAs in the feces of VB12-deficient mice (Fig. 4 G), as propionate and butyrate specifically inhibited SPI-I gene expression in similar assays (Fig. S5 C). Altogether, our results suggest that VB12 supports iEC-mitochondrial and microbiota metabolic functions to suppress STm virulence and reduce epithelial oxygenation, contributing to the control of aerobic STm infection.
Figure S5.
Microbiota-associated products of VB12-deficient mice favor the expression of STm virulence genes in vitro. (A) STm growth in LB medium containing water extracts of the feces collected from VB12-deficient mice gavaged with PBS or VB12 (8 wk; n = 5 extracts/group). Each dot represents one extract using feces pooled from one female and one male mouse. (B) qPCR analysis of STm SPI-1 (invA, sipA, sopA) and SPI-2 (sifA, sseF) genes after 24 h of growth. hilA, a transcriptional regulator of SPI-1 genes, was also analyzed. (C) Expression of the SPI-1 and SPI-2 genes in STm cultures supplemented with 10 nM VB12, 10 mM sodium propionate, or 10 mM sodium butyrate (n = 5 cultures/group). Data are from one experiment (C) or combined from two experiments (A and B). *, P < 0.05; **, P < 0.01; ***, P < 0.001; two-tailed unpaired t test (B) or one-way ANOVA plus Tukey’s posttest (C).
Discussion
The maintenance of intestinal homeostasis requires a delicate balance between the epithelium, gut microbiota, and dietary factors. Deficiency in key dietary micronutrients such as VB12 may functionally compromise the molecular and metabolic cell machinery and critically deteriorate the gut microbial consortium, potentially increasing the risk of illness or death from infectious diseases (Black, 2003). Thus, understanding how micronutrients, particularly VB12, modulate intestinal epithelial and microbial function is of critical importance to design preventive and therapeutic interventions for intestinal disorders. Here, we demonstrated that VB12 oral supplementation contributed to the regulation of molecular and metabolic activities of iECs and gut microbiota and how these complex interplays cooperatively promoted the mitochondrial oxygen-consuming reactions of iECs to control STm infection.
The ileal epithelium is a multicellular interface located in close proximity to a dense microbial milieu that undergoes rapid renewal every 3–5 d (Barker, 2014). These continuous steps of cellular proliferation, differentiation, and programmed anoikis are highly associated with changes in cellular metabolism driving mitochondrial activity that supplies energy and essential metabolites for host dynamic development. Disturbances of such a mitochondrial metabolic homeostasis due to VB12 deficiency are integral aspects of the intestinal pathological process, potentially resulting in intestinal disorders, including inflammatory bowel disease and colon cancer (Rath et al., 2018). Thus, mitochondrial function is a vital cellular checkpoint that critically sustains the functions of iECs and highly affects disease outcome (Berger et al., 2016).
In this process, VB12 regulates cellular levels of homocysteine, a risk marker of cardiovascular diseases (Hayden and Tyagi, 2004), and restricts the accumulation of methylmalonate, an inhibitor of mitochondrial respiration and the TCA cycle (Okun et al., 2002). Thus far, only a few studies have documented the potential association between VB12 and mitochondrial DNA variation in the elderly population (Praveen et al., 2020). Interestingly, recent studies demonstrated that a VB12-restricted diet activates the mitochondrial unfolded protein response, and thus promotes the lifespan extension of Caenorhabditis elegans (Amin et al., 2020). In contrast, studies from another group showed that VB12 deficiency disrupts mitochondrial homeostasis and increases pathogen sensitivity in C. elegans without affecting lifespan (Revtovich et al., 2019). Thus, it is still unclear how this vitamin is involved in mitochondrial regulation, particularly in gut ECs. In this study, we examined VB12’s roles in maintaining the functional homeostasis of iECs not only in steady state, but also during STm infection. Obtained results demonstrated that VB12 supplementation increased the transcription of mitochondrially encoded genes and the metabolic activity of the carnitine shuttle and TCA cycle in iECs. Importantly, such a VB12-dependent regulation was seen only in iECs, whereas no mitochondrial activation was observed in cecal or colonic ECs. This might be due to the receptor-dependent activation of lipid absorption and cellular signaling (Christensen and Birn, 2002), as cubilin is expressed by iECs rather than cecal or colonic ECs (Andersen et al., 2010).
Methionine and lysine are important substrates for carnitine biosynthesis (Krajcovicova-Kudlackova et al., 2000). Carnitine and acetylcarnitine, along with Cpt1a and Cpt2 genes, are critical components of carnitine shuttle. These were all reduced in iECs owing to VB12 deficiency. Mechanistically, carnitine shuttle is responsible for transporting LCFAs into mitochondria, which are subsequently degraded by β-oxidation to generate an important source of energy through the TCA cycle and ETC (Indiveri et al., 2011). Accordingly, carnitine deficiency has been reported in infants with cardiomyopathy or hypoketotic hypoglycemia, and genetic deficiency of the carnitine transporter OCTN2 is tightly linked to progressive cardiomyopathy and skeletal muscle weakness in children (Stanley, 2004). Decreased cellular carnitine may result from impaired epithelial biosynthetic pathways and/or intestinal absorption associated with gut microbiota or diets, as VB12 deficiency reduced the absorptive pathways of iECs during intestinal STm infection, shown by RNA-seq.
VB12 is synthesized exclusively by bacteria and archaea, but not mammals or plants (Roth et al., 1996). In the human gut, >80% of sequenced gut microbes are predicted to use corrinoids such as VB12 and its precursors, whereas <25% of them possess the genetic capacity to biosynthesize these molecules (Degnan et al., 2014). Additionally, when microbially released in the gut, VB12 may be minimally absorbed by the host because the receptor necessary for its absorption is expressed in the ileum, but the majority of VB12 and its precursors are detected in the colon, wherein microbial abundancy is the highest (Degnan et al., 2014). Thus, dietary VB12, as the primary source in humans, is required for a variety of microbial metabolic functions and may mediate microbial interactions. However, it is still controversial whether VB12 shapes the gut microbial community. In this respect, we demonstrated that VB12 sustained gut microbial diversity and composition, and more importantly, the functional microbially associated metabolites. For instance, nicotinate and nicotinamide, two precursors of nicotinamide adenine dinucleotide (NAD) essential for multiple metabolic pathways, including TCA cycle and oxidative phosphorylation (Zapata-Perez et al., 2021), were augmented in intestinal lumens of VB12-gavaged mice. NAD, acting as a signaling molecule, may support the activation of NAD-consuming enzymes, particularly PPARs (Hayashida et al., 2010), to regulate epithelial lipid metabolism in response to STm infection. Additionally, it has been reported that Turicibacter controls host lipid metabolic process in the ileum and increases the cellular levels of LCFAs, including arachidonate (Fung et al., 2019). Here, VB12 supplementation increased the abundance of Turicibacter, which was associated with modifications of fatty acid activation and biosynthesis pathways, specifically in the ileal lumen during STm infection, resulting in increased bioavailability of luminal free fatty acids such as γ-linolenic acid and 13(S)-HODE that may serve as ligands to activate PPARs in iECs. In VB12-deficient mice, STm infection inhibited Pparg expression and impaired lipid metabolic process in iECs, and pharmacological activation of PPARs promoted host protection against STm colonization. These observations, along with previous findings demonstrating the impairment of myocardial and hepatic PPARs (PPARα and PPARγ) in rats experiencing VB12 deficiency (Ahmad et al., 2013; Garcia et al., 2011), indicate that VB12 instructs microbially associated metabolisms to sustain the PPAR signaling in iECs and protect mice against STm infection. Additionally, the absence of VB12 may impact general host health and development and thus reduce host capacity to support a complex and diverse microbiota ecosystem, influencing host susceptibility to pathogen infection (Libertucci and Young, 2019).
Bacteroidetes are the most abundant Gram-negative bacteria in the human gut, and members of this phylum, including Bacteroides thetaiotaomicron, are genetically deficient for VB12 biosynthesis. Instead, they rely on multiple transporters to capture VB12 from the environment (Wexler et al., 2018) and use it as a cofactor for the bacterial MCM that catalyzes the interconversion of R-methylmalonyl-CoA and succinyl-CoA, a step in the metabolism of branched amino acids and fatty acids to propionate (Degnan et al., 2014). Here, we provide additional in vivo evidence that VB12 treatment leads to increased levels of Bacteroidetes (e.g., S24-7 and Bacteroides) and the associated propionate in the colon during STm infection. It has been reported that Bacteroidetes-produced propionate directly limits STm growth through acidification of the bacterial milieu that robustly disrupts intracellular pH homeostasis (Jacobson et al., 2018). In this regard, we posit that VB12-supported Bacteroidetes promoted the production of SCFAs, particularly propionate, to protect mice orally infected with STm. Additionally, VB12 induced the expansion of Clostridia (e.g., Roseburia and Butyricicoccus) and, as a result, led to the enrichment of cecal butyrate, which is known to fortify the intestinal barrier function and enhance EC oxygen consumption through β-oxidation, thereby limiting oxygen-dependent STm growth (Rivera-Chavez et al., 2016). Furthermore, both propionate and butyrate can directly inhibit STm virulence gene expression (Gantois et al., 2006; Lawhon et al., 2002). Using in vitro assays, we demonstrated that fecal water extracts of VB12-gavaged mice suppressed the expression of SPI-1 genes (e.g., invA, sipA, and sopA) and thus may limit systemic STm infection in these mice. Based on these results, we conclude that VB12 oral supplementation favors a protective line of the gut microbiota and associated metabolites to synergistically resist intestinal STm infection.
Luminal oxygenation along the length of the mammalian intestine is nonuniform and is likely to be a functional combination of the biomass of gut microbiota, the local metabolism, and the anatomy of blood flow (Friedman et al., 2018). Oxygen levels in the lumen of the ileum are higher than those in the cecum and colon, where a hypoxic environment is maintained in a healthy gut (Albenberg et al., 2014). Although the ileal lumen is not physiologically hypoxic (Zheng et al., 2015), aberrant levels of luminal molecular oxygen can be implicated in pathogen-induced intestinal pathology (Rivera-Chavez et al., 2016). In this scenario, SCFAs, particularly butyrate, decrease colonic epithelial oxygenation, and antibiotic depletion of butyrate-producing Clostridia drives luminal oxygen-dependent STm expansion (Rivera-Chavez et al., 2016). To expand on these findings, we demonstrated that while oxygen-mediated STm growth was reduced by VB12 supplementation, potentially through increased SCFAs produced by cecal and colonic microbiota, those metabolites were not impacted by VB12 in the ileum. Further, elegant studies have shown that activation of PPAR signaling by gut microbiota limits luminal oxygen bioavailability by driving epithelial β-oxidation (Byndloss et al., 2017). Here, VB12 sustained not only PPAR signaling but also supported the mitochondrial respiration of iECs, which together regulated the epithelial oxygenation to limit CydA-dependent aerobic STm growth.
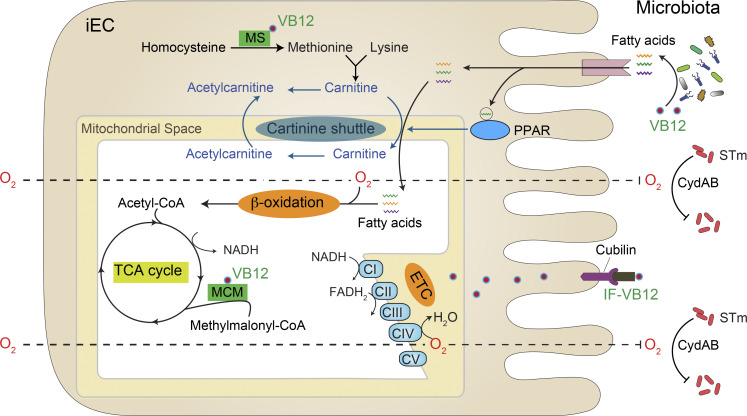
In summary, our results support a novel prospective model by which VB12 coordinates epithelial and microbial responses to intestinal STm infection (Fig. 7). The novel mechanisms demonstrating this complex interplay at the intestinal mucosal surface may provide promising targets to maximize the contribution of crucial cofactors, such as VB12, to fortify the host and microbiota homeostasis, collectively contributing to the health of the host.
Figure 7.
Conceptual model of VB12-dependent mitochondrial regulation in iECs and the consequences for colonization resistance against STm. In the ileum region, VB12 supports the function of microbiota and promotes the production of fatty acids. When transported into iECs, these fatty acids may act as ligands for PPAR, which activates genes related to carnitine shuttle and the lipid metabolic process. Meanwhile, VB12 is absorbed and internalized into iECs by binding to the receptor cubilin. The cytoplasmic VB12 acts as a cofactor for MS converting homocysteine to methionine, and therefore sustains the carnitine biosynthetic pathway. Gene and metabolic activation of carnitine shuttle facilitates the mitochondrial translocation of conjugated fatty acyl-CoA, which can be subsequently used for mitochondrial β-oxidation. VB12 also functions as a cofactor for the mitochondrial MCM, generating succinyl-CoA to support the TCA cycle and the coupled ETC. Elevated mitochondrial β-oxidation and ETC consume significant amounts of oxygen (O2) that diffuses across the ileal epithelial layer, leading to reduced oxygen bioavailability in the ileum lumen. The dampened ileal epithelial oxygenation therefore controls oxygen-dependent Salmonella growth.
Materials and methods
Mice and VB12 treatments
C57BL/6 mice were obtained from The Jackson Laboratory and maintained under specific pathogen–free and Helicobacter-free conditions. These mice were used to establish a murine model of VB12 deficiency by feeding them a diet depleted of any traces of VB12 (TD.180321; ENVIGO), including brewer’s yeast, which is rich in vitamins (e.g., VB12) and minerals. Mice were kept on the VB12-deficient diet for at least two generations to deplete VB12 in newborns (Ghosh et al., 2016; Roman-Garcia et al., 2014). To better control nutritional intake, we compared the VB12-deficient mice that were orally gavaged with VB12 (cyanocobalamin; Sigma-Aldrich) versus PBS to elucidate VB12-dependent effects, instead of having a control group fed regular chow (Teklad global 18% protein rodent diet, catalog no. 2918; ENVIGO) that contains VB12 but also brewer’s yeast. The amount of VB12 used for rescuing VB12 deficiency was determined based on daily food consumption: one mouse typically consumes ∼5 g of regular chow containing 0.08 mg VB12/kg diet. This corresponds to a VB12 intake of ∼400 ng. Considering that breastfeeding may provide an additional source of VB12 and the limited food consumption by newborns, we designed a regimen for VB12 supplementation: 40 ng/mouse/twice a week before weaning and 400 ng/mouse/twice a week after weaning. Given that VB12-deficiency from birth can have long-lasting repercussions (Ghosh et al., 2016; Roman-Garcia et al., 2014), we applied this regimen to gavage mice kept on a VB12-deficient diet for at least two generations to alleviate the effects of VB12 deficiency. Subsequently, sex- and age-matched mice from VB12 and PBS groups were used for the experiments in our studies. All experiments were conducted according to protocols approved by the Institutional Animal Care and Usage Committee of the University of Florida under protocol number 202008484 and the Department of Laboratory Animal Resources of the University of Texas Health at San Antonio under protocol numbers 20210045AB and 20210065AR.
Isolation of ECs
ECs were isolated as previously described (Haber et al., 2017), with modifications. Briefly, the ileal, cecal, and colonic tissues of VB12- and PBS-gavaged mice were dissected and washed with cold PBS. The tissues were opened longitudinally and cut into small fragments (2–3 cm in length), followed by incubation with 30 mM EDTA-PBS on ice for 30 min, during which the tissues were shaken vigorously every 8–10 min. Isolated crypts were washed once with cold PBS and dissociated with TrypLE Express (Invitrogen) for 10 min at room temperature. The single-cell suspension was passed through a 70-μm cell strainer and used for FACS. To FACS-sort the cells, cell suspensions were labeled with a cocktail of fluorescent antibodies specific for APC-CD45 (catalog 17-0451-83; Invitrogen), FITC-CD31 (catalog 102406; BioLegend), PE/Cy7-TER-119 (catalog 116222; BioLegend), and PE-EpCAM (catalog 12-5791-83; Invitrogen). Dead cells were excluded using LIVE/DEAD Fixable Violet Dead Cell Stain (Invitrogen). CD45− CD31− TER-119− EpCAM+ ECs were sorted using a SONY SH800S Cell Sorter or a CytoFLEX SRT Cell Sorter.
RNA-seq
RNA-seq libraries were sequenced as previously described (Ge et al., 2020). Obtained raw reads were aligned to the mouse reference genomes (GRCm38) using STAR v2.7.5c. Normalized transcripts per million (TPM) were generated using RSEM v1.3.3. DESeq2 was used to determine significantly expressed genes based on the criteria (TPM >1, false discovery rate <0.05, fold-change >1.5). TPM values were used for principal component analysis (PCA) and plotting heat maps in R. Gene set enrichment analysis (GSEA) was performed using DAVID (https://david.ncifcrf.gov/), and representative GSEA plots were generated using GSEA v4.1.0 based on 1,000 permutations.
Metabolomic analysis
Metabolites were analyzed at the Southeast Center for Integrated Metabolomics (http://www.secim.ufl.edu; Choi et al., 2020). Samples were run in both positive and negative ionization with a mass resolution of 35,000 at m/z 200, as separate injections. Feature alignment and curation were performed by MZmine through an automated routine developed in-house. After normalizing to total ion chromatogram, intensities were tested for group significance using Student’s t test or Mann–Whitney U test. Metabolites were identified by comparison to metabolomic library of purified standards. Metabolic pathway analysis was performed using Mummichog (Li et al., 2013), with default parameters. The pathways represented by at least two significant metabolites in positive or negative mode are presented. Additional metabolites (e.g., γ-linoleic acid and 13[S]-HODE) were annotated based on their enrichment in significant Mummichog pathways and by the m/z match with an accuracy of 10 ppm. These metabolites are indicated with asterisks (*) in heatmaps. Propionate and butyrate were validated by comparing to the peaks of their respective standards.
Microbiota analysis
16S ribosomal RNA (rRNA) libraries were constructed and sequenced as described previously (Colliou et al., 2017; Kumar et al., 2021). A minimal sequence depth of 34,553 reads/sample was obtained. The 2× 300-bp paired-end reads were processed using QIIME 2 (v2020.8). Briefly, reads were merged, quality trimmed, and clustered into operational taxonomic units at 97% sequence similarity. Taxonomy was assigned using Greengenes 13_8. To estimate within-sample richness, α-diversity was determined using Shannon and Chao1 indexes. To evaluate differences in diversity across samples, β-diversity was determined using unweighted UniFrac distances. Differences in relative abundance at different taxonomic levels were determined using Student’s t test or Mann–Whitney U test.
Measurement of mitochondrial contents, membrane potential, and mROS
The epithelial single-cell suspension was incubated with 100 nM MitoTracker Green, 5 μM MitoSOX Red, and 2 μM MitoProbe JC-1 (Invitrogen) for 30 min at 37°C to measure mitochondrial mass, mROS, and membrane potential, respectively. Dead cells were detected with LIVE/DEAD Fixable Violet Dead Cell Stain kit (Invitrogen), followed by incubation with mouse Fc blocking reagent (Miltenyi Biotec) and APC/Cy7-EpCAM antibody (BioLegend). Mean fluorescence intensity in EpCAM+ iECs was quantified by flow cytometry. mtDNA copies were measured by quantitative real-time PCR (qPCR) comparing copies of mitochondrially encoded mt-Nd1 to those of chromosomally encoded β-globin.
Seahorse analysis
iECs (∼5 × 105 cells) were isolated from VB12-deficient and -supplemented mice 2 d after STm infection. OCRs and extracellular acidification rates of iECs were analyzed using an XFe96 Extracellular Flux Analyzer (Seahorse Bioscience) as previously described (Ge et al., 2020).
Bacterial strains and culture conditions
The WT STm strain SL1344 (streptomycin-resistant) was kindly provided by Dr. Roy Curtiss at the University of Florida (Table S1). The STm WT strain was grown at 37°C in Luria-Bertani (LB) medium supplemented with 200 μg/ml streptomycin, and the STm mutants were grown in LB medium supplemented with 200 μg/ml streptomycin and 25 μg/ml chloramphenicol.
For analyzing the effects of fecal contents of VB12-deficient and -gavaged mice on STm growth and virulence gene expression, four to five feces were collected from one female and one male mouse of each group, respectively. The fecal samples (8–10 pellets) were then pooled and homogenized in water (100 mg feces/ml water), and fecal water extracts (Gratton et al., 2016) were obtained by collecting the supernatants after centrifugation. For each experiment, five samples were generated using feces from five female and five male mice in each group. To begin the growth experiments, a single colony of STm WT strain was grown in LB medium for 24 h under anaerobic conditions and diluted 1:100 into a 1:1 mixture of fecal-water extracts and 2× LB medium. Samples were taken at 6 and 24 h of growth and plated to determine CFU/ml, and experiments were terminated at 24 h to analyze gene expression. Similarly, STm cultures were diluted 1:100 in LB medium supplemented with 10 nM VB12, 10 mM sodium propionate, or 10 mM sodium butyrate (Sigma-Aldrich). Samples were collected at 24 h of growth to analyze gene expression.
RNA isolation and gene expression
Total bacterial RNA was extracted from STm cultures using Trizol LS reagent (Invitrogen), and RNA mixtures were further purified using the Aurum Total RNA Mini Kit (Bio-Rad). cDNA was synthesized using SuperScript IV VILO Master Mix (Thermo Fisher Scientific). For validating DEGs identified by RNA-seq, total RNA was isolated using the RNeasy Micro Plus kit (Qiagen), and cDNA was generated using SMART-Seq HT Kit (Takara). qPCR was performed with PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) on a QuantStudio 6 Pro real-time PCR system (Thermo Fisher Scientific). The relative quantification (2−ΔCt) was used to determine the expression level of the target genes normalized to rpoD (STm) or Gapdh (mouse). Primer sequences can be found in Table S2.
Construction of STm mutants
Plasmids and primers used for STm gene deletion are listed in Tables S1 and S2, respectively. The cydA, cyoA, or cyxA gene was deleted from the chromosome of STm using the Lambda Red system (Datsenko and Wanner, 2000). PCR was used to amplify the chloramphenicol-resistance flippase recognition target cassette with 5′ and 3′ arms homologous to DNA flanking the targeted genes using plasmid pKD3 as the template. The respective PCR products were treated with DpnI and purified. STm SL1344 containing the Red helper plasmid pKD46 was grown in LB medium supplemented with ampicillin (100 μg/ml) and L-arabinose (1 mM) to an OD600 of ∼0.6 and electroporated with the purified PCR products. The chloramphenicol-resistant colonies obtained were selected to confirm the gene deletion by PCR and sequencing. The Red plasmid was then eliminated by growing the bacterial strains in LB medium without ampicillin at 37°C, resulting in ampicillin-sensitive and chloramphenicol-resistant STm mutants.
STm infection and CFU determination
Before infection, mice were fasted for 4 h with unlimited access to water. Mice were orally infected with ∼2 × 108 CFU of STm in 200 μl PBS. For CI experiments, mice were orally infected with a 1:1 mixture of 108 CFU of each STm strain. For treatment with PPARα/γ dual agonist tesaglitazar (Tocris), VB12-deficient mice were orally gavaged with tesaglitazar (1.5 mg/kg body weight/d) diluted in 100 μl of sterile PBS for 4 d (days 0, 1, 2, and 3). Mice were euthanized on day 4 to collect ileal, cecal, and colonic contents. In the streptomycin-treated mouse model, food was withdrawn for 4 h before mice were orally treated with 20 mg/mouse of streptomycin; afterward, mice were supplied with food. 20 h after streptomycin treatment, food was withdrawn again 4 h before the mice were infected with 2 × 105 CFU of STm in 200 μl PBS. To determine the STm burdens, VB12- and PBS-gavaged mice infected with STm were euthanized at the indicated time points to collect intestinal tissues, fecal samples, and organs. Homogenized samples were plated on LB plates containing the appropriate antibiotics. Additionally, Peyer’s patches were treated with 100 µM gentamicin for 1 h at 37°C to remove extracellular bacteria and washed three times with PBS before homogenizing and plating. STm CFUs were counted after overnight incubation at 37°C and normalized to the initial weights of the samples. For competition assays, the CI was determined based on the relative recovery of WT and the respective mutant strains. If no STm was detected, samples or mice were excluded from CI calculation.
Statistical analysis
Statistical analyses were performed using GraphPad Prism v9.1.2. Before statistical analysis, normality was tested using the Shapiro–Wilk normality test. Where the groups followed a Gaussian distribution, two-tailed unpaired Student’s t test was performed. Where the groups did not follow a Gaussian distribution, Mann–Whitney U test was performed. For experiments with three or more variables, one-way ANOVA followed by Tukey’s posttest was performed. P < 0.05 was considered significant: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the differential impacts of VB12 oral supplementation on the transcriptomes of ileal, cecal, and colonic ECs. Fig. S2 shows transcriptomic responses of iECs to STm infection. Fig. S3 shows differentially enriched metabolites in the ileal, cecal, and colonic contents during STm infection. Fig. S4 shows the gut microbiota and transcriptomic profile of iECs in streptomycin-treated mice. Fig. S5 shows the impact of microbiota-associated fecal products on the expression of STm virulence genes. Table S1 shows STm strains and plasmids used in this study. Table S2 shows primer sequences used for qPCR and constructing STm mutants.
Supplementary Material
lists Salmonella strains and plasmids used in this study.
lists primers used in this study.
Acknowledgments
We thank Dr. Roy Curtiss for fruitful discussion and providing Salmonella strain and plasmids.
This research was supported by National Institutes of Health (NIH) R01 DK109560, NIH R01 DK109560C, and NIH R01AI154630-01 to M. Mohamadzadeh.
Author contributions: M. Mohamadzadeh directed the studies and supervised data analyses. Y. Ge performed cellular and molecular studies and bioinformatically analyzed the data. M. Zadeh generated the mouse model. Y. Ge and M. Zadeh performed animal experiments. Y. Ge and M. Mohamadzadeh interpreted the data and wrote the manuscript.
Data availability
All data associated with this study are present in the paper or the supplemental materials. The transcriptomic raw FastQ files are available under the accession no. PRJNA750489 at the NCBI BioProject.
References
- Ahmad, S., Kumar K.A., Basak T., Bhardwaj G., Yadav D.K., Lalitha A., Chandak G.R., Raghunath M., and Sengupta S.. 2013. PPAR signaling pathway is a key modulator of liver proteome in pups born to vitamin B(12) deficient rats. J. Proteonomics. 91:297–308. 10.1016/j.jprot.2013.07.027 [DOI] [PubMed] [Google Scholar]
- Albenberg, L., Esipova T.V., Judge C.P., Bittinger K., Chen J., Laughlin A., Grunberg S., Baldassano R.N., Lewis J.D., Li H., et al. 2014. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 147:1055–1063.e8. 10.1053/j.gastro.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, M.R., Mahmud S.A., Dowgielewicz J.L., Sapkota M., and Pellegrino M.W.. 2020. A novel gene-diet interaction promotes organismal lifespan and host protection during infection via the mitochondrial UPR. PLoS Genet. 16:e1009234. 10.1371/journal.pgen.1009234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, C.B.F., Madsen M., Storm T., Moestrup S.K., and Andersen G.R.. 2010. Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes. Nature. 464:445–448. 10.1038/nature08874 [DOI] [PubMed] [Google Scholar]
- Barker, N. 2014. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15:19–33. 10.1038/nrm3721 [DOI] [PubMed] [Google Scholar]
- Benight, N.M., Stoll B., Chacko S., da Silva V.R., Marini J.C., Gregory J.F., Stabler S.P., and Burrin D.G.. 2011. B-vitamin deficiency is protective against DSS-induced colitis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 301:G249–G259. 10.1152/ajpgi.00076.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, E., Rath E., Yuan D., Waldschmitt N., Khaloian S., Allgauer M., Staszewski O., Lobner E.M., Schottl T., Giesbertz P., et al. 2016. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nat. Commun. 7:13171. 10.1038/ncomms13171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy, K., Wang T., Chen W.W., Freinkman E., Abu-Remaileh M., and Sabatini D.M.. 2015. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 162:540–551. 10.1016/j.cell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, R. 2003. Micronutrient deficiency: An underlying cause of morbidity and mortality. Bull. World Health Organ. 81:79. [PMC free article] [PubMed] [Google Scholar]
- Boran, P., Baris H.E., Kepenekli E., Erzik C., Soysal A., and Dinh D.M.. 2020. The impact of vitamin B12 deficiency on infant gut microbiota. Eur. J. Pediatr. 179:385–393. 10.1007/s00431-019-03517-2 [DOI] [PubMed] [Google Scholar]
- Byndloss, M.X., Olsan E.E., Rivera-Chavez F., Tiffany C.R., Cevallos S.A., Lokken K.L., Torres T.P., Byndloss A.J., Faber F., Gao Y., et al. 2017. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic enterobacteriaceae expansion. Science. 357:570–575. 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, S.E., Mehan K.A., Tunstall R.J., Febbraio M.A., and Cameron-Smith D.. 2003. 17beta-estradiol upregulates the expression of peroxisome proliferator-activated receptor alpha and lipid oxidative genes in skeletal muscle. J. Mol. Endocrinol. 31:37–45. 10.1677/jme.0.0310037 [DOI] [PubMed] [Google Scholar]
- Cevallos, S.A., Lee J.Y., Tiffany C.R., Byndloss A.J., Johnston L., Byndloss M.X., and Baumler A.J.. 2019. Increased epithelial oxygenation links colitis to an expansion of tumorigenic bacteria. mBio. 10:e02244-19. 10.1128/mBio.02244-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.C., Brown J., Gong M., Ge Y., Zadeh M., Li W., Croker B.P., Michailidis G., Garrett T.J., Mohamadzadeh M., and Morel L.. 2020. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci. Transl. Med. 12:eaax2220. 10.1126/scitranslmed.aax2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, E.I., and Birn H.. 2002. Megalin and cubilin: Multifunctional endocytic receptors. Nat. Rev. Mol. Cell Biol. 3:256–266. 10.1038/nrm778 [DOI] [PubMed] [Google Scholar]
- Cimen, I., Astarci E., and Banerjee S.. 2011. 15-lipoxygenase-1 exerts its tumor suppressive role by inhibiting nuclear factor-kappa B via activation of PPAR gamma. J. Cell. Biochem. 112:2490–2501. 10.1002/jcb.23174 [DOI] [PubMed] [Google Scholar]
- Colliou, N., Ge Y., Sahay B., Gong M., Zadeh M., Owen J.L., Neu J., Farmerie W.G., Alonzo F. 3rd, Liu K., et al. 2017. Commensal propionibacterium strain UF1 mitigates intestinal inflammation via Th17 cell regulation. J. Clin. Invest. 127:3970–3986. 10.1172/JCI95376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko, K.A., and Wanner B.L.. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan, P.H., Taga M.E., and Goodman A.L.. 2014. Vitamin B12 as a modulator of gut microbial ecology. Cell Metabol. 20:769–778. 10.1016/j.cmet.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien, M., Belzer C., and de Vos W.M.. 2017. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 106:171–181. 10.1016/j.micpath.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Elias-Arnanz, M. 2020. Anaerobic bacteria need their vitamin B-12 to digest estrogen. Proc. Natl. Acad. Sci. USA. 117:1833–1835. 10.1073/pnas.1921340117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, N.P., Misyak S.A., Schmelz E.M., Guri A.J., Hontecillas R., and Bassaganya-Riera J.. 2010. Conjugated linoleic acid ameliorates inflammation-induced colorectal cancer in mice through activation of PPARgamma. J. Nutr. 140:515–521. 10.3945/jn.109.115642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech, M. 2001. The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutat. Res. 475:57–67. 10.1016/s0027-5107(01)00079-3 [DOI] [PubMed] [Google Scholar]
- Fenech, M., Aitken C., and Rinaldi J.. 1998. Folate, vitamin B12, homocysteine status and DNA damage in young Australian adults. Carcinogenesis. 19:1163–1171. 10.1093/carcin/19.7.1163 [DOI] [PubMed] [Google Scholar]
- Forman, B.M., Chen J., and Evans R.M.. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA. 94:4312–4317. 10.1073/pnas.94.9.4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Lopez, J., Duplessis M., Bui A., Reymond C., Poisson W., Blais L., Chong J., Gervais R., Rico D.E., Cue R.I., et al. 2020. Correlations between the composition of the bovine microbiota and vitamin B-12 abundance. Msystems. 5:e00107-20. 10.1128/mSystems.00107-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, E.S., Bittinger K., Esipova T.V., Hou L., Chau L., Jiang J., Mesaros C., Lund P.J., Liang X., FitzGerald G.A., et al. 2018. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc. Natl. Acad. Sci. USA. 115:4170–4175. 10.1073/pnas.1718635115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, T.C., Vuong H.E., Luna C.D.G., Pronovost G.N., Aleksandrova A.A., Riley N.G., Vavilina A., McGinn J., Rendon T., Forrest L.R., and Hsiao E.Y.. 2019. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 4:2064–2073. 10.1038/s41564-019-0540-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantois, I., Ducatelle R., Pasmans F., Haesebrouck F., Hautefort I., Thompson A., Hinton J.C., and Van Immerseel F.. 2006. Butyrate specifically down-regulates salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72:946–949. 10.1128/AEM.72.1.946-949.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M.M., Gueant-Rodriguez R.M., Pooya S., Brachet P., Alberto J.M., Jeannesson E., Maskali F., Gueguen N., Marie P.Y., Lacolley P., et al. 2011. Methyl donor deficiency induces cardiomyopathy through altered methylation/acetylation of PGC-1α by PRMT1 and SIRT1. J. Pathol. 225:324–335. 10.1002/path.2881 [DOI] [PubMed] [Google Scholar]
- Ge, Y., Gong M., Zadeh M., Li J., Abbott J.R., Li W., Morel L., Sonon R., Supekar N.T., Azadi P., et al. 2020. Regulating colonic dendritic cells by commensal glycosylated large surface layer protein A to sustain gut homeostasis against pathogenic inflammation. Mucosal Immunol. 13:34–46. 10.1038/s41385-019-0210-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., Sinha J.K., Putcha U.K., and Raghunath M.. 2016. Severe but not moderate vitamin B12 deficiency impairs lipid profile, induces adiposity, and leads to adverse gestational outcome in female C57BL/6 mice. Front. Nutr. 3:1. 10.3389/fnut.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginocchio, C., Pace J., and Galan J.E.. 1992. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc. Natl. Acad. Sci. USA. 89:5976–5980. 10.1073/pnas.89.13.5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelz, S.E., Vogelstein B., Hamilton S.R., and Feinberg A.P.. 1985. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 228:187–190. 10.1126/science.2579435 [DOI] [PubMed] [Google Scholar]
- Gratton, J., Phetcharaburanin J., Mullish B.H., Williams H.R., Thursz M., Nicholson J.K., Holmes E., Marchesi J.R., and Li J.V.. 2016. Optimized sample handling strategy for metabolic profiling of human feces. Anal. Chem. 88:4661–4668. 10.1021/acs.analchem.5b04159 [DOI] [PubMed] [Google Scholar]
- Haber, A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. 2017. A single-cell survey of the small intestinal epithelium. Nature. 551:333–339. 10.1038/nature24489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida, S., Arimoto A., Kuramoto Y., Kozako T., Honda S., Shimeno H., and Soeda S.. 2010. Fasting promotes the expression of SIRT1, an NAD+ -dependent protein deacetylase, via activation of PPARalpha in mice. Mol. Cell. Biochem. 339:285–292. 10.1007/s11010-010-0391-z [DOI] [PubMed] [Google Scholar]
- Hayden, M.R., and Tyagi S.C.. 2004. Homocysteine and reactive oxygen species in metabolic syndrome, type 2 diabetes mellitus, and atheroscleropathy: The pleiotropic effects of folate supplementation. Nutr. J. 3:4. 10.1186/1475-2891-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiveri, C., Iacobazzi V., Tonazzi A., Giangregorio N., Infantino V., Convertini P., Console L., and Palmieri F.. 2011. The mitochondrial carnitine/acylcarnitine carrier: Function, structure and physiopathology. Mol. Aspect. Med. 32:223–233. 10.1016/j.mam.2011.10.008 [DOI] [PubMed] [Google Scholar]
- Intrasuksri, U., Rangwala S.M., O’Brien M., Noonan D.J., and Feller D.R.. 1998. Mechanisms of peroxisome proliferation by perfluorooctanoic acid and endogenous fatty acids. Gen. Pharmacol. 31:187–197. 10.1016/s0306-3623(98)00029-9 [DOI] [PubMed] [Google Scholar]
- Jacobson, A., Lam L., Rajendram M., Tamburini F., Honeycutt J., Pham T., Van Treuren W., Pruss K., Stabler S.R., Lugo K., et al. 2018. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe. 24:296–307.e7. 10.1016/j.chom.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer, I., Ringseis R., Biemann R., Wen G., and Eder K.. 2011. 13-hydroxy linoleic acid increases expression of the cholesterol transporters ABCA1, ABCG1 and SR-BI and stimulates apoA-I-dependent cholesterol efflux in RAW264.7 macrophages. Lipids Health Dis. 10:222. 10.1186/1476-511X-10-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, D., Shi B., Erfe M.C., Craft N., and Li H.. 2015. Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl. Med. 7:293ra103. 10.1126/scitranslmed.aab2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, C.J., Alexeev E.E., Farb L., Vickery T.W., Zheng L., Eric L.C., Kitzenberg D.A., Battista K.D., Kominsky D.J., Robertson C.E., et al. 2019. Oral vitamin B12 supplement is delivered to the distal gut, altering the corrinoid profile and selectively depleting Bacteroides in C57BL/6 mice. Gut Microb. 10:654–662. 10.1080/19490976.2019.1597667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J., Wilson K.E., Glover L.E., Kominsky D.J., Magnuson A., et al. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 17:662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovicova-Kudlackova, M., Simoncic R., Bederova A., Babinska K., and Beder I.. 2000. Correlation of carnitine levels to methionine and lysine intake. Physiol. Res. 49:399–402 [PubMed] [Google Scholar]
- Kumar, A., Priyamvada S., Ge Y., Jayawardena D., Singhal M., Anbazhagan A.N., Chatterjee I., Dayal A., Patel M., Zadeh K., et al. 2021. A novel role of SLC26A3 in the maintenance of intestinal epithelial barrier integrity. Gastroenterology. 160:1240–1255.e3. 10.1053/j.gastro.2020.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon, S.D., Maurer R., Suyemoto M., and Altier C.. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451–1464. 10.1046/j.1365-2958.2002.03268.x [DOI] [PubMed] [Google Scholar]
- Li, J., Ge Y., Zadeh M., Curtiss R. 3rd, and Mohamadzadeh M.. 2020. Regulating vitamin B12 biosynthesis via the cbiMCbl riboswitch in Propionibacterium strain UF1. Proc. Natl. Acad. Sci. USA. 117:602–609. 10.1073/pnas.1916576116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Park Y., Duraisingham S., Strobel F.H., Khan N., Soltow Q.A., Jones D.P., and Pulendran B.. 2013. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 9:e1003123. 10.1371/journal.pcbi.1003123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberato, M.V., Nascimento A.S., Ayers S.D., Lin J.Z., Cvoro A., Silveira R.L., Martinez L., Souza P.C.T., Saidemberg D., Deng T., et al. 2012. Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) gamma activators and pan-PPAR partial agonists. PLoS One. 7:e36297. 10.1371/journal.pone.0036297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libertucci, J., and Young V.B.. 2019. The role of the microbiota in infectious diseases. Nat. Microbiol. 4:35–45. 10.1038/s41564-018-0278-4 [DOI] [PubMed] [Google Scholar]
- Litvak, Y., Mon K.K.Z., Nguyen H., Chanthavixay G., Liou M., Velazquez E.M., Kutter L., Alcantara M.A., Byndloss M.X., Tiffany C.R., et al. 2019. Commensal enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe. 25:128–139.e5. 10.1016/j.chom.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, P., and Flint H.J.. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19:29–41. 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- Lurz, E., Horne R.G., Maattanen P., Wu R.Y., Botts S.R., Li B., Rossi L., Johnson-Henry K.C., Pierro A., Surette M.G., and Sherman P.M.. 2020. Vitamin B12 deficiency alters the gut microbiota in a murine model of colitis. Front. Nutr. 7:83. 10.3389/fnut.2020.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich, A., Tian N., Byan L., and Bilder G.. 2001. A potent PPARalpha agonist stimulates mitochondrial fatty acid beta-oxidation in liver and skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 280:E270–E279. 10.1152/ajpendo.2001.280.2.E270 [DOI] [PubMed] [Google Scholar]
- Molloy, A.M., Kirke P.N., Brody L.C., Scott J.M., and Mills J.L.. 2008. Effects of folate and vitamin B12 deficiencies during pregnancy on fetal, infant, and child development. Food Nutr. Bull. 29:S101–S111; discussion S112–S115. 10.1177/15648265080292S114 [DOI] [PubMed] [Google Scholar]
- Nicholson, J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., and Pettersson S.. 2012. Host-gut microbiota metabolic interactions. Science. 336:1262–1267. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- O’Leary, F., and Samman S.. 2010. Vitamin B12 in health and disease. Nutrients. 2:299–316. 10.3390/nu2030299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, R., and Brown D.L.. 2003. Vitamin B12 deficiency. Am. Fam. Physician. 67:979–986. [PubMed] [Google Scholar]
- Okun, J.G., Horster F., Farkas L.M., Feyh P., Hinz A., Sauer S., Hoffmann G.F., Unsicker K., Mayatepek E., and Kolker S.. 2002. Neurodegeneration in methylmalonic aciduria involves inhibition of complex II and the tricarboxylic acid cycle, and synergistically acting excitotoxicity. J. Biol. Chem. 277:14674–14680. 10.1074/jbc.M200997200 [DOI] [PubMed] [Google Scholar]
- Parada Venegas, D., De la Fuente M.K., Landskron G., Gonzalez M.J., Quera R., Dijkstra G., Harmsen H.J.M., Faber K.N., and Hermoso M.A.. 2019. Corrigendum: Short chain fatty acids (SCFAs)-Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:1486. 10.3389/fimmu.2019.01486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen, G., Shalini T., Sivaprasad M., and Reddy G.B.. 2020. Relative telomere length and mitochondrial DNA copy number variation with age: Association with plasma folate and vitamin B12. Mitochondrion. 51:79–87. 10.1016/j.mito.2020.01.007 [DOI] [PubMed] [Google Scholar]
- Rath, E., Moschetta A., and Haller D.. 2018. Mitochondrial function - gatekeeper of intestinal epithelial cell homeostasis. Nat. Rev. Gastroenterol. Hepatol. 15:497–516. 10.1038/s41575-018-0021-x [DOI] [PubMed] [Google Scholar]
- Revtovich, A.V., Lee R., and Kirienko N.V.. 2019. Interplay between mitochondria and diet mediates pathogen and stress resistance in Caenorhabditis elegans. PLoS Genet. 15:e1008011. 10.1371/journal.pgen.1008011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez, F., Zhang L.F., Faber F., Lopez C.A., Byndloss M.X., Olsan E.E., Xu G., Velazquez E.M., Lebrilla C.B., Winter S.E., and Baumler A.J.. 2016. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 19:443–454. 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cuenca, S., Monjo M., Gianotti M., Proenza A.M., and Roca P.. 2007. Expression of mitochondrial biogenesis-signaling factors in brown adipocytes is influenced specifically by 17beta-estradiol, testosterone, and progesterone. Am. J. Physiol. Endocrinol. Metab. 292:E340–E346. 10.1152/ajpendo.00175.2006 [DOI] [PubMed] [Google Scholar]
- Roman-Garcia, P., Quiros-Gonzalez I., Mottram L., Lieben L., Sharan K., Wangwiwatsin A., Tubio J., Lewis K., Wilkinson D., Santhanam B., et al. 2014. Vitamin B12-dependent taurine synthesis regulates growth and bone mass. J. Clin. Invest. 124:2988–3002. 10.1172/JCI72606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, J.R., Lawrence J.G., and Bobik T.A.. 1996. Cobalamin (coenzyme B12): Synthesis and biological significance. Annu. Rev. Microbiol. 50:137–181. 10.1146/annurev.micro.50.1.137 [DOI] [PubMed] [Google Scholar]
- Schirmer, M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Horst R.T., Jansen T., Jacobs L., Bonder M.J., et al. 2016. Linking the human gut microbiome to inflammatory Cytokine production capacity. Cell. 167:1897. 10.1016/j.cell.2016.11.046 [DOI] [PubMed] [Google Scholar]
- Selhub, J. 2002. Folate, vitamin B12 and vitamin B6 and one carbon metabolism. J. Nutr. Health Aging. 6:39–42 [PubMed] [Google Scholar]
- Sokolovskaya, O.M., Shelton A.N., and Taga M.E.. 2020. Sharing vitamins: Cobamides unveil microbial interactions. Science. 369:eaba0165. 10.1126/science.aba0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler, S.P. 2013. Vitamin B12 deficiency. N. Engl. J. Med. 368:2041–2042. 10.1056/NEJMc1304350 [DOI] [PubMed] [Google Scholar]
- Stabler, S.P., and Allen R.H.. 2004. Vitamin B12 deficiency as a worldwide problem. Annu. Rev. Nutr. 24:299–326. 10.1146/annurev.nutr.24.012003.132440 [DOI] [PubMed] [Google Scholar]
- Stanley, C.A. 2004. Carnitine deficiency disorders in children. Ann. N. Y. Acad. Sci. 1033:42–51. 10.1196/annals.1320.004 [DOI] [PubMed] [Google Scholar]
- Toh, B.H., van Driel I.R., and Gleeson P.A.. 1997. Pernicious anemia. N. Engl. J. Med. 337:1441–1448. 10.1056/NEJM199711133372007 [DOI] [PubMed] [Google Scholar]
- Visconti, A., Le Roy C.I., Rosa F., Rossi N., Martin T.C., Mohney R.P., Li W., de Rinaldis E., Bell J.T., Venter J.C., et al. 2019. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 10:4505. 10.1038/s41467-019-12476-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler, A.G., Schofield W.B., Degnan P.H., Folta-Stogniew E., Barry N.A., and Goodman A.L.. 2018. Human gut Bacteroides capture vitamin B12 via cell surface-exposed lipoproteins. Elife. 7:e37138. 10.7554/eLife.37138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahn, G.B., Abato J.E., and Jadavji N.M.. 2021. Role of vitamin B12 deficiency in ischemic stroke risk and outcome. Neural Regen. Res. 16:470–474. 10.4103/1673-5374.291381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata-Perez, R., Wanders R.J.A., van Karnebeek C.D.M., and Houtkooper R.H.. 2021. NAD(+) homeostasis in human health and disease. EMBO Mol. Med. 13:e13943. 10.15252/emmm.202113943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Kelly C.J., and Colgan S.P.. 2015. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: Cellular responses to hypoxia. Am. J. Physiol. Cell Physiol. 309:C350–C360. 10.1152/ajpcell.00191.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Xiang S., Feng X., Wang H., Tian S., Xu Y., Shi L., Yang L., Li M., Shen Y., et al. 2019. Impact of cyanocobalamin and methylcobalamin on inflammatory bowel disease and the intestinal microbiota composition. J. Agric. Food Chem. 67:916–926. 10.1021/acs.jafc.8b05730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists Salmonella strains and plasmids used in this study.
lists primers used in this study.
Data Availability Statement
All data associated with this study are present in the paper or the supplemental materials. The transcriptomic raw FastQ files are available under the accession no. PRJNA750489 at the NCBI BioProject.