Wang et al. report that the SARS-CoV-2 ORF10 promotes IFT46 degradation via stimulating CUL2ZYG11B activity, thereby impairing both cilia biogenesis and maintenance. The study provides a pathological mechanism connecting COVID-19 symptoms with cilia dysfunction.
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causal pathogen of the ongoing global pandemic of coronavirus disease 2019 (COVID-19). Loss of smell and taste are symptoms of COVID-19, and may be related to cilia dysfunction. Here, we found that the SARS-CoV-2 ORF10 increases the overall E3 ligase activity of the CUL2ZYG11B complex by interacting with ZYG11B. Enhanced CUL2ZYG11B activity by ORF10 causes increased ubiquitination and subsequent proteasome-mediated degradation of an intraflagellar transport (IFT) complex B protein, IFT46, thereby impairing both cilia biogenesis and maintenance. Further, we show that exposure of the respiratory tract of hACE2 mice to SARS-CoV-2 or SARS-CoV-2 ORF10 alone results in cilia-dysfunction-related phenotypes, and the ORF10 expression in primary human nasal epithelial cells (HNECs) also caused a rapid loss of the ciliary layer. Our study demonstrates how SARS-CoV-2 ORF10 hijacks CUL2ZYG11B to eliminate IFT46 and leads to cilia dysfunction, thereby offering a powerful etiopathological explanation for how SARS-CoV-2 causes multiple cilia-dysfunction-related symptoms specific to COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a single-stranded RNA betacoronavirus of the family Coronaviridae, and it is the cause of the ongoing global pandemic of Coronavirus disease 19 (COVID-19; Wang et al., 2020; Zhu et al., 2020b). SARS-CoV-2 is transmitted more rapidly compared to other coronaviruses. Knowledge about the pathogenicity and biology of SARS-CoV-2 that can aid the prevention and treatment of COVID-19 is urgently sought. The common symptoms of COVID-19 are fever, cough, and myalgia or fatigue; some patients display dyspnea accompanied by hyperinflammation and acute respiratory distress syndrome (ARDS; Huang et al., 2020; Zhu et al., 2020b). Recently, loss of smell and taste have been reported to be strongly associated with COVID-19 (Bagheri et al., 2020; Makaronidis et al., 2020; Yan et al., 2020a). Numerous odorant receptors accumulate on sensory cilia in the olfactory epithelium, and cilia dysfunction impacts lung diseases and anosmia (Li et al., 2020c; Reiter and Leroux, 2017), so COVID-19 symptoms may be related to cilia dysfunction. However, any pathological mechanisms connecting COVID-19 symptoms with cilia dysfunction remain unknown.
Protein ubiquitination has been reported to be essential for the infection of some viruses, and viruses have evolved to hijack the ubiquitin system to increase viral pathogenesis (Gu and Jan Fada, 2020; Hershko and Ciechanover, 1998; Pickart, 2001). For example, human immunodeficiency virus type I (HIV-1) encodes VPU, VPR, and VIF proteins which hijack five types of Cullin-RING E3 ligase (CRLs) complexes (CRLs 1, 2, 4A/B & 5). This hijacking promotes infections by degrading host antiviral proteins (Liu and Tan, 2020). A recent proteomics analysis of SARS-CoV-2-infected host cells implicated ubiquitin–proteasome machinery components in virus-related processes, and a small-molecule inhibitor of the ubiquitin-proteasome system protein AAA ATPase p97 was shown to inhibit SARS-CoV-2 replication (Bojkova et al., 2020). Additionally, ORF9b of SARS-CoV-2 was shown to interrupt K63-linked polyubiquitination of NEMO, thereby inhibiting the canonical NF-κB pathway and subsequent IFN production in infected cells (Wu et al., 2021).
SARS-CoV-2 ORF10 (henceforth “ORF10”) is a 38 amino acid protein encoded by the SARS-CoV-2 genome (Wu et al., 2020); the origin of ORF10 can be traced to a 25 amino acid ORF that is present in the SARS-CoV genome (Michel et al., 2020). Intriguingly, genomic alignment analysis has indicated that ORF10 is highly conserved and that it is evolving under positive selection (Cagliani et al., 2020; Tsai et al., 2020), suggesting it could be responsible for some novel infection processes and/or symptoms of COVID-19. A SARS-CoV-2 protein interactome study revealed that ORF10 interacts with members of a cullin-2 (CUL2) RING E3 ligase complex, CUL2ZYG11B (Gordon et al., 2020; Mena et al., 2021; Timms et al., 2019). Cullin 2-RING (CUL2) is a ubiquitin E3 ligase comprising the subunits Cullin-2, RBX1, Elongin B, Elongin C, and ZYG11B. CUL2ZYG11B regulates protein degradation by targeting the N-terminal glycine of its substrate proteins. ZYG11B is the substrate receptor of CUL2 and is responsible for recognition of the N-terminal glycine degrons (Eldeeb et al., 2019; Timms et al., 2019). Thus, we were motivated to investigate the idea that ORF10 might somehow hijack CUL2ZYG11B-modulated ubiquitination and thereby influence SARS-CoV-2–host interactions.
Pursuing this line of inquiry in the present study, we found that SARS-CoV-2 ORF10 could interact with the substrate adaptor of the Cullin–RING E3 ligase complex (ZYG11B); we show that this binding substantially enhances the E3 ligase activity of CUL2ZYG11B. This ORF10-mediated enhancement of CUL2ZYG11B activity may promote the degradation of an intraflagellar transport (IFT) complex B protein, IFT46, which impairs the delivery of new axonemal components and perturbs both cilia formation and maintenance. We also conducted in vivo experiments wherein the respiratory tract of hACE2 humanized mice was exposed to Spike-lentivirus-ORF10, which caused multiple pathogenic phenotypes. Our study thus demonstrates a mechanism through which the SARS-CoV-2 ORF10 cause cilia dysfunction, which helps explain symptoms specific to COVID-19.
Results
ORF10 stimulates CUL2ZYG11B’s ubiquitination activity
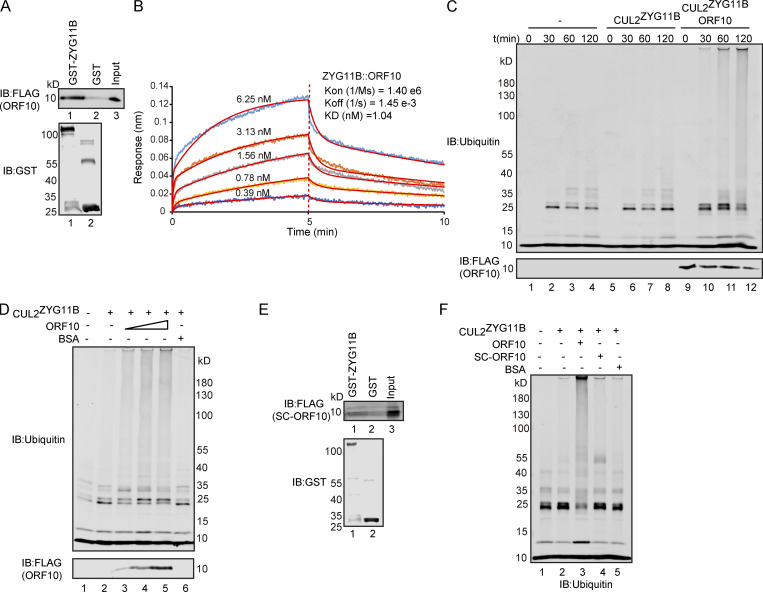
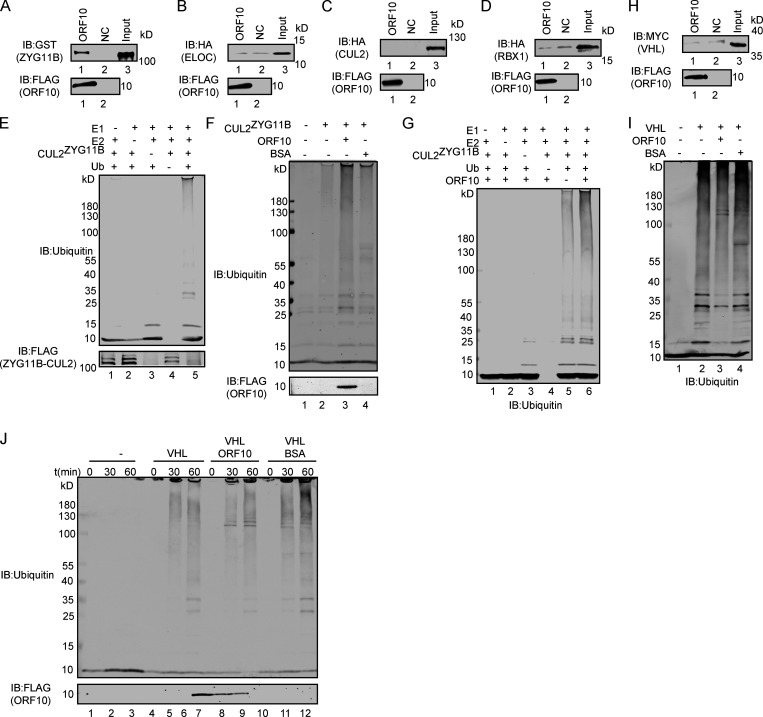
It has been reported that ORF10 might interact with ZYG11B, which is known to function as a substrate adaptor of the Cullin2-RING E3 ligase complex (Gordon et al., 2020) that regulates protein degradation via the ubiquitin–proteasome pathway (Timms et al., 2019). To explore the functional roles of ORF10 in the SARS-CoV-2–host interaction, we first examined the interaction between ORF10 and CUL2ZYG11B by using GST- and FLAG-pulldown experiments. ORF10 could directly bind ZYG11B but did not interact with other CUL2ZYG11B components (Fig. 1 A and Fig. S1, A–D). To detect the binding affinity, we performed Bio-Layer Interferometry (BLI) experiment and found that ORF10-ZYG11B had pretty high binding affinity (Fig. 1 B).
Figure 1.
ORF10 stimulates CUL2ZYG11B E3 ligase activity by interacting with ZYG11B. (A) SARS-CoV-2 ORF10 directly interacts with ZYG11B. Purified GST-ZYG11B and GST were used to pull down ORF10. The number below Western blot bands presents the lane number. (B) ORF10-ZYG11B interaction shows a high binding affinity. Bio-Layer interferometry (BLI) was performed to quantify the binding affinity of the ORF10-ZYG11B interaction. Data analysis was performed using the 2:1 heterogenous ligand-binding model. (C) ORF10 stimulates CUL2ZYG11B E3 ligase activity. ORF10 was added to an in vitro CUL2ZYG11B ubiquitination system containing ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin, ATP, and the ZYG11B-CUL2 immunocomplex. The reaction products were collected at the indicated times, and reaction products were assessed with immunoblotting using anti-ubiquitin and anti-FLAG antibodies. (D) ORF10 stimulates the CUL2ZYG11B ubiquitination activity in a concentration-dependent manner. ORF10 was added to the CUL2ZYG11B ubiquitination system from C as a concentration gradient and reaction products were assessed with immunoblotting using anti-ubiquitin and anti-FLAG antibodies. (E) ZYG11B does not bind with ORF10 from SARS-CoV (SC-ORF10). Purified GST-ZYG11B and GST were used to pull down SC-ORF10. (F) SARS-ORF10 has no effect on the ubiquitination activity of CUL2ZYG11B. ORF10, SC-ORF10, or BSA was added to the CUL2ZYG11B ubiquitination system from C. The reaction products were assessed with immunoblotting using an anti-ubiquitin antibody. Source data are available for this figure: SourceData F1.
Figure S1.
ORF10 specifically stimulates CUL2ZYG11B activity. (A) ZYG11B could interact with ORF10. Purified ORF10-FLAG was used to pull down GST-ZYG11B. (B) ELOC could not bind with ORF10. Purified ORF10-FLAG was used to pull down 6 × His-ELOC-HA. (C) CUL2 could not interact with ORF10. Purified ORF10-FLAG was used to pull down CUL2-HA. (D) RBX1 could not bind with ORF10. Purified ORF10-FLAG was used to pull down 6 × His-RBX1-HA. (E) Reconstitution of the CUL2ZYG11B ubiquitination system. Either E1, E2, ZYG11B-CUL2 immunocomplex, or Ub were withheld from the CUL2ZYG11B ubiquitination system in vitro. The reaction products were detected by immunoblotting with anti-ubiquitin and FLAG antibodies. (F) ORF10 stimulates the ubiquitination activity of CUL2ZYG11B. Either ORF10 or BSA was added to the CUL2ZYG11B ubiquitination system in vitro, which contains E1, E2, ubiquitin, ATP and ZYG11B-CUL2 immunocomplex. The reaction products were detected by immunoblotting with anti-ubiquitin and FLAG antibodies. (G) ORF10 is essential for the stimulation of CUL2ZYG11B ubiquitination activity. Either E1, E2, ZYG11B-CUL2 immunocomplex, Ub or ORF10 were withheld from the CUL2ZYG11B ubiquitination system in vitro. The reaction products were detected by immunoblotting with anti-ubiquitin antibody. (H) VHL could not bind with ORF10. Purified ORF10-FLAG was used to pull down 6 × His-VHL-MYC. (I) ORF10 could not stimulate the ubiquitination activity of CUL2VHL. Either ORF10 or BSA was added to the CUL2VHL ubiquitination system in vitro, which contains E1, E2, ubiquitin, ATP, and CUL2VHL. The reaction products were detected by immunoblotting with anti-ubiquitin antibody. (J) ORF10 has no effect on the ubiquitination activity of CUL2VHL. Either ORF10 or BSA was added to the CUL2VHL ubiquitination system in vitro, which contains E1, E2, ubiquitin, ATP, and CUL2VHL. The reaction products were collected at the indicated times, and reaction products were detected by immunoblotting with anti-ubiquitin and FLAG antibodies. Source data are available for this figure: SourceData FS1.
As ZYG11B is a substrate adaptor in CUL2ZYG11B, the interaction between ORF10 and ZYG11B could in theory disrupt or promote the ubiquitination of host and/or viral proteins. To test the effects of ORF10 on CUL2ZYG11B, we reconstituted the CUL2ZYG11B immunocomplex in vitro to assess CUL2ZYG11B-mediated ubiquitination (Fig. S1 E). Upon the addition of ORF10 to this CUL2ZYG11B ubiquitination system, we were surprised to find that ORF10 did not inhibit, but rather greatly enhanced, the E3 ligase activity of CUL2ZYG11B (Fig. 1 C; and Fig. S1, F and G). Moreover, titrating ORF10 showed that this stimulation occurred in a concentration-dependent manner (Fig. 1 D). These results support that ORF10 directly interacts with ZYG11B and that ORF10 can stimulate the E3 ligase activity of CUL2ZYG11B.
Given that a previous study identified CUL2ZYG11B but no other CUL2 RING E3 ligase complex as part of the ORF10 interactome (Gordon et al., 2020), we examined the specificity of ORF10’s stimulation of CUL2ZYG11B E3 ligase activity. In vitro experiments with the CUL2VHL complex (with its distinct substrate adaptor, von Hippel-Lindau protein; VHL) showed that ORF10 does not interact with VHL and that the presence of ORF10 does not influence the E3 ligase activity of CUL2VHL (Fig. S1, H–J).
ORF10’s origin can be traced to a Bat-CoV and SARS-CoV sequence encoding a 25 amino acid gene product (“SC-ORF10”; Michel et al., 2020); we used these 25 residues protein to assess the specificity of ORF10’s binding with ZYG11B. That is, immunoprecipitation (IP) experiments showed that ORF10, but not SC-ORF10, can physically interact with ZYG11B (Fig. 1, A, B, and E). Moreover, we confirmed that adding SC-ORF10 did not affect the E3 ligase activity of CUL2ZYG11B (Fig. 1 F). Collectively, these results demonstrate that ORF10 binding with ZYG11B specifically enhances CUL2ZYG11B activity by directly interacting with ZYG11B.
ORF10 expression decreases the stability of multiple cilium-related proteins
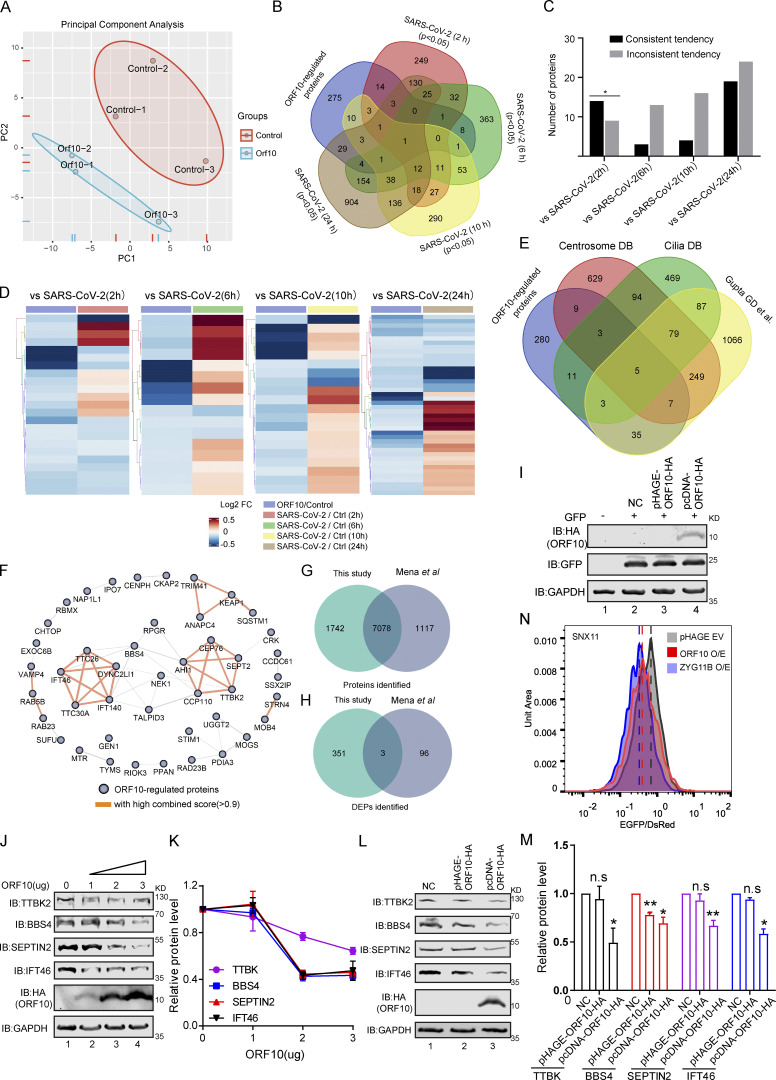
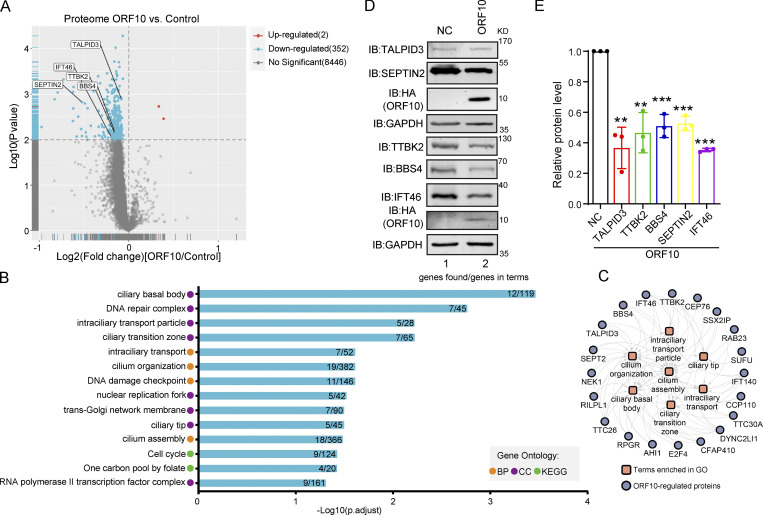
Given that SARS-CoV-2 ORF10 can stimulate the E3 ligase activity of CUL2ZYG11B, we investigated how ORF10 expression affects the stability of CUL2ZYG11B target proteins. Specially, we conducted Tandem Mass Tag (TMT)-based proteomics analysis to profile the change of global protein levels between ORF10-expressing and mock-transfected HEK293T cells. Across three replicates for each group, reproducibility was high (Fig. S2 A), and we obtained quantification data for a total of 8,820 proteins (Table S1), among which 352 downregulated and 2 upregulated proteins were detected in the ORF10-expressing cells (Fig. 2 A). These results clearly indicate that ORF10 affects protein stability in HEK293T cells. A comparison of these proteins with data from a proteomics study of SARS-CoV-2 infection (Bojkova et al., 2020) showed ∼22% (79/354) overlap between our proteomics data from ORF10-expressing cells and the SARS-CoV-2 infected proteome (Fig. S2 B). Among these overlapped proteins, some proteins showed downregulated tendency in both ORF10 expression and SARS-CoV-2 infection (2 h; Fig. S2, C and D), indicating the downregulation of some proteins after SARS-CoV-2 infection may be partially accounting for the expression of ORF10.
Figure S2.
Overlap of different expression proteins (DEPs) in ORF10 expressed and SARS-CoV-2 infected cells. (A) Principal component analysis (PCA) of proteome of control and ORF10 expression group. (B) Overlap between the DEPs in ORF10 expressed cells and previously identified SARS-CoV-2 infected proteome (Bojkova et al., 2020). (C) Number of proteins in both DEPs of ORF10 expressed cells and SARS-CoV-2 infected proteome. (D) A heatmap of downregulated proteins in ORF10 expressed cells and previously reported SARS-CoV-2 infected cells (Bojkova et al., 2020). (E) Overlap between the DEPs in ORF10 expressed cells and a published cilia-related proteome (Alves-Cruzeiro et al., 2014; Gupta et al., 2015; van Dam et al., 2013). (F) The network of protein–protein interactions between ORF10-regulated cilium proteins according to the String database (Szklarczyk et al., 2017). (G) Overlap between the identified protein in this study and Mena et al. (2021) study. (H) Overlap between the DEPs in this study and Mena et al. (2021) study. (I) Immunoblotting analysis of the expression of pcDNA-ORF10-HA and pHAGE-ORF10-HA plasmids in transfected HEK293T cells within 24 h. (J) The protein level of TTBK2, BBS4, SEPTIN2, and IFT46 was dramatically decreased along with the gradient increased ORF10 expression. Immunoblotting analysis of TTBK2, BBS4, SEPTIN2, IFT46, and ORF10 levels in ORF10-expressing and empty vector (NC) control HEK293T cells. GAPDH served as a loading control. (K) Odyssey-based quantification of protein levels in J (n = 3 independent experiments). Data are presented as the mean ± SEM. Statistical analysis was performed with two-tailed unpaired Student’s t test. (L) Cells transfected with pcDNA-ORF10-HA display reduced level of multiple cilium-related proteins. Immunoblotting analysis of TTBK2, BBS4, SEPTIN2, IFT46, and ORF10 levels in ORF10-expressing and empty vector (NC) control HEK293T cells. GAPDH served as a loading control. (M) Odyssey-based quantification of protein levels in L (n = 3 independent experiments). Data are presented as the mean ± SEM. * indicates P < 0.05, ** indicates P < 0.01. Statistical analysis was performed with two-tailed unpaired Student’s t test. (N) Stability analysis of the SNX11 degron-fused EGFP with exogenous expression of ORF10 and ZYG11B proteins. overexpression of ZYG11B substantially reduced the EGFP/DsRed ratio by accelerated degradation of the SNX11 degron-fused EGFP protein. Source data are available for this figure: SourceData FS2.
Figure 2.
ORF10 downregulates cilium-related proteins. (A) Volcano plots of differentially expressed proteins (DEPs) were detected in a quantitative proteomic analysis of ORF10-expressing and empty vector control HEK293T cells. The red spots and blue spots indicate significantly upregulated and downregulated proteins, respectively. (B) Functional classification analysis of DEPs according to enrichment analysis of the biological process (BP) and cellular component (CC) terms and KEGG pathways. (C) The network of DEPs and enriched cilium-related gene ontology terms. (D) Cells expressing ORF10 display reduced accumulation of multiple cilium-related proteins. Immunoblotting analysis of TALPID3, TTBK2, BBS4, SEPTIN2, IFT46, and ORF10 levels in ORF10-expressing and empty vector (NC) control HEK293T cells. GAPDH served as a loading control. (E) Odyssey-based quantification of protein levels in D (n = 3 independent experiments). Data are presented as the mean ± SEM. ** indicated P < 0.01, *** indicated P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F2.
GO and KEGG analyses indicated that many of the proteins with decreased levels in ORF10-expressing cells have annotated functions related to ciliogenesis (e.g., “cilia basal body,” “intraciliary transport particle,” and “cilium organization”; Fig. 2, B and C; and Fig. S2, E and F; and Table S2). This was striking, as COVID-19 symptoms relate to cilia dysfunction (Li et al., 2020c). We again analyzed ORF10-expressing and empty vector (NC) transfected HEK293T cells with immunoblotting and thus confirmed that the presence of ORF10 significantly decreased the levels of proteins with known cilium-related functions such as TTBK2 (cilia initiation; Lo et al., 2019), BBS4 (basal body maintenance; Uytingco et al., 2019), SEPTIN2 (regulating ciliary length; Ghossoub et al., 2013), TALPID3 (cilia gating; Yan et al., 2020b), and IFT46 (intraflagellar transport complex B proteins required for cilia biogenesis and maintenance; Lee et al., 2015; Park et al., 2016; Shi et al., 2018; Fig. 2, D and E). Beyond demonstrating that the presence of ORF10 in cells results in decreased levels of several hundred proteins, these results support that SARS-CoV-2 ORF10 may help explain some of the cilium-related clinical features of COVID-19.
A recent study also performed TMT-based proteomics analysis of HEK293T cells after ORF10 expression (Mena et al., 2021). Although the identified proteins in HEK293T cells are very similar between our data and that study (Fig. S2 G), there is little overlap between the differentially expressed proteins (DEPs; Fig. S2 H), and they did not observe differences in the cilia-related proteins mentioned in our study. To address the basis of this difference between the two data sets, we carefully compared the experimental materials used in the two studies and found that the ORF10 expression plasmid backbone used in our study (pcDNA) differed from their study (pHAGE). Further, it is notable that the study by Mena et al. (2021) did not include data for ORF10 levels in HEK293T cells. We monitored ORF10 levels upon transfecting HEK293T cells to the pcDNA-ORF10-HA or pHAGE-ORF10-HA plasmid and found that ORF10-HA was successfully detected in cells harboring the pcDNA-ORF10-HA plasmid, but was not detected in cells harboring pHAGE-ORF10-HA plasmids (Fig. S2 I).
Thus, we speculate that the levels of ORF10 might cause the observed difference in the cilium-related protein of the two studies. Pursuing this, we used immunoblotting to measure the protein levels of four cilium-related proteins in HEK293T cells sample having a gradient of increasing ORF10 expression. We found that the protein level of these cilium-related proteins was dramatically decreased as ORF10 expression increased (Fig. S2, J and K), supporting that the protein level of ORF10 does affect the stability of these cilium-related proteins. We also monitored the protein levels of the cilium-related proteins in HEK293T cells transfected with pcDNA-ORF10-HA plasmids or pHAGE-ORF10-HA plasmids, and found the protein levels of cilium-related proteins was dramatically decreased in the cells transfected with pcDNA-ORF10-HA compared with that of cells transfected with pHAGE-ORF10-HA (Fig. S2, L and M).
Mena et al. (2021) reported that ORF10 expression does not affect the degradation of known substrate proteins of the CUL2ZYG11B complexes. To test whether differential ORF10 levels may have affected the data for CUL2ZYG11B ligase activity in the two studies, we reconstituted a Global Protein Stability (GPS) system with reporters to detect CUL2ZYG11B activity. We found that HEK293T cells transfected with pcDNA-ORF10-HA had a reduced EGFP: DsRed ratio value compared with the empty vector control cells (Fig. S2 N), which was similar to the effects observed for cells overexpressing ZYG11B (Fig. S2 N). These results indicate that the ORF10 level can account for the differences between the two studies and additionally support that ORF10 does affect the stability of these cilium-related proteins.
ORF10 impairs cilium biogenesis and maintenance
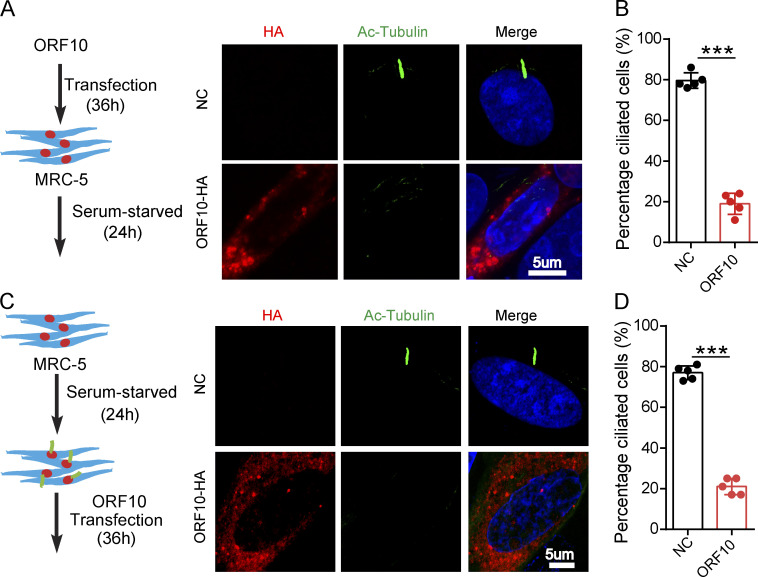
To investigate the consequences of ORF10 expression in cilium biogenesis and maintenance, we transfected pcDNA-ORF10-HA and empty vector into MRC-5 cells (initially isolated from normal lung tissue of a 14-wk-old male fetus) and NIH3T3 cells (initially isolated from mouse embryonic fibroblast cells) before subsequently inducing primary cilium biogenesis with a serum starvation protocol (Li et al., 2020b; Nandadasa et al., 2019; Fig. 3 A and Fig. S3 A). Further, staining of both cell types with an antibody against the canonical cilia marker acetylated alpha-tubulin (Ac-Tubulin; Nekooki-Machida and Hagiwara, 2020) revealed a significant reduction in primary cilium assembly (Fig. 3, A and B; and Fig. S3, A and B). Both of these findings support that ORF10 can impair primary cilium biogenesis.
Figure 3.
ORF10 impairs cilium biogenesis and maintenance. (A) ORF10 inhibits cilium biogenesis. Immunofluorescence analysis of Ac-Tubulin (green) and HA (red) in ORF10-expressing and empty vector control MRC-5 cells. Nuclei were stained with DAPI (blue). (B) Quantification of the distribution of ciliated cells among the ORF10-expressing and empty-vector control MRC-5 cells (A; n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (C) ORF10 perturbs cilium maintenance. Immunofluorescence analysis of Ac-Tubulin (green) and HA (red) in ORF10-expressing and empty-vector control MRC-5 ciliated cells. Nuclei were stained with DAPI (blue). (D) Quantification of the ratio of ciliated cells in ORF10 expressed and empty-vector transfected ciliated cells (C;n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F3.
Figure S3.
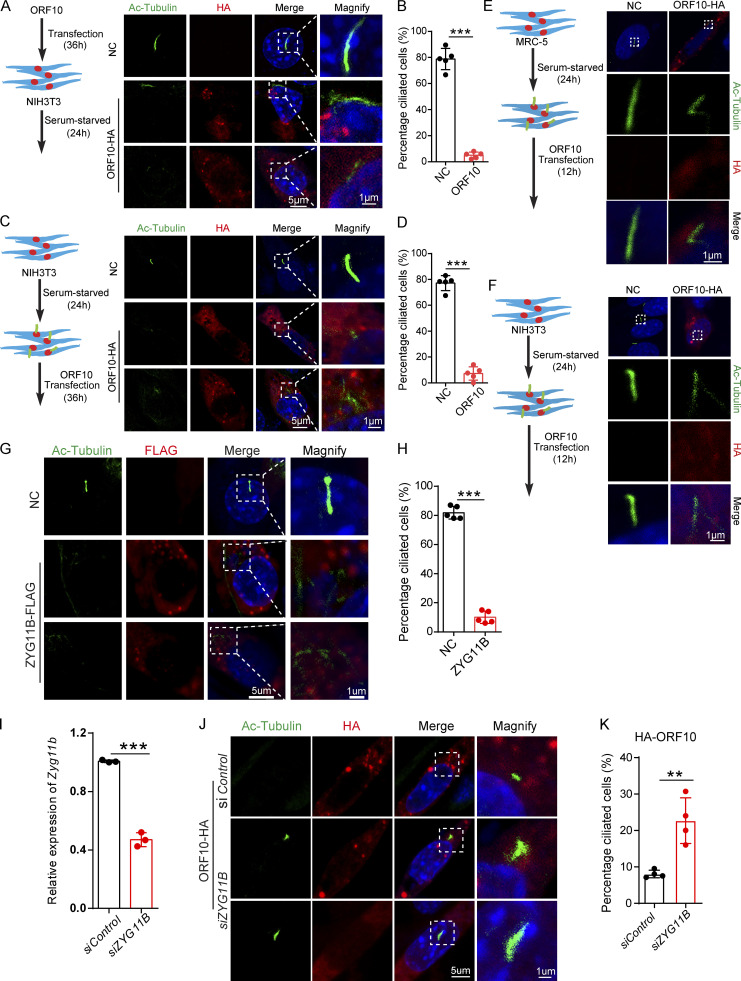
Cilia dysfunction in ORF10 expressing cells is related to ZYG11B. (A) ORF10 inhibits cilia biogenesis in mouse NIH3T3 cells. Immunofluorescence analysis for Ac-Tubulin (green) and HA (red) was performed in ORF10-expressing and empty-vector transfected NIH3T3 cells. Nuclei were stained with DAPI (blue). (B) Quantification ratio of ciliated cells in ORF10-expressing and empty-vector transfected cells (A) (n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (C) ORF10 perturbs cilia maintenance in mouse NIH3T3 cells. Immunofluorescence analysis for Ac-Tubulin (green) and HA (red) was performed in ORF10-expressing and empty-vector transfected NIH3T3 ciliated cells. Nuclei were stained with DAPI (blue). (D) Quantification of the ratio of ciliated cells in ORF10-expressing and empty-vector transfected cells (C; n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (E and F) ORF10 is localized in the cytoplasm of MRC-5 cells (E) and NIH3T3 cells (F). Immunofluorescence analysis for Ac-Tubulin (green) and HA (red) was performed in ORF10-expressing and empty-vector transfected MRC-5 cells or NIH3T3 cells for 12 h. Nuclei were stained with DAPI (blue). (G) ZYG11B overexpression perturbs cilia biogenesis. Immunofluorescence analysis for Ac-Tubulin (green) and FLAG (red) was performed in ZYG11B overexpressing and empty-vector transfected NIH3T3 cells. Nuclei were stained with DAPI (blue). (H) Quantification ratio of ciliated cells in ZYG11B overexpressing and empty-vector transfected cells (G; n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (I) The knockdown efficiency of Zyg11b. Relative mRNA level of Zyg11b in siControl or siZYG11B transfected NIH3T3 cells. Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (J) Knockdown of Zyg11b partially rescues the ciliogenesis defects in ORF10-expressing cells (siControl or siZYG11B). Nuclei were stained with DAPI (blue). (K) Distribution of ciliated cells among ORF10-expressing cells co-transfected with vectors for expression of siControl or siZYG11B (I; n = 4 independent experiments). Data are presented as the mean ± SEM. ** indicates P < 0.01. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData FS3.
We next assessed the effect of ORF10 expression in cilium maintenance. After the starvation-induced initiation of primary cilium biogenesis in MRC-5 cells and NIH3T3 cells, we transfected pcDNA-ORF10-HA into these ciliated cells (Fig. 3 C and Fig. S3 C). Disrupted cilium was observed in ORF10-expressing but not empty vector control cells (Fig. 3, C and D and Fig. S3, C and D), indicating that ORF10 also perturbs cilium maintenance. These results collectively indicate that ORF10 somehow impairs both cilium biogenesis and maintenance in cells.
ORF10 impairs cilium biogenesis and maintenance by promoting IFT46 degradation through CUL2ZYG11B
To explore the molecular mechanism underlying ORF10 expression in cilium dysfunction, we first examined whether ORF10 directly localized in the cilium region to destroyed cilium biogenesis and maintenance. We found ORF10 is actually localized in the cytoplasm of NIH3T3 cells and MRC-5 cells (Fig. S3, A and C), which is consistent with recently reported results (Zhang et al., 2020). To more accurately profile the localization of ORF10 in primary ciliated cells, we detected the localization of ORF10 prior to the loss of cilium in NIH3T3 cells and MRC-5 cells, and found that ORF10 did not show any obvious co-localization with cilium (Fig. S3, E and F). Thus, ORF10 may impair cilium biogenesis and maintenance in some indirect manner.
Given our results showing that ORF10 stimulates the E3 ligase activity of CUL2ZYG11B and that some cilium-related proteins are downregulated upon ORF10 expression, we speculate that ORF10 may impair cilium biogenesis and maintenance by promoting the degradation of cilium-related proteins through CUL2ZYG11B. To test it, we overexpressed ZYG11B in NIH3T3 cells and subsequently induced primary cilium biogenesis with a serum starvation protocol (Fig. S3 G). The ZYG11B-overexpressing cells displayed significant defects in primary cilium assembly (Fig. S3, G and H). Further, knockdown of Zyg11b partially rescued the ciliogenesis defects of ORF10-expressing cell (after serum starvation; Fig. S3, I–K). These results functionally implicate the ZYG11B-ORF10 interaction in the cilium dysfunction that we observed in ORF10-expressing cells.
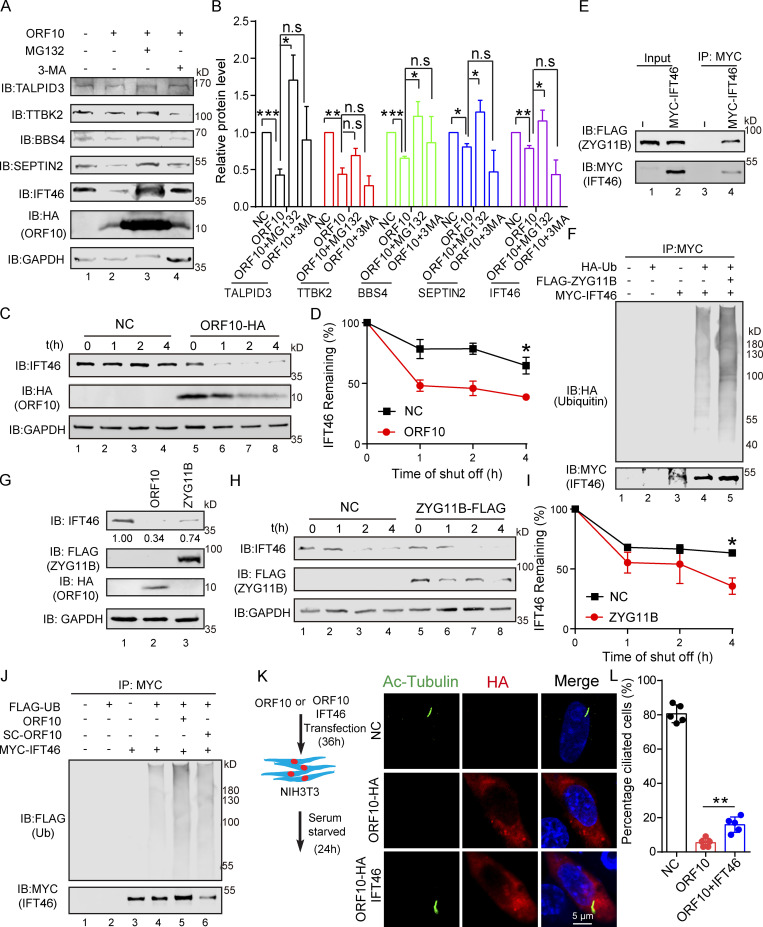
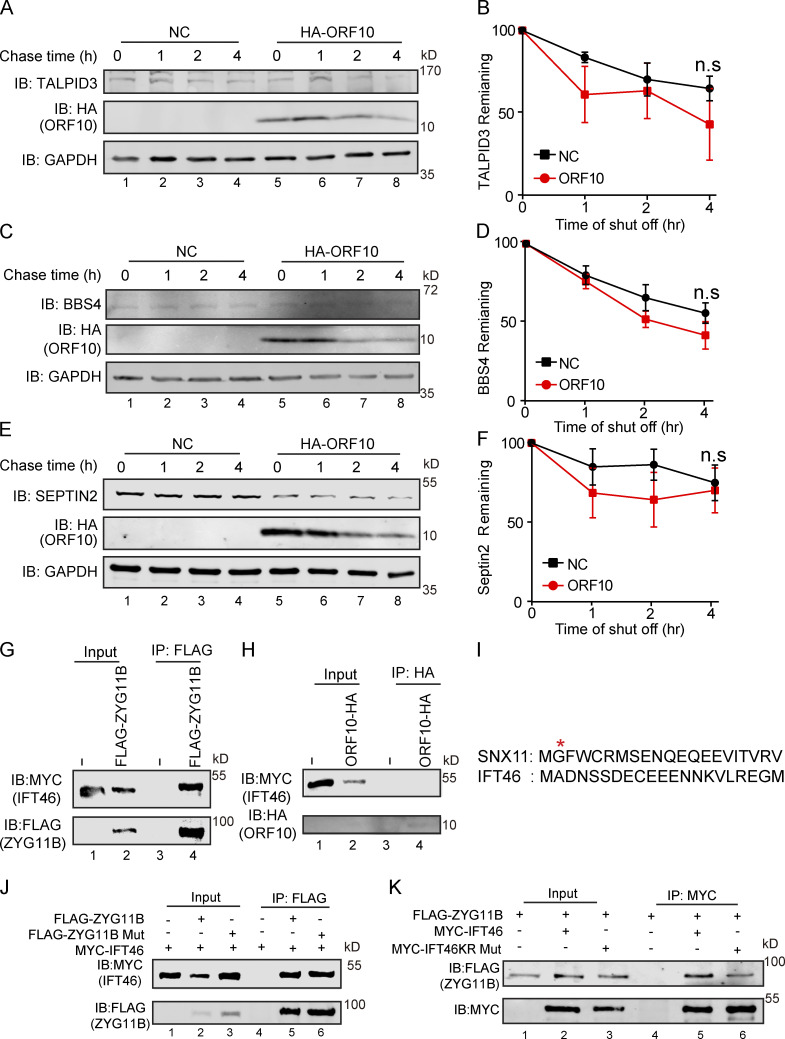
We next explored ORF10-CUL2ZYG11B substrates that may affect cilium biogenesis and maintenance when ORF10 is present in cells. Many cilium-related proteins, such as TALPID3, BBS4, SEPTIN2, and IFT46, are degraded by the ubiquitin-proteasome pathway in ORF10-expressing cells except TTBK2. Proteasome inhibition (with MG132) partially rescued the ORF10-mediated promotion of protein degradation, whereas the inhibition of autophagy-lysosome degradation system (with 3-MA) exerted no rescue effect (Fig. 4, A and B). Further, cycloheximide (CHX) chase assays showed that ORF10 strongly promoted IFT46 degradation compared with the control group; less pronounced impacts from ORF10 were observed on the extent of TALPID3, TTBK2, and SEPTIN2 degradation (Fig. S4, A–F). These findings narrowed our focus to the required cilium biogenesis protein IFT46.
Figure 4.
ORF10 promotes IFT46 degradation by stimulating CUL2ZYG11B activity. (A) ORF10 reduces TALPID3, TTBK2, BBS4, SEPTIN2, and IFT46 levels in a ubiquitin–proteasome pathway-dependent manner. Immunoblotting analysis of TALPID3, TTBK2, BBS4, SEPTIN2, IFT46, and ORF10 levels in ORF10-expressing and empty vector control HEK293T cells that were treated with or without the proteasome inhibitor MG-132 or the autophagy-lysosome degradation inhibitor 3-MA. GAPDH served as a loading control. (B) Odyssey-based quantification of protein levels in A (n = 3 independent experiments). Data are presented as the mean ± SEM. * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (C) ORF10 promotes the degradation of IFT46. A cycloheximide chase (CHX) assay of IFT46 was performed in ORF10-expressing and empty vector control HEK293T cells. Samples were taken at 0, 1, 2, and 4 h after the addition of CHX. The IFT46 and ORF10 protein levels were detected by immunoblotting using rabbit IFT46 antibody and rabbit HA antibody. GAPDH served as a loading control. (D) Quantification of the IFT46 levels in C. The protein level of IFT46 was quantified using Odyssey software. (n = 3 independent experiments). Data are presented as the mean ± SEM. * indicates P < 0.05. Statistical analysis was performed with two-tailed unpaired Student’s t test. (E) ZYG11B physically interacts with IFT46. MYC-IFT46 and ZYG11B-FLAG (or empty vector) were co-transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-MYC antibody, followed by analysis using anti-FLAG and anti-MYC antibodies. (F) CUL2ZYG11B promotes the ubiquitination of IFT46. MYC-IFT46, ZYG11B-FLAG, and HA-Ub were transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-MYC antibody, followed by analysis using anti-HA and anti-MYC antibodies. (G) The IFT46 level is dramatically decreased in ZYG11B-overexpressing HEK293T cells. Immunoblotting analysis against IFT46, FLAG, and HA for control, ORF10-expressing, and ZYG11B-overexpressing HEK293T cells. GAPDH served as a loading control. (H) Overexpression of ZYG11B promotes IFT46 degradation. A cycloheximide chase (CHX) assay of IFT46 was performed ZYG11B-overexpressing HEK293T cells. Samples were taken at 0, 1, 2, and 4 h after the addition of CHX. The IFT46 and ZYG11B protein levels were assessed with immunoblotting. GAPDH served as a loading control. (I) Quantification of the relative IFT46 levels in H. The protein level of IFT46 were quantified using Odyssey software. (n = 3 independent experiments). Data are presented as the mean ± SEM. * indicates P < 0.05. Statistical analysis was performed with two-tailed unpaired Student’s t test. (J) ORF10 promotes the ubiquitination of IFT46. MYC-IFT46 and FLAG-Ub were transfected into HEK293T cells. 24 h after transfection, cells were collected and added with ORF10 or SARS-ORF10 for immunoprecipitation (IP) with an anti-MYC antibody, followed by analysis using anti-FLAG and anti-MYC antibodies. (K) IFT46 overexpression partially rescues the cilia biogenesis defects of ORF10-expressing NIH3T3 cells. Immunofluorescence analysis for Ac-Tubulin (green) and HA (red) in ORF10-expressing, ORF10/IFT46 co-transfected, and empty-vector cells. Nuclei were stained with DAPI (blue). (L) Distribution of ciliated cells among the ORF10-expressing, ORF10/IFT46 co-transfected, and empty-vector cells (K; n = 5 independent experiments). Data are presented as the mean ± SEM. ** indicates P < 0.01. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F4.
Figure S4.
The interaction of ZYG11B and IFT46 is not based on a Gly/N-degron-based mechanism. (A, C, and E) A cycloheximide chase (CHX) assay of TALPID3, BBS4, and SEPTIN2 was performed in ORF10-expressing and empty vector control HEK293T cells. Samples were taken at 0, 1, 2, and 4 h after the addition of CHX. The TALPID3, BBS4, SEPTIN2, and ORF10 protein levels were detected by immunoblotting using TALPID3, BBS4, SEPTIN2 antibodies, and HA antibody. GAPDH served as a loading control. (B, D, and F) Quantification of the TALPID3, BBS4, and SEPTIN2 levels in (A, C, and E). The protein level of TALPID3, BBS4 and SEPTIN2 was quantified using Odyssey software. (n = 3 independent experiments). Data are presented as the mean ± SEM. Statistical analysis was performed with two-tailed unpaired Student’s t test. (G) ZYG11B physically interacts with IFT46. MYC-IFT46 and ZYG11B-FLAG (or empty vector) were co-transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-FLAG antibody, followed by analysis using anti-FLAG and anti-MYC antibodies. (H) ORF10 could not interacts with IFT46. MYC-IFT46 and ORF10-HA (or empty vector) were co-transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-HA antibody, followed by analysis using anti-HA and anti-MYC antibodies. (I) The protein sequences of SNX11 and IFT46. (J) ZYG11B-Mut did not affect the interaction of IFT46 and ZYG11B. MYC-IFT46 and ZYG11B-FLAG (or ZYG11B-Mut-FLAG) were co-transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-FLAG antibody, followed by analysis using anti-FLAG and anti-MYC antibodies. (K) IFT46 KR Mut did not affect the interaction of IFT46 and ZYG11B. ZYG11B-FLAG and MYC-IFT46 (or IFT46 KR Mut) were co-transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-MYC antibody, followed by analysis using anti-FLAG and anti-MYC antibodies. Source data are available for this figure: SourceData FS4.
Recalling that ORF10 stimulates the E3 ligase activity of CUL2ZYG11B (Fig. 1), it was unsurprising that ORF10 expression enhanced the ubiquitination level of IFT46 (Fig. 4 J), while SC-ORF10 has little effect on it. Co-immunoprecipitation (co-IP) experiments also showed that IFT46 can physically interact with ZYG11B, but not ORF10 (Fig. 4 E; and Fig. S4, G and H) and that overexpression of ZYG11B in HEK293T cells increased the extent of IFT46 ubiquitination and decreased the overall IFT46 level (Fig. 4, F and G). In addition, overexpressing ZYG11B in HEK293T cells significantly promoted IFT46 degradation (Fig. 4, H and I), indicating that CUL2ZYG11B catalyzes the ubiquitination of IFT46 to induce its degradation.
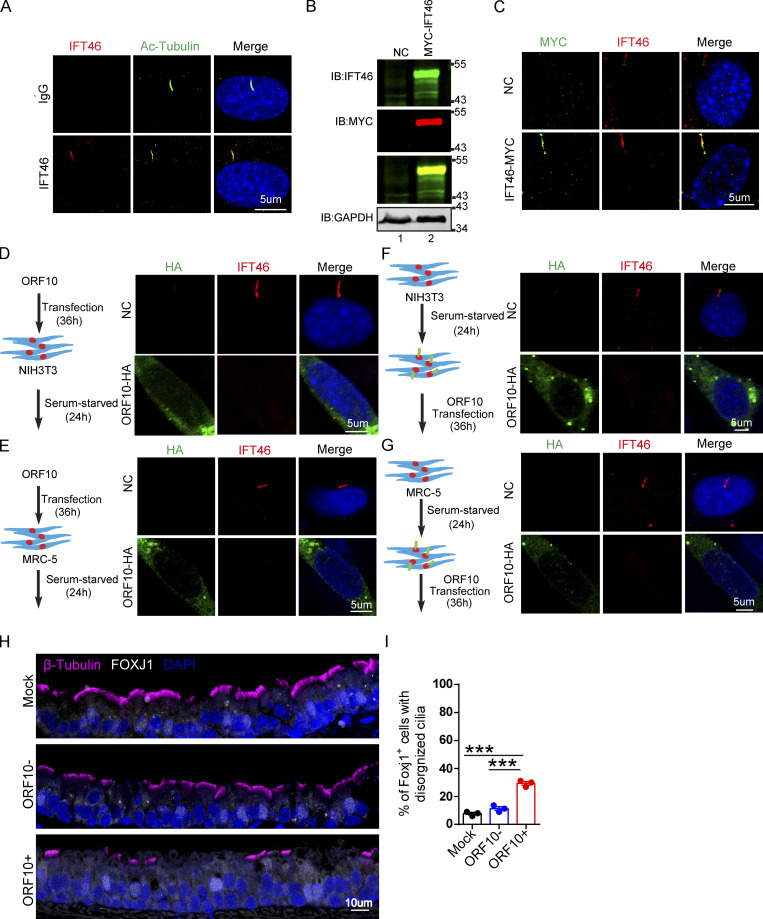
IFT46 is one of the highly conserved intraflagellar transport (IFT) complex B proteins, which deliver new axonemal components to their assembly site and are essential for cilium biogenesis and maintenance (Silverman and Leroux, 2009). As the knockdown of Ift46 was previously shown to cause ciliary defects (Lee et al., 2015), we reasoned that ORF10 may perturb cilium formation and maintenance by promoting IFT46 degradation. We examined the localization of IFT46 in NIH3T3 and MRC-5 cells expressing ORF10 and found that the IFT46 signal intensity was dramatically reduced compared with control cells; we also noted that the cells with ORF10 did not form cilium (Fig. S5, A–E). The introduction of ORF10 into NIH3T3 and MRC-5 cells after primary cilium biogenesis caused dysregulated cilium localization of IFT46 (Fig. S5, F and G). In addition, IFT46 overexpression partially rescued the ciliogenesis defects in ORF10-expressing cells. There was a significantly increased proportion of ciliated cells among ORF10-IFT46-expressing cells compared with ORF10-expressing cells (Fig. 4, K and L). Thus, ORF10 impairs cilium biogenesis and maintenance by promoting IFT46 degradation through CUL2ZYG11B.
Figure S5.
ORF10 perturbs the cilia localization of IFT46 during cilia formation and maintenance. (A) Immunofluorescence analysis for Ac-Tubulin (green) and IFT46 (red) was performed in NIH3T3 ciliated cells. Nuclei were stained with DAPI (blue). (B) Immunoblotting analysis of IFT46 and MYC was performed in IFT46-MYC expressing and empty-vector transfected cells. GAPDH served as a loading control. (C) Immunofluorescence analysis for MYC (green) and IFT46 (red) was performed in IFT46-MYC expressing and empty-vector transfected ciliated cells. Nuclei were stained with DAPI (blue). (D and E) ORF10 perturbs the cilium localization of IFT46. Immunofluorescence analysis for IFT46 (red) and HA (green) was performed in ORF10-expressing and empty-vector transfected NIH3T3 cells (D) or MRC-5 cells (E). Nuclei were stained with DAPI (blue). (F and G) The cilium localization of IFT46 is dysregulated in ORF10-expressing ciliated cells. Immunofluorescence analysis for HA (green) and IFT46 (red) was performed in ORF10-expressing and empty-vector transfected NIH3T3 cells (F) or MRC-5 cells (G). Nuclei were stained with DAPI (blue). (H) Immunofluorescence analysis for cilia in FOXJ1 positive epithelial cells. FOXJ1 is the marker of epithelial ciliated cells. (I) Quantification of FOXJ1 positive epithelial cells with disorganized cilia. n = 3 independent experiments. Data are presented as the mean ± SD. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData FS5.
CUL2ZYG11B promoting IFT46 degradation to impair cilium biogenesis and maintenance via the C2 domain of IFT46
ZYG11B is the substrate receptor of CUL2 and is responsible for the recognition of the N-terminal glycine degrons (Yan et al., 2021). Given that IFT46 does not have any previously identified ZYG11B degron sequences (Fig. S4 I), IFT46 may interact with ZYG11B through some other mechanism(s). Pursuing this, we mutated three residues (ZYG11B- W522A, D526A, N567A; henceforth “ZYG11B-Mut”) of ZYG11B known as essential for CUL2ZYG11B recognition of Gly/N-degron sequences in its substrates (Yan et al., 2021) and monitored interactions between ZYG11B and IFT46 in HEK293T cells. Co-IP showed no difference in the extent of the ZYG11B-IFT46 interaction between WT ZYG11B and the ZYG11B variant incapable of recognizing Gly/N-degron sequences (Fig. S4 J), indicating that the interaction between ZYG11B and IFT46 is not based on a Gly/N-degron-based mechanism.
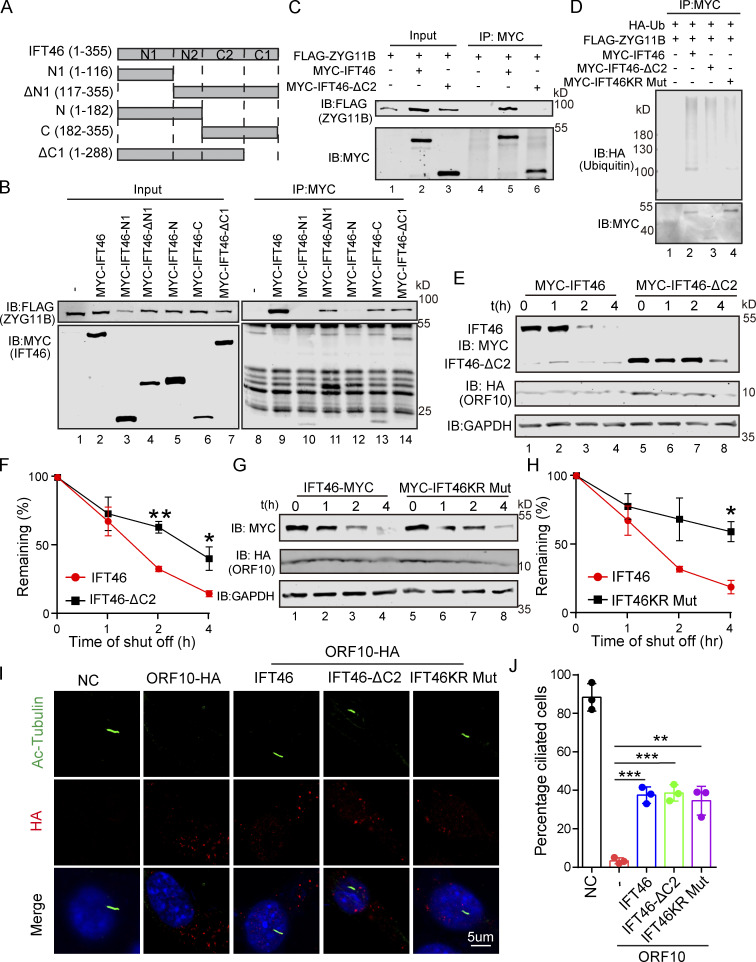
We also generated a series of internal deletion mutants of IFT46 to map the binding regions for ZYG11B (Fig. 5 A) and found that the C-terminal domain (amino acids 182–288, IFT46-C2) of IFT46 was sufficient for binding to ZYG11B (Fig. 5, B and C). Further, an IFT46 mutant variant lacking its C2 region (IFT46-△C2) carried no ubiquitination signal in HEK293T cells and had a much longer half-life in cells than WT IFT46 after ORF10 expression, clearly supporting that disrupting the IFT46-ZYG11B interaction perturbs the degradation of IFT46 (Fig. 5, D–F). To further experimentally validate that the effect of ZYG11B-IFT46 interaction on ORF10 caused cilium dysfunction, we overexpressed IFT46-△C2 in ORF10-expressing NIH3T3 cells. We found that IFT46–△C2 expression rescued the ciliogenesis defects in ORF10-expressing cells (Fig. 5, I and J). These results show that CUL2ZYG11B promoted IFT46 degradation to impair cilium biogenesis and its maintenance via a non-classical way, which recognized the C2 domain of IFT46.
Figure 5.
CUL2ZYG11B promoting IFT46 degradation to impair cilium biogenesis and maintenance via the C2 domain of IFT46. (A) Schematic overview of IFT46 truncation mutants. (B) Interactions between ZYG11B and truncation mutants of IFT46. Immunoprecipitation (IP) of FLAG-tagged ZYG11B and MYC-tagged truncation mutants of IFT46 followed by immunoblotting to detect the interaction in HEK293T cells. (C) C2 domain (amino acids 182–288, IFT46-C2) of IFT46 was sufficient for binding to ZYG11B. MYC-IFT46 (or IFT46-△C2, empty vector) and ZYG11B-FLAG were co-transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-MYC antibody, followed by analysis using anti-FLAG and anti-MYC antibodies. (D) CUL2ZYG11B promotes the ubiquitination of IFT46 via the C2 domain of IFT46. HA-Ub, ZYG11B-FLAG, and MYC-IFT46 or IFT46-△C2 were transfected into HEK293T cells. 24 h after transfection, cells were collected for immunoprecipitation (IP) with an anti-MYC antibody, followed by analysis using anti-HA and anti-MYC antibodies. (E) ORF10 promotes the degradation of IFT46 via the C2 domain of IFT46. A cycloheximide chase (CHX) assay of IFT46 or IFT46-△C2 was performed in ORF10-expressing HEK293T cells. Samples were taken at 0, 1, 2, and 4 h after the addition of CHX. The IFT46, IFT46-△C2 and ORF10 protein levels were detected by immunoblotting using MYC and HA antibodies. GAPDH served as a loading control. (F) Quantification of the IFT46 and IFT46-△C2 levels in E. The protein level of IFT46 and IFT46-△C2 was quantified using Odyssey software. (n = 3 independent experiments). Data are presented as the mean ± SEM. ** indicates P < 0.01, * indicates P < 0.05. Statistical analysis was performed with two-tailed unpaired Student’s t test. (G) IFT46 KR Mut blocked the degradation of IFT46. A cycloheximide chase (CHX) assay of IFT46 or IFT46 KR Mut was performed in ORF10-expressing HEK293T cells. Samples were taken at 0, 1, 2, and 4 h after the addition of CHX. The IFT46, IFT46 KR Mut and ORF10 protein levels were detected by immunoblotting using MYC and HA antibody. GAPDH served as a loading control. (H) Quantification of the IFT46 and IFT46 KR Mut levels in G. The protein level of IFT46 and IFT46 KR Mut was quantified using Odyssey software. (n = 3 independent experiments). Data are presented as the mean ± SEM. * indicates P < 0.05. Statistical analysis was performed with two-tailed unpaired Student’s t test. (I) IFT46-△C2 and IFT46 KR Mut overexpression partially rescues the cilia biogenesis defects of ORF10-expressing NIH3T3 cells. Immunofluorescence analysis for Ac-Tubulin (green) and HA (red) in ORF10-expressing, ORF10/IFT46, ORF10/IFT46△C2, ORF10/IFT46 KR Mut co-transfected, and empty-vector cells. Nuclei were stained with DAPI (blue). (J) Quantification of ciliated cells in I (n = 3 independent experiments). Data are presented as the mean ± SEM. ** indicates P < 0.01, *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F5.
To further confirm whether ORF10 impaired cilium biogenesis and maintenance via facilitating IFT46 degradation through the ubiquitin–proteasome pathway, we generated a variant of IFT46 in which all K residues were mutated to R (IFT46 KR Mut), which blocks the potential CUL2ZYG11B-mediated ubiquitination but did not disrupt the IFT46-ZYG11B interaction (Fig. S4 K). In addition, the CHX chase assays showed that the IFT46 KR Mut blocked the degradation of IFT46 after ORF10 expression (Fig. 5, G and H). IFT46 KR Mut expression also rescued the ciliogenesis defects in ORF10-expressing cells (Fig. 5, I and J). Taken together, these results support that ORF10 caused cilium dysfunction via stimulating CUL2ZYG11B-mediated IFT46 degradation through the ubiquitin–proteasome pathway.
SARS-CoV-2 ORF10 impairs epithelium of respiratory tract cilia
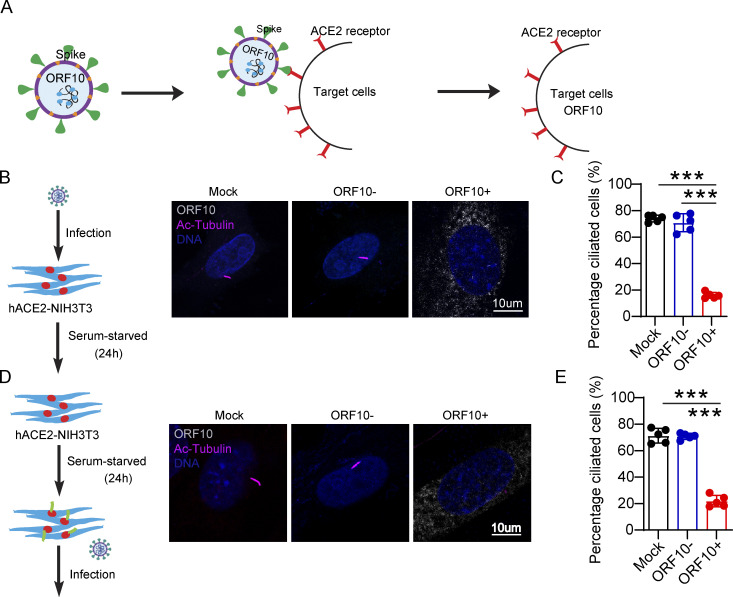
Having found that ORF10 can impair ciliogenesis by promoting IFT46 degradation through CUL2ZYG11B, we next explored potential impacts of ORF10 on the pathobiology of SARS-CoV-2 infection. Lacking a direct way to examine the pathogenic effect(s) of ORF10 in humans, we developed a lentivirus assay to mimic SARS-CoV-2 infection and exclude the potential effect(s) of other SARS-CoV-2 ORFs (Fig. 6 A). Given that SARS-CoV-2 infects host cells by binding the cellular receptor ACE2 through its Spike (S) protein (Hoffmann et al., 2020) to better simulate the process of SARS-CoV-2 infection target cells, we expressed SARS-CoV-2 Spike protein in lentiviral pseudotype particles and then exposed NIH3T3 cells expressing hACE2 to these particles (Fig. 6 A). Then, we infected hACE2 expressed NIH3T3 cells with these lentivirus particles pseudotyped carrying a DNA sequence encoding SARS-CoV-2 ORF10 (“Spike-lentivirus-ORF10”) or not. The ORF10 was expressed in hACE2-NIH3T3 cells exposed to Spike-lentivirus-ORF10, and the infected cells displayed a significant reduction in the proportion of ciliated cells compared to the Spike-lentivirus group, a finding that supported the success of the lentivirus experimental strategy and confirmed an impact of ORF10 in perturbation of cilia biogenesis and maintenance (Fig. 6, B–E).
Figure 6.
Spike-lentivirus-ORF10 perturbs cilia biogenesis and maintenance. (A) Schematic diagram of lentivirus infection of target cells. (B) Spike-lentivirus-ORF10 perturbs cilia biogenesis. Immunofluorescence analysis for Ac-Tubulin (magenta) and HA (white) was performed in mock, Spike-lentivirus, and Spike-lentivirus-ORF10 infected NIH3T3 cells. Nuclei were stained with DAPI (blue). (C) Quantification ratio of ciliated cells in mock, Spike-lentivirus, and Spike-lentivirus-ORF10 infected cells (B; n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (D) Spike-lentivirus-ORF10 perturbs cilia maintenance. Immunofluorescence analysis for Ac-Tubulin (magenta) and HA (white) was performed in mock, Spike-lentivirus, and Spike-lentivirus-ORF10 infected NIH3T3 ciliated cells. Nuclei were stained with DAPI (blue). (E) Quantification ratio of ciliated cells in mock, Spike-lentivirus, and Spike-lentivirus-ORF10 infected cells (D; n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F6.
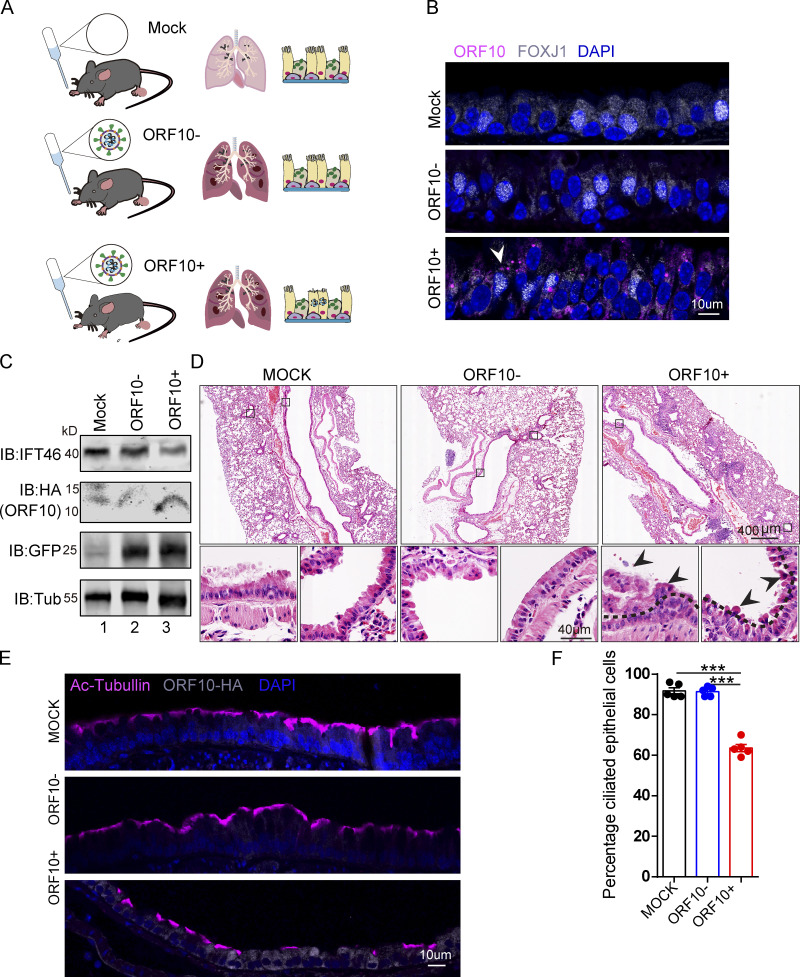
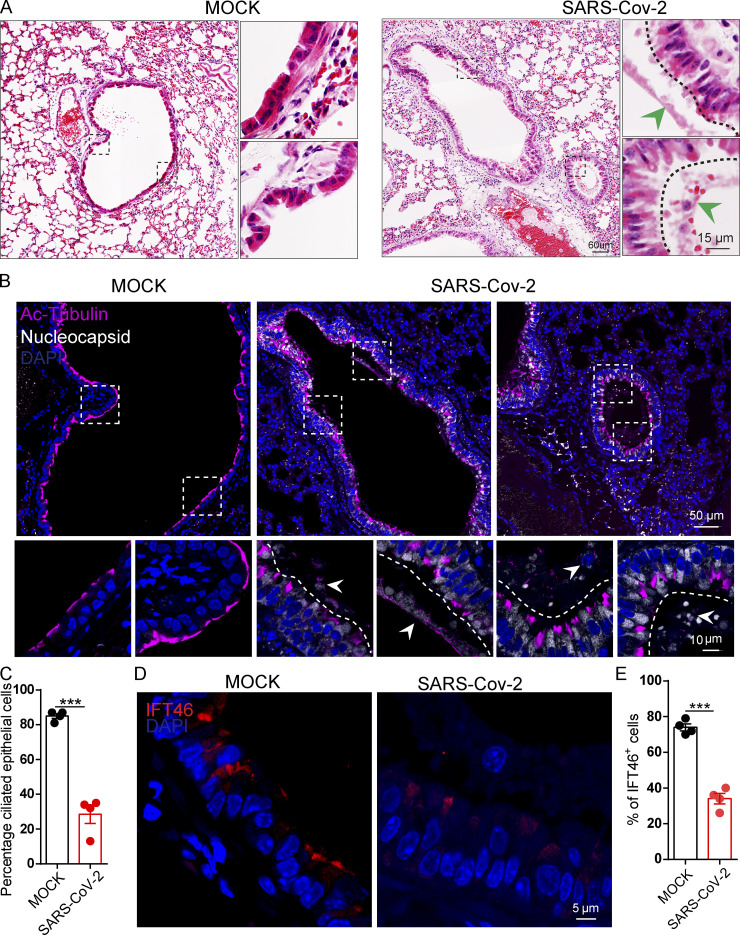
We then exposed hACE2 mice to 107 PFU lentivirus pseuodotyped particles (intranasally, Spike-lentivirus, ORF10-; Spike-lentivirus-ORF10, ORF10+; Fig. 7 A). Immunofluorescence analysis of infected mouse lungs showed that ORF10 was clearly expressed in epithelial ciliated cells of the trachea (Fig. 7, B and C) and the percentage of FOXJ1 positive cells with disorganized cilia was increased (Fig. S5, H and I). Further histological analysis of mice intranasally exposed to Spike-lentivirus-ORF10 revealed epithelial damage (Fig. 7 D). These phenotypes are similar to observations of SARS-CoV-2 infected hACE2 mice (Fig. 8 A). In addition, immunoblotting of lung tissue extracts revealed a decrease in the IFT46 level upon exposure to Spike-lentivirus-ORF10 compared with the Spike-lentivirus and control group (Fig. 7 C).
Figure 7.
SARS-CoV-2 ORF10 infection impairs trachea cilia. (A) Schematic of the experimental design for the SARS-CoV-2 ORF10 infection system used in the present study. The dark pink lung symbols in rows two and three means virus infection. (B) ORF10 expression was detected in the trachea of hACE2 mice. Immunofluorescence analysis for HA (magenta) and FOXJ1 (white) was performed in Spike-lentivirus-ORF10 infected trachea. Nuclei were stained with DAPI (blue). The white arrowhead indicates FOXJ1 and ORF10 positive cells. (C) IFT46 was downregulated in mouse lungs after Spike-lentivirus-ORF10 infection. Immunoblotting analysis of IFT46, HA, and GFP was performed in mock, Spike-lentivirus, and Spike-lentivirus-ORF10 infected hACE2 mouse lungs. Tubulin served as a loading control. (D) Haematoxylin and eosin (H&E) staining of hACE2 lungs after mock, Spike-lentivirus (ORF10−), and Spike-lentivirus-ORF10 (ORF10+) infection. The black arrows indicate epithelial damage and the dashed lines indicate the boundary of the cilia layers. (E) ORF10 damaged the cilia layer in mouse trachea. Immunofluorescence analysis for Ac-Tubulin (magenta) and HA (white) was performed in mock, Spike-lentivirus, and Spike-lentivirus-ORF10 infected mice. Nuclei were stained with DAPI (blue). (F) Quantification ratio of ciliated epithelial cells in E (n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F7.
Figure 8.
SARS-CoV-2 infection impairs bronchiole cilia. (A) H&E staining of hACE2 lungs after mock, and SARS-CoV-2 infection. The green arrowheads indicate epithelial damage. (B) SARS-CoV-2 infection damaged cilia layer in mouse bronchioles. Immunofluorescence analysis for Ac-Tubulin (magenta) and Nucleocapsid (white) of SARS-CoV-2 was performed in mock, and SARS-CoV-2 infected mice. Nuclei were stained with DAPI (blue). The white arrows indicate epithelial damage. (C) Quantification ratio of ciliated epithelial cells in B (n = 4 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (D) SARS-CoV-2 infection influence the level IFT46 in mouse bronchioles. Immunofluorescence analysis for IFT46 (red) was performed in mock, and SARS-CoV-2 infected mice. Nuclei were stained with DAPI (blue). (E) Quantification of IFT46 positive cells in D. n = 4 independent experiments. Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F8.
We also detected damaged cilia layers in the trachea of mice exposed to Spike-lentivirus-ORF10 (Fig. 7, E and F). Given that the immunofluorescence analysis of SARS-CoV-2 infection in hACE2 mice displayed damaged cilia layers (Fig. 8, B and C) and reduced IFT46 (Fig. 8, D and E), these results show that ORF10 can impair bronchiole cilia and suggest that such damage may contribute to the pathobiology of SARS-CoV-2 infection by facilitating the spread of SARS-CoV-2 deeper into the lung parenchyma.
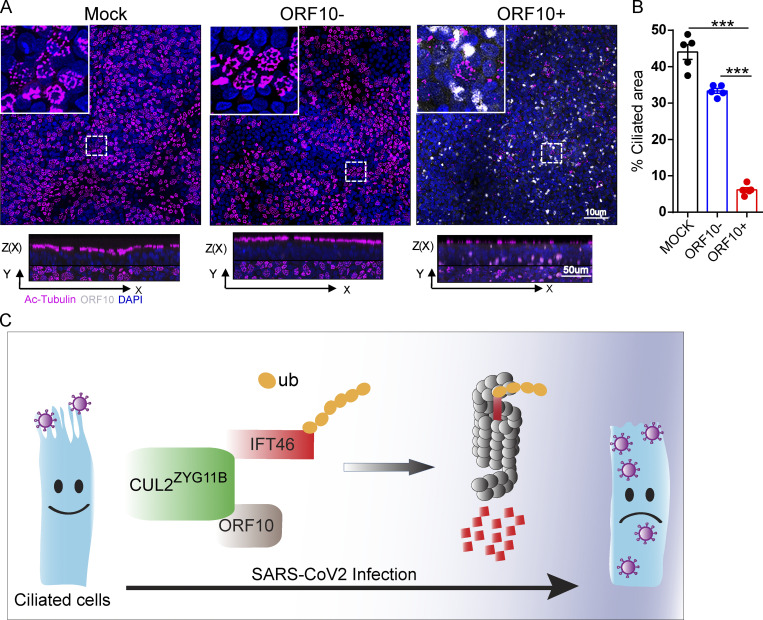
Cilium shrinking has been detected in SARS-CoV-2 infected human airway epithelial (HAE) cells (Zhu et al., 2020a). To confirm the potential for pathogenic effect(s) of ORF10 in humans, we infected primary human nasal epithelial cells (HNECs) with Spike-lentivirus-ORF10. The multiciliated HNECs exposed to Spike-lentivirus-ORF10 displayed a significant reduction in the proportion of ciliated cells compared to the Spike-lentivirus and the mock controls (Fig. 9, A and B), confirming an impact of ORF10 on cilia biogenesis and maintenance in human. Further, these findings suggest that the 13 additional amino acids of SARS-CoV-2 ORF10 can potentially explain the cilia dysfunction clinical features of COVID-19, and perhaps even account for the exacerbated respiratory infectivity of SARS-CoV-2.
Figure 9.
Spike-lentivirus-ORF10 perturbs cilia biogenesis in multiciliated primary human nasal epithelial cells (HNECs). (A) Spike-lentivirus-ORF10 perturbs cilia biogenesis in multiciliated primary human nasal epithelial cells (HNECs). Confocal imaging of mock, Spike-lentivirus, and Spike-lentivirus-ORF10 infected epithelium for 2 dpi (days post infection). (B) Quantification ratio of ciliated area in A (n = 5 independent experiments). Data are presented as the mean ± SEM. *** indicates P < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (C) The model of the effect of SARS-CoV-2 infection on the ciliated cell. Source data are available for this figure: SourceData F9.
Discussion
Cilia are microtubule-based cellular organelles that support a wide variety of essential functions in diverse cellular processes (Reiter and Leroux, 2017; Spassky and Meunier, 2017). Cilia dysfunction is associated with human diseases including anosmia, hearing loss, bronchiectasis, and asthma, among others (Reiter and Leroux, 2017). Cilium shrinking has been detected in SARS-CoV-2 infected HAE cells (Zhu et al., 2020a); there are also reports that mucus accumulation and smell and taste impairments—which could be caused by cilia dysfunction—are common clinical and pathological features of COVID-19 (Bagheri et al., 2020; He et al., 2020; Li et al., 2020c; Makaronidis et al., 2020; Yan et al., 2020a). A recent study of SARS-CoV-2 infection within a reconstituted human bronchial epithelium model showed a rapid loss of the ciliary layer, with axoneme loss and misorientation of the remaining basal bodies. Further analysis of SARS-CoV-2 infection in Syrian hamsters revealed the loss of motile cilia in vivo (Robinot et al., 2021).
We also found that the SARS-CoV-2 infection in hACE2 mice displayed damaged cilia layers in bronchioles (Fig. 8, A and B). A genome-wide association study (GWAS) of 1,778 COVID-19 cases showed that variants in two genes required for ciliogenesis (DNAH7 and CLUAP1) are genetic risk loci for increased COVID-19 mortality (Hu et al., 2021). Further, the levels of master regulators of cilia biogenesis and maintenance (e.g., FOXJ1, RFX3, NEK10, and DNAI2) are aberrantly downregulated in ciliated cells upon SARS-CoV-2 infection (He et al., 2020; Robinot et al., 2021). In the present study, we demonstrate that SARS-CoV-2 ORF10 interacts with ZYG11B, which stimulates the ubiquitination activity of CUL2ZYG11B (Fig. 1). Enhanced CUL2ZYG11B activity causes increased ubiquitination and subsequent proteasome-mediated degradation of multiple cilium-related proteins (including IFT46), thereby impairing the cilia biogenesis and maintenance in cells (Figs. 2 and 3).
Recently, Yan et al. (2021) reported that three residues (W522, D526, and N567) of substrates are required for specific recognition of the Gly/N-degron by CRL2ZYG11B (Yan et al., 2021). We found that the mutation of key recognition elements in ZYG11B did not affect the interaction of ZYG11B and IFT46 (Fig. S4 J). CUL2ZYG11B’s interaction with the C2 domain of IFT46 promotes IFT46 ubiquitination (Fig. 5 B). Therefore, the interaction of ZYG11B with IFT46 was independent on its recognition of the Gly/N-degron. In addition, the exposure of the respiratory tract of hACE2-humanized mice to Spike-lentivirus-ORF10 results in multiple pathogenic phenotypes, which are similar to the phenomena of SARS-CoV-2 infection in hACE2 mice (Figs. 7 and 8). Our study therefore demonstrates that SARS-CoV-2 ORF10 hijacks CUL2ZYG11B to cause cilia dysfunction through the elimination of IFT46 and uncovers a pathological mechanism of SARS-CoV-2 that connects cilia dysfunction to reported COVID-19 symptoms (Fig. 9 C).
A recent study of HEK293T cells reported that ORF10 expression does not affect the degradation of known substrate proteins of the CUL2ZYG11B or CUL2ZER1 complexes, and that study dismissed the idea of a pathogenic impact from ORF10 study (Mena et al., 2021). We found the ORF10 level can account for the differences between our study and Mena et al. (2021) results (Fig. S2, I–K), and the protein level of ORF10 does affect the stability of some cilium-related proteins (Fig. S2, L and M). The Global Protein Stability (GPS) system also showed that the high level of ORF10 indeed enhanced the CUL2ZYG11B activity (Fig. S2 N). In addition, we confirmed that the 38-residue SARS-CoV-2 ORF10 can bind ZYG11B, and our study advances further to show that SARS-CoV-2 ORF10 can stimulate the E3 ligase activity of CUL2ZYG11B. The binding of ORF10 and ZYG11b presents a plausible explanation for the proteasomal degradation hijacking we observed to cause severe defects in cilia biogenesis and maintenance.
The impact of ORF10 in promoting the ubiquitination activity of the CUL2ZYG11B complex deepens our understanding of the molecular mechanisms underlying the interaction between host cells and SARS-CoV-2, and additionally presents a candidate target for developing potential therapies to treat COVID-19. Since the knockdown of Zyg11b and the overexpression of IFT46 could partially rescue the ciliogenesis defects in ORF10 expressed cells (Fig. 4 K and Fig. S3, I–K), so it is worth noting that there may be other substrates or pathways of ORF10 participating in ciliogenesis. Inhibiting CUL2ZYG11B by some drugs and/or supplementing IFT46 (or other cilia related proteins) might be useful in fighting this coronavirus. Given that the ORF10-ZYG11B interaction is essential for the cilia-dysfunction-related symptoms of COVID-19, some specific targeting agents that disrupt the interaction between ORF10 and ZYG11B might be considered as potential treatments.
Materials and methods
Antibodies and proteins
Mouse antibody to FLAG (1:2,000, M20008L), mouse antibody to GST (1:2,000, M20007L), mouse antibody to HA (1:1,000, M20003L), mouse antibody to GFP (1:1,000, M20004L), and mouse antibody to MYC (1:1,000, M20002M) antibodies were purchased from Abmart. Rabbit antibodies to GAPDH (1:10,000, A19056), rabbit antibodies to Tubulin (1:10,000, A6830), and mouse antibody to HA (1:200, AE008) were purchased from Abclonal. Mouse anti-6 × HIS (1:1,000, AB18184) were purchased from Abcam. Mouse antibody to Ubiquitin (1:100, sc-8017) was purchased from Santa Cruz Biotechnology. Rabbit antibody to HA (1:200, 51064-2-AP), rabbit antibodies to SEPTIN2 (1:1,000, 11397-1-AP), rabbit antibodies to TALPID3 (1:1,000, 24421-1-AP), rabbit antibodies to BBS4 (1:1,000, 12766-1-AP), and rabbit antibodies to IFT88 (1:200, 13967-1-AP) were purchased from Proteintech Group. Rabbit antibody to IFT46 (1:100, CSB-PA873621LA01HU) was purchased from Cusabio. Rabbit antibodies to TTBK2 (1:1,000, bs-11771R) and rabbit antibodies to Cytokeratin5 (1:200, bs-1208) were purchased from Bioss. Mouse antibody to FOXJ1 (1:200, 14-9965-82) was purchased from Thermo Fisher Scientific. Mouse antibody to Ac-Tubulin (1:400, T7451) was purchased from Sigma-Aldrich. Rabbit antibody to Coronavirus Nucleocapsid (1:500, 40143-R001-50) was purchased from Sino Biological. The E1 enzyme and Ubiquitin were purchased from Boston Biochem. The VHL-ELOB/C-CUL2-RBX1 protein was purchased from Abcam (ab271787).
Protein purification
ORF10, ZYG11B, ELOC, CUL2, RBX1, and VHL were expressed as N-terminal hexahistidine-tagged or Glutathione S-transferase (GST)-tagged fusion proteins in pET21a, pET28a, or pGEX-4t-1 vectors. Briefly, the plasmid was introduced into Rosetta (DE3) cells and grown in Terrific Broth at 37°C to an optical density of 0.8. The temperature of the culture was then shifted to 16°C and cells were induced with 0.25 mM isopropyl-D-thiogalactoside (IPTG) for 16 h. Cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris, pH 7.4/300 mM NaCl/10 mM imidazole/10% glycerol for hexahistidine-tagged fusion protein; 50 mM Tris, pH 7.4/300 mM NaCl/2 mM MgCl2/5% glycerol) with 1 mM PMSF. After lysis by sonication and high-speed centrifugation of lysates, the supernatant was incubated with Ni Sepharose 6 Fast Flow (GE Healthcare) or Glutathione Sepharose 4B (GE Healthcare) for 2 h at 4°C. The resin was washed and the protein was eluted using the lysis buffer supplemented with 250 mM imidazole or 10 mM glutathione. Purified proteins were dialyzed in the aforementioned protein purification buffer (50 mM Tris, pH 7.4/300 mM NaCl/2 mM MgCl2/5% glycerol).
In vitro ubiquitination assay
The ubiquitination experiments were conducted as follows: E1 (60 nM), E2 (200 nM), and ZYG11B-CUL2 immunocomplex were incubated with Ub (10 nM) at 30°C in a buffer containing 25 mM Tris-HCl (pH 7.4), 2 mM magnesium/ATP, and 0.1 mM DTT. The immunoblotting was performed using a fluorescent dye-labeled secondary antibody (Invitrogen), and the blots were scanned using an Odyssey infrared imager.
GST and FLAG pull-down experiments
GST-ZYG11B were incubated with Glutathione Sepharose 4B (GE Healthcare) at 4°C for 2 h. The GST protein was used as a negative control. Beads were centrifuged and washed with an IP buffer (50 mM Tris, pH 7.4, 50 mM NaCl, 2 mM magnesium chloride, 0.1% Triton). Then, ORF10-FLAG was added and the beads were incubated at 4°C for 2 h. The beads were centrifuged and washed three times with IP buffer and SDS loading buffer was added. For FLAG pull-down, ORF10-FLAG was incubated in anti-FLAG M2 Affinity Gel (A2220; Sigma-Aldrich). The empty beads were used as a negative control. Beads were centrifuged and washed with IP buffer, GST-ZYG11B was added, and the beads were incubated at 4°C for 2 h. The beads were washed three times with IP buffer and SDS loading buffer was added. The samples were analyzed by immunoblotting with anti-GST and anti-FLAG antibodies. The binding kinetic and dissociation constant of ZYG11B-ORF10 were performed using an Octet Red 96 (Forté Bio) in assay buffer (PBS, 0.002% [vol/vol] Tween 20). ZYG11B was immobilized to a level of 0.5 nM on streptavidin-coated biosensors (Forté Bio). To verify that no nonspecific binding was present during the assay, non-functionalized biosensors were used as a control by measuring in parallel all ligand concentrations as well as running buffer. For ORF10, a twofold dilution series from 6.25 to 0.39 nM was employed. Data analysis was performed using Data Analysis software 9.0.0.14 (Forté Bio) using the 2:1 heterogenous ligand-binding model. The binding curves were exported to excel for plotting of curves.
Cell culture, transfection, and knockdown
HEK293T, MRC-5, and NIH-3T3 cells (FuHeng Cell Center) were cultured in DMEM (c11995500bt; Gibco) with 10% FBS and 1% penicillin/streptomycin (15140122; Gibco) under 5% CO2 and detached by incubating in trypsin (25200072; Gibco). Expression plasmids were transfected using Lipofectamine 2000 reagent (11668019; Thermo Fisher Scientific) according to the manufacturer’s instructions. For knockdown experiments, siRNA duplexes (150 pmol) siRNA with 6 μl lipofectamine 2000 reagent targeting Zyg11b were used in 6-well plates along with nontargeting control siRNA duplexes, to transfect NIH-3T3 cells for 24 h. The sequences of siRNA duplexes were as follows: siZyg11b#1 sense, 5′-GCUUGUCAUGCAGUGGCUUTT-3′ (Zyg11b-Mus-1520; GenePharma) and siZyg11b#1 antisense, 5′-AAGCCACUGCAUGACAAGCTT-3′ (Zyg11b-Mus-1520; GenePharma). Nontargeting siRNA duplex (GenePharma, Negative control) served as the negative control. MRC-5 and NIH-3T3 cells were serum-starved for 24 h to induce cilium formation (Li et al., 2020b; Nandadasa et al., 2019). After cilium formation upon serum starvation for 24 h, MRC-5 and NIH-3T3 cells were harvested 2 d after transfection of plasmid DNA or siRNA duplexes for fluorescence microscopy and preparation of lysates.
Lentivirus production and transduction of NIH-3T3 cells
The lentiviral vectors containing cytomegalovirus (CMV) were used in this study to drive the expression of ORF10. HEK293T cells were transfected with the target vector and the other two plasmids (pCAGGS-Spike, psPAX) and were incubated in a fresh medium after 6 h of transfection. Supernatants containing lentivirus were collected and stored at −80°C after 24, 48, and 72 h of transfection. Lentivirus titers were identified by infecting HEK293T cells with viral supernatant for 24 h. The NIH-3T3 cells were transfected with pDC316-human ACE2 plasmids using Lipofectamine 2000 reagent according to the manufacturer’s instructions for 48 h. Cells were transduced with 107 MOI (multiplicity of infection) Lentiviral Vector Pseudotypes to induce the expression of ORF10. Cells were serum-starved for 24 h to induce cilium formation. After cilium formation upon serum starvation for 24 h, MRC-5 and NIH-3T3 cells were harvested 2 d after transfection of Lentiviral Vector Pseudotypes for fluorescence microscopy.
Subjects
Five subjects undergoing paranasal sinus surgery for noninflammatory diseases (i.e., cerebrospinal fluid leak, septal deviation, and bullous middle turbinate) were recruited in this study. Nasal mucosa from the middle turbinate or the uncinate process was obtained during surgery. This study was approved by the Ethics Committee of Beijing Tongren Hospital (TRECKY2019-050), and all subjects signed informed consent before recruitment in the study.
Cell culture of primary human nasal epithelial cells (HNECs)
Freshly obtained nasal mucosa was dissociated in the 0.1% protease type XIV solution (p5147; Sigma-Aldrich) in DMEM at 4°C. The epithelial cells were released from their tissue matrix by vigorous shaking after overnight incubation. Then the epithelial cells were plated on a plastic culture dish to remove fibroblasts at 37°C in a 5% CO2 atmosphere for 1 h. The purified epithelial cells were seeded on transwell (Corning Costar 3470; Corning Inc.) with human placental collagen (C7521; Sigma-Aldrich). Cells were cultured at 37°C in a 5% CO2 atmosphere until establishing air–liquid interface (ALI) culture by removing the apical culture medium. The DMEM: BEGM (1:1) culture medium containing 50 nM all-trans retinoic acid (R2625; Sigma-Aldrich) was replaced every 2 d. The differentiated HNECs were treated with lentivirus particles and harvested after 2 d for fluorescence microscopy.
Generation of human ACE2 knock-in mouse model
The human ACE2 gene knock-in mouse model was prepared by using the CRISPR/Cas9 method by the Institute of Zoology, Chinese Academy of Sciences (IOZ, CAS). The donor vector containing the homology arm sequence and the human ACE2 cDNA (with BGH PolyA sequence), the specific sgRNA (5′-GAAAGATGTCCAGCTCCTCCTGG-3′), and Cas9 mRNA were mixed and microinjected into the pronuclei of C57BL/6 zygotes, then the microinjected zygotes were transferred into oviducts of pseudopregnant foster mother mice. The human ACE2 cDNA was inserted into the first coding sequence of mouse ACE2 (mACE2), so its expression was under the control of the mouse ACE2 promoter. The genotyping primers were as follows: forward: 5′-TAAGCTCGCGAGAAGCAAGGAGATCACAGATTTCTGT-3′ and reverse: 5′-TGCACTAAGCCAAACCCCAACATCATAATAATCTCAGAGATGG-3′. The hACE2 humanized mice exposed to the lentivirus particles pseudotyped with SARS-CoV-2 spike glycoprotein (Spike-lentivirus-ORF10) and the mock group were maintained in an animal biosafety level 2 (ABSL2) laboratory. All animals were allowed free access to water and diet and provided with a 12-h light/dark cycle. For intranasal infection, hACE2 mice were anesthetized with sodium pentobarbital at a dose of 50 mg/kg through the intraperitoneal route, and then intranasally infected with 107 PFU of lentivirus particles pseudotyped with SARS-CoV-2 spike glycoprotein (Genewiz) or PBS, respectively. Mice were sacrificed on day 3 post-infection for tissue processing. All of the animal experiments were performed according to approved institutional animal care and use committee (IACUC) protocols (#2021-002) of the Institute of Zoology, Chinese Academy of Sciences.
Mouse challenge experiments
For intranasal infection, hACE2 mice were anesthetized with sodium pentobarbital at a dose of 50 mg/kg though the intraperitoneal route and then intranasally infected with 105 pfu of SARS-CoV-2. Mice were sacrificed on day 4 post-infection for tissue processing. Animals were housed in an isolator in BioSafety Level 3 animal facilities at the Institute of Microbiology, Chinese Academy of Sciences. The organs and fluids were recovered from the animals infected with live SARS-CoV-2 following the approved standard operating procedures of these facilities.
Tissue collection and histological analysis
For histological examination, the lungs from at least three mice for each group were dissected and fixed with 4% paraformaldehyde (P1110; Solarbio) for up to 24 h. Next, the lungs were dehydrated using graded ethanol and embedded in paraffin. Then, the lungs were cut into 5-μm sections using microtome-cryostat (CM1950; Leica) and mounted on glass slides. After deparaffinization, the slides were stained with hematoxylin and eosin (H&E) for histological analysis. The slides were scanned by Leica Aperio VESA8 equipped with a 40 × objective lens (HC Plan-Apochromat; Leica) and acquired a camera (Grasshopper 3 color Bayer Camera, Point Grey) at RT. Images were analyzed by using Aperio ImageScope software (v12.3.2.7001) and aligned by Adobe Illustrator (CS4).
Immunostaining and fluorescence microscopy
Samples were fixed with 4% paraformaldehyde for 5 min and washed in PBS three times (pH 7.4) at room temperature (RT). Then, the cells were treated with 0.5% Triton X-100 for 10 min and washed in PBS three times. After blocking with 5% bovine serum albumin for 30 min, the slides were incubated with a primary antibody in 1% BSA containing 0.3% Triton X-100 at 4°C overnight. The next day, samples were washed three times with PBS and incubated with FITC- or TRITC-conjugated secondary antibody at a dilution of 1:200 for 1 h at 37°C. Next, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Goat anti-rabbit FITC, goat anti-mouse FITC, goat anti-rabbit TRITC, and goat anti-mouse TRITC-conjugated secondary antibodies, which were purchased from Zhong Shan Jin Qiao. Images were collected immediately using a confocal microscope (LSM 880 Meta plus Zeiss Axiovert Zoom; Zeiss) equipped with a 63 × (NA = 1.40) oil-immersion objective lens (Plan-Apochromat; Zeiss) and acquired by highly sensitive PMT at RT. Images were analyzed by using ZEN software (2011_Lite_×64) and aligned by Adobe Illustrator (CS4).
Sample preparation for mass spectrometry
Cell pellets were resuspended in protein extraction buffer (8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.2, 1% [vol/vol] EDTA-free protease inhibitor, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF) and homogenized by sonication. The supernatants were collected by centrifugation at 40,000 g for 60 min and then the protein concentration was determined by the Bradford method. The prepared proteins were reduced with 5 mM DTT for 25 min at 56°C, alkylated with 14 mM iodoacetamide for 30 min, and then quenched with 5 mM DTT for 15 min. Then the protein mixtures were diluted in 25 mM Tris-HCl, pH 8.2. After digestion with trypsin, the peptide mixtures were desalted in an OASIS HLB 1cc Vac cartridge (Waters) and the eluates were dried using a SpeedVac concentrator (Labconco).
A total of 40 μg peptides of each sample dissolved in 200 mM triethylammonium bicarbonate (TEAB) were subjected to 6-plex tandem mass tags (TMT) labeling, as previously described (Fan et al., 2020; Li et al., 2020a). The TMT-labeled peptides were then mixed and fractionated with a BEH C18 Column (300 μm × 150 mm, 1.7 μm; Waters) with the ACQUITY UPLC M-class system (Waters). A total of 20 fractions were generated and then lyophilized using a SpeedVac concentrator.
Mass spectrometry analysis
Mass spectrometry data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to a Proxeon Easy-nLC 1,200 system. Peptides were separated on an analytical column (75 μm × 160 mm, 1.9 μm, Dr. Maisch) using a 95 min gradient (3–5% buffer B for 5 s, 5–15% buffer B for 40 min, 15–28% buffer B for 34 min and 50 s, 28–38% buffer B for 12 min, 38–100% buffer B for 5 s, 100% buffer B for 8 min) at 300 nl/min. MS1 data were obtained in the Orbitrap using 60k resolution, and MS/MS was performed by high energy dissociation (HCD) in the Orbitrap with a resolution of 15k.
Processing of mass spectrometry data
Raw files were searched using the MaxQuant software (version: 1.6.5.0) against the human UniProt sequence database (release 2020_01). Searches were performed with a 4.5 ppm precursor mass tolerance and 20 ppm product ion mass tolerance. Trypsin/P enzyme specificity was selected by allowing for up to two missed cleavages simultaneously. TMT tags on lysine residues and peptide N termini (+229.1629 Da) and carbamidomethylation of cysteine residues (+57.0215 Da) were set as static modifications. Oxidation of methionines and acetylation of protein N termini were searched as variable modifications. False discovery rates (FDRs) of the identified peptides and proteins were estimated by searching against the database with the amino acid sequence reversed. Only peptides containing at least seven amino acids and 1% FDR were considered to be successfully identified. The relative protein abundance ratios between the two groups were calculated based on TMT reagent reporter ion intensities from HCD spectra by MaxQuant. Student’s t tests were used to calculate significant differences in abundance among groups. The change of a protein with a P value <0.01 was considered significant.
Bioinformatics analysis
Gene ontology and KEGG annotations of differentially expressed proteins were analyzed by clusterProfiler R package (Yu et al., 2012). Heatmaps of differentially expressed proteins were generated using the ComplexHeatmap R package (Gu et al., 2016). Networks were constructed with Cytoscape (v.3.7.2; Shannon et al., 2003), and protein–protein interaction networks were analyzed using the STRING database (Szklarczyk et al., 2017).
Immunoprecipitation
Transfected cells were lysed in TAP lysis buffer (50 mM Hepes-KOH, pH 7.5, 100 mM KCl, 2 mM EDTA, 10% glycerol, 0.1% NP-40 10 mM NaF, 0.25 mM Na3VO4, 50 mM β-glycerolphosphate) plus protease inhibitors (04693132001; Roche) for 30 min on ice, and centrifuged at 13,000 g for 15 min. For immunoprecipitation, cell lysates were incubated with anti-FLAG antibody overnight at 4°C and then incubated with protein A-Sepharose (17-1279-03; GE) for 2 h at 4°C. Thereafter, the precipitants were washed two times with IP buffer (20 mM Tris, pH 7.4, 2 mM EGTA, 1% NP-40), and the immune complexes were eluted with sample buffer containing 1% SDS for 10 min at 55°C and analyzed by immunoblotting.
Chase assays
HEK293T cells were plated 1 d before the experiment and cycloheximide (R750107; CHX; Sigma-Aldrich) was added to the culture at 100 μg/ml to block new protein synthesis. Samples were taken at 0 h, 1 h, 2 h, and 4 h after the addition of CHX for 2 h. Protein levels were determined by immunoblotting. To distinguish lysosomal vs. proteasomal degradation, the cells were treated with 20 μM MG132 (sc-201270; Santa Cruz) for 12 h to inhibit proteasomal degradation or 10 mM 3-MA (M9281; Sigma-Aldrich) for 12 h to inhibit lysosomal degradation.
Statistical analysis
All experiments were repeated at least three times, and all data were expressed as mean ± SEM. SPSS software (IBM SPSS statistics 26) was used for testing the normality of the data. The statistical significance of the differences between the mean values for the different genotypes was measured by two-tailed unpaired Student’s t test. Statistical analysis and figures were made with GraphPad Prism 9. The data were considered significant when the P value was <0.05 (*), 0.01 (**), or 0.001 (***).
Online supplemental material
Fig. S1 shows ORF10 specifically stimulates CUL2ZYG11B activity. Fig. S2 shows overlap of different expression proteins (DEPs) in ORF10 expressed and SARS-CoV-2 infected cells. Fig. S3 shows cilia dysfunction in ORF10 expressing cells is related to ZYG11B. Fig. S4 shows the interaction of ZYG11B and IFT46 is not based on a Gly/N-degron-based mechanism. Fig. S5 shows ORF10 perturbs the cilia localization of IFT46 during cilia formation and maintenance. Table S1 is an identification and quantification of 8820 proteins in HEK293T cells. Table S2 lists gene ontology terms and KEGG pathways significant enriched in DEPs between ORF10-overexpressed and Mock groups. Data S1 contains Fig. 2 E relative protein level (/GAPDH); Fig. 3, B and D percentage ciliated cells; Fig. 4 A relative protein level (/GAPDH); Fig. 4, D and I IFT46 remaining (%); Fig. 4 L percentage ciliated cells; Fig. 5, F and H remaining (%); Fig. 5 J and Fig. 6, C and E percentage ciliated cells; Fig. 7 F and Fig. 8 C percentage ciliated epithelial cells; Fig. 8 E % of IFT46+ cells; Fig. 9 B quantification ratio of ciliated area; Fig. S2, K and M relative protein level (/GAPDH); Fig. S2 N EGFP/DsRed; Fig. S3, B, D, H, and K percentage ciliated cells; Fig. S3 I relative expression of Zyg11b; Fig. S4 B TALPID3 remaining (%); Fig. S4 D BBS4 remaining (%); Fig. S3 F SEPTIN2 remaining (%).
Supplementary Material
is a identification and quantification of 8820 proteins in HEK293T cells.
lists gene ontology terms and KEGG pathways significant enriched in DEPs between ORF10-overexpressed and Mock groups.
contains all statistical data.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
We thank Shiwen Li, Xili Zhu, Hua Qin, and Yue Wang for their help with confocal laser scanning microscopy.
This work was supported by the National Science Fund for Distinguished Young Scholars (81925015), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16020700) and the Youth Innovation Promotion Association CAS (2018109).
The authors declare no competing financial interests.
Author contributions: L. Wang, C. Liu, B. Yang, H. Zhang and J. Jiao: conceptualization, investigation, data curation, validation, project administration, methodology, formal analysis, and writing-original draft. R. Zhang, S. Xiao and Y. Chen: investigation and methodology. S. Liu and Y. Guo: investigation, methodology and formal analysis. B. Liu and Y. Ma: methodology. X. Duan: resources. M. Guo and B. Wu: investigation. X. Wang and Y. Gui: resources. X. Huang and H. Yang: supervision. M. Fang: resources and supervision. L. Zhang and S. Duo: conceptualization, resources and supervision. X. Guo: conceptualization, data curation, methodology, resources and supervision. W. Li: conceptualization, resources, supervision, funding acquisition, writing—original draft, review, and editing.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Uytingco et al., 2019) with the dataset identifier PXD031842. The authors declare that all other data and materials supporting the findings of this study are available within the article and should be addressed to the corresponding author.
References
- Alves-Cruzeiro, J.M., Nogales-Cadenas R., and Pascual-Montano A.D.. 2014. CentrosomeDB: A new generation of the centrosomal proteins database for human and Drosophila melanogaster. Nucleic Acids Res. 42:D430–D436. 10.1093/nar/gkt1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri, S.H., Asghari A., Farhadi M., Shamshiri A.R., Kabir A., Kamrava S.K., Jalessi M., Mohebbi A., Alizadeh R., Honarmand A.A., et al. 2020. Coincidence of COVID-19 epidemic and olfactory dysfunction outbreak in Iran. Med. J. Islam. Repub. Iran. 34:62. 10.34171/mjiri.34.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova, D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., and Munch C.. 2020. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 583:469–472. 10.1038/s41586-020-2332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani, R., Forni D., Clerici M., and Sironi M.. 2020. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 83:104353. 10.1016/j.meegid.2020.104353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldeeb, M., Esmaili M., and Fahlman R.. 2019. Degradation of proteins with N-terminal glycine. Nat. Struct. Mol. Biol. 26:761–763. 10.1038/s41594-019-0291-1 [DOI] [PubMed] [Google Scholar]
- Fan, Y., Cheng Y., Li Y., Chen B., Wang Z., Wei T., Zhang H., Guo Y., Wang Q., Wei Y., et al. 2020. Phosphoproteomic analysis of neonatal regenerative myocardium revealed important roles of checkpoint Kinase 1 via activating mammalian target of rapamycin C1/ribosomal protein S6 Kinase b-1 pathway. Circulation. 141:1554–1569. 10.1161/CIRCULATIONAHA.119.040747 [DOI] [PubMed] [Google Scholar]
- Ghossoub, R., Hu Q., Failler M., Rouyez M.C., Spitzbarth B., Mostowy S., Wolfrum U., Saunier S., Cossart P., Jamesnelson W., and Benmerah A.. 2013. Septins 2, 7 and 9 and MAP4 colocalize along the axoneme in the primary cilium and control ciliary length. J. Cell Sci. 126:2583–2594. 10.1242/jcs.111377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 583:459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H., and Jan Fada B.. 2020. Specificity in ubiquitination triggered by virus infection. Int. J. Mol. Sci. 21:E4088. 10.3390/ijms21114088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Eils R., and Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32:2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Gupta, G.D., Coyaud E., Goncalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W.T., et al. 2015. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 163:1484–1499. 10.1016/j.cell.2015.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J., Cai S., Feng H., Cai B., Lin L., Mai Y., Fan Y., Zhu A., Huang H., Shi J., et al. 2020. Single-cell analysis reveals bronchoalveolar epithelial dysfunction in COVID-19 patients. Protein Cell. 11:680–687. 10.1007/s13238-020-00752-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover A.. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Li C., Wang S., Li T., and Zhang H.. 2021. Genetic variants are identified to increase risk of COVID-19 related mortality from UK Biobank data. Hum. Genom. 15:10. 10.1186/s40246-021-00306-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395:497–506. 10.1016/s0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.S., Hwang K.S., Oh H.W., Ji-Ae K., Kim H.T., Cho H.S., Lee J.J., Yeong Ko J., Choi J.H., Jeong Y.M., et al. 2015. IFT46 plays an essential role in cilia development. Dev. Biol. 400:248–257. 10.1016/j.ydbio.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zhu S., Shu W., Guo Y., Guan Y., Zeng J., Wang H., Han L., Zhang J., Liu X., et al. 2020a. Characterization of metabolic patterns in mouse oocytes during meiotic maturation. Mol. Cell. 80:525–540.e9. 10.1016/j.molcel.2020.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Wei Z., Li G., Zhang Q., Niu S., Xu D., Mao N., Chen S., Gao X., Cai W., et al. 2020b. Silica perturbs primary cilia and causes myofibroblast differentiation during silicosis by reduction of the KIF3A-repressor GLI3 complex. Theranostics. 10:1719–1732. 10.7150/thno.37049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Li M., and Ou G.. 2020c. COVID-19, cilia, and smell. FEBS J. 287:3672–3676. 10.1111/febs.15491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., and Tan X.. 2020. Viral manipulations of the cullin-RING ubiquitin ligases. Adv. Exp. Med. Biol. 1217:99–110. 10.1007/978-981-15-1025-0_7 [DOI] [PubMed] [Google Scholar]
- Lo, C.H., Lin I.H., Yang T.T., Huang Y.C., Tanos B.E., Chou P.C., Chang C.W., Tsay Y.G., Liao J.C., and Wang W.J.. 2019. Phosphorylation of CEP83 by TTBK2 is necessary for cilia initiation. J. Cell Biol. 218:3489–3505. 10.1083/jcb.201811142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makaronidis, J., Mok J., Balogun N., Magee C.G., Omar R.Z., Carnemolla A., and Batterham R.L.. 2020. Seroprevalence of SARS-CoV-2 antibodies in people with an acute loss in their sense of smell and/or taste in a community-based population in London, UK: An observational cohort study. PLoS Med. 17:e1003358. 10.1371/journal.pmed.1003358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena, E.L., Donahue C.J., Vaites L.P., Li J., Rona G., O’Leary C., Lignitto L., Miwatani-Minter B., Paulo J.A., Dhabaria A., et al. 2021. ORF10-Cullin-2-ZYG11B complex is not required for SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA. 118:e2023157118. 10.1073/pnas.2023157118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, C.J., Mayer C., Poch O., and Thompson J.D.. 2020. Characterization of accessory genes in coronavirus genomes. Virol. J. 17:131. 10.1186/s12985-020-01402-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandadasa, S., Kraft C.M., Wang L.W., O’Donnell A., Patel R., Gee H.Y., Grobe K., Cox T.C., Hildebrandt F., and Apte S.S.. 2019. Secreted metalloproteases ADAMTS9 and ADAMTS20 have a non-canonical role in ciliary vesicle growth during ciliogenesis. Nat. Commun. 10:953. 10.1038/s41467-019-08520-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekooki-Machida, Y., and Hagiwara H.. 2020. Role of tubulin acetylation in cellular functions and diseases. Med. Mol. Morphol. 53:191–197. 10.1007/s00795-020-00260-8 [DOI] [PubMed] [Google Scholar]
- Park, I., Lee H.K., Kim C., Ismail T., Kim Y.K., Park J.W., Kwon O.S., Kang B.S., Lee D.S., Park T.J., et al. 2016. IFT46 plays crucial roles in craniofacial and cilia development. Biochem. Biophys. Res. Commun. 477:419–425. 10.1016/j.bbrc.2016.06.083 [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533. 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Reiter, J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinot, R., Hubert M., de Melo G.D., Lazarini F., Bruel T., Smith N., Levallois S., Larrous F., Fernandes J., Gellenoncourt S., et al. 2021. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 12:4354. 10.1038/s41467-021-24521-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L., Shi X., and Shen Y.. 2018. Intraflagellar transport 46 (IFT46) is essential for trafficking IFT proteins between cilia and cytoplasm in Paramecium. Sci. Rep. 8:9259. 10.1038/s41598-018-27050-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, M.A., and Leroux M.R.. 2009. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 19:306–316. 10.1016/j.tcb.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Spassky, N., and Meunier A.. 2017. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18:423–436. 10.1038/nrm.2017.21 [DOI] [PubMed] [Google Scholar]
- Szklarczyk, D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. 2017. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45:D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms, R.T., Zhang Z., Rhee D.Y., Harper J.W., Koren I., and Elledge S.J.. 2019. A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science. 365:eaaw4912. 10.1126/science.aaw4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, P.H., Wang M.L., Yang D.M., Liang K.H., Chou S.J., Chiou S.H., Lin T.H., Wang C.T., and Chang T.J.. 2020. Genomic variance of open reading frames (ORFs) and spike protein in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Chin. Med. Assoc. 83:725–732. 10.1097/JCMA.0000000000000387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uytingco, C.R., Williams C.L., Xie C., Shively D.T., Green W.W., Ukhanov K., Zhang L., Nishimura D.Y., Sheffield V.C., and Martens J.R.. 2019. BBS4 is required for intraflagellar transport coordination and basal body number in mammalian olfactory cilia. J. Cell Sci. 132:jcs222331. 10.1242/jcs.222331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam, T.J., Wheway G., Slaats G.G., Group S.S., Huynen M.A., and Giles R.H.. 2013. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2:7. 10.1186/2046-2530-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Horby P.W., Hayden F.G., and Gao G.F.. 2020. A novel coronavirus outbreak of global health concern. Lancet. 395395:470496–473496. 10.1016/s0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. 2020. A new coronavirus associated with human respiratory disease in China. Nature. 579:265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Shi Y., Pan X., Wu S., Hou R., Zhang Y., Zhong T., Tang H., Du W., Wang L., et al. 2021. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 34:108761. 10.1016/j.celrep.2021.108761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C.H., Faraji F., Prajapati D.P., Boone C.E., and DeConde A.S.. 2020a. Association of chemosensory dysfunction and COVID-19 in patients presenting with influenza-like symptoms. Int. Forum Allergy Rhinol. 10:806–813. 10.1002/alr.22579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H., Chen C., Chen H., Hong H., Huang Y., Ling K., Hu J., and Wei Q.. 2020b. TALPID3 and ANKRD26 selectively orchestrate FBF1 localization and cilia gating. Nat. Commun. 11:2196. 10.1038/s41467-020-16042-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., Li Y., Wang G., Zhou Z., Song G., Feng Q., Zhao Y., Mi W., Ma Z., and Dong C.. 2021. Molecular basis for recognition of Gly/N-degrons by CRL2(ZYG11B) and CRL2(ZER1). Mol. Cell. 81:3262–3274.e3. 10.1016/j.molcel.2021.06.010 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Cruz-Cosme R., Zhuang M.W., Liu D., Liu Y., Teng S., Wang P.H., and Tang Q.. 2020. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct. Target. Ther. 5:269. 10.1038/s41392-020-00372-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Wang W., Liu Z., Liang C., Wang W., Ye F., Huang B., Zhao L., Wang H., Zhou W., et al. 2020a. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 11:3910. 10.1038/s41467-020-17796-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang D., Wang W.L., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. 2020b. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382:727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is a identification and quantification of 8820 proteins in HEK293T cells.
lists gene ontology terms and KEGG pathways significant enriched in DEPs between ORF10-overexpressed and Mock groups.
contains all statistical data.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Uytingco et al., 2019) with the dataset identifier PXD031842. The authors declare that all other data and materials supporting the findings of this study are available within the article and should be addressed to the corresponding author.