Abstract
Studies estimating the human health impact of the foodborne disease often include estimates of the number of gastroenteritis hospitalisations. The aims of this study were to examine the degree to which hospital discharge data underreport hospitalisations due to bacterial gastroenteritis and to estimate the frequency of stool sample submission among patients presenting with gastroenteritis. Using linked laboratory and hospital discharge data from a healthcare organisation and its affiliated hospital, we examined the International Classification of Disease (ICD-9-CM) diagnosis codes assigned to hospitalised adults with culture-confirmed Campylobacter, Salmonella, or Escherichia coli O157 infections and determined the frequency of stool sample submission. Among 138 hospitalised patients with culture-confirmed infections, 43% of Campylobacter patients, 56% of Salmonella patients and 35% of E. coli O157 patients had that pathogen-specific code listed on the discharge record. Among patients without their infection listed as a diagnosis, 65% were assigned a nonspecific gastroenteritis code. Submitting a specimen for culture ⩾3 days before discharge was significantly associated with having the pathogen-specific diagnosis listed. Of 6181 patients assigned a nonspecific gastroenteritis code, 69% had submitted a stool sample for bacterial culture. This study can be used to understand differences and adjust for the underreporting and underdiagnosed of Campylobacter, Salmonella and E. coli O157 in hospital discharge and surveillance data, respectively.
Key words: Campylobacter, Escherichia coli (E. coli), gastroenteritis, hospital discharge data, Salmonella, surveillance
Foodborne diseases are a continuing public health problem worldwide [1]. Campylobacter, Salmonella and Escherichia coli O157 are common causes of bacterial foodborne illness [2]. Acute gastroenteritis is the clinical presentation for the vast majority of cases. Many countries track the occurrence of pathogens commonly transmitted by food by requiring physicians or laboratories to report individual cases of illness caused by particular pathogens or outbreaks to public health authorities [3]. Hospitals also list diagnoses on discharge summaries [4, 5]. These data provide important information on the pathogens causing illness and trends in the incidence of disease; however, only a small fraction of illnesses are diagnosed and reported [3]. Therefore, to estimate the overall burden of foodborne disease in the population, investigators must find ways to extrapolate from surveillance and hospital discharge data [5–9].
Studies estimating the overall burden of foodborne diseases typically estimate the numbers of diagnosed and undiagnosed illnesses caused by specific pathogens or syndromes and their related hospitalisations and deaths [6, 7, 10]. Estimating the number of episodes resulting in hospitalisation, the focus of this paper, is important for assessing the full economic and human health impact from foodborne diseases; however, accurately estimating hospitalisations is challenging. Hospital discharge data are one source of relevant information, but they may underestimate diagnosed illnesses. For an illness caused by a pathogen to be recorded on the discharge record, a physician must consider the diagnosis and order the appropriate diagnostic tests. The pathogen must be detected before discharge and coding must be accurate. Without pre-discharge identification of a pathogen, infections producing signs and symptoms of gastroenteritis may be coded as nonspecific or noninfectious conditions. Laboratory-based surveillance conducted by public health departments is another source of information on infection caused by specific reportable pathogens, but hospitalisation status is not routinely captured.
In the USA, the approach to estimating the total number of foodborne illness hospitalisations has been to use hospital discharge survey data to estimate the total number of gastroenteritis hospitalisations and to use data from laboratory-based surveillance to estimate the number of hospitalisations caused by specific pathogens, adjusting for underdiagnosis [7, 10]. The goal of this study was to explore whether linked laboratory and hospital discharge data from a healthcare organisation could better inform foodborne illness estimates by describing underreporting and underdiagnosis in hospital discharge data and determining the frequency with which patients hospitalised with gastroenteritis submit a stool sample for bacterial culture.
Methods
We used laboratory and hospital discharge data from the Marshfield Clinic and St. Joseph's Hospital, its affiliated hospital, to conduct two separate analyses. In the first analysis, we linked laboratory and hospital discharge data and examined the International Classification of Disease (ICD-9-CM) diagnosis codes assigned to hospitalised adults with culture-confirmed Campylobacter, Salmonella, or E. coli O157 infections. We wanted to determine the percentage of patients with culture-confirmed Salmonella, Campylobacter, or E. coli O157 infections who had that specific ICD-9-CM intestinal infectious diseases diagnosis code listed on the hospital discharge record and to describe the use of nonspecific ICD-9-CM gastroenteritis diagnosis codes for these patients. In the second analysis, we estimated the frequency of stool sample submission for culture among hospitalised adults with ICD-9-CM intestinal infectious diseases diagnosis codes and nonspecific gastroenteritis codes assigned at discharge.
Study population
This study was conducted using data from Marshfield, Wisconsin, a community where residents in Marshfield and the surrounding area (population about 60 000) receive nearly all outpatient and in-patient care from the Marshfield Clinic (a network of integrated outpatient clinics) and St. Joseph's Hospital. Marshfield Clinic Laboratories routinely tests stool specimens submitted for bacterial culture for Salmonella, Campylobacter and E. coli O157. Inpatient and outpatient diagnoses and laboratory data are accessible through a combined electronic medical record. Laboratory results from specimens submitted as part of outpatient visits are available to in-hospital physicians and vise-versa; however, laboratory data do not automatically populate the hospital record. The electronic medical record captures 93% of all outpatient visits and 97% of all hospital discharges for residents in this area [11].
Using electronic medical records from 1 January 2004–31 December 2014, we identified hospitalised patients ⩾18 years of age with culture-confirmed Salmonella, Campylobacter, or E. coli O157 infections who had submitted a specimen for bacterial culture during or ⩽7 days before hospital admission. For E. coli O157 infections, we restricted the period to 1 January 2004–31 December 2013 because Marshfield Clinic Laboratories switched to non-culture-based diagnostic tests for Shiga toxin-producing E. coli in 2014, so we were then unable to identify E. coli O157 infections. We defined hospital admission as an overnight admission to St. Joseph's Hospital. If a patient had multiple hospital admissions within 7 days of the culture-confirmed laboratory result, we included the first admission. For patients with multiple positive cultures, we used the first positive test. We categorised specimens by source (stool, blood, etc.). Hospital discharge data and laboratory data were linked using the unique patient identification number.
Hospital discharge diagnosis codes
We reviewed the International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM) diagnosis codes assigned to each patient on the discharge record by the attending physician(s) and used for the purposes of reimbursement. We determined the percentage of patients who had an ICD-9-CM diagnosis code for intestinal infectious diseases specifying Salmonella, Campylobacter, or E. coli O157 infection. These included typhoid or paratyphoid fevers (002.0–002.9) or other Salmonella infections (003.0–003.9); intestinal infection due to Campylobacter (008.43); intestinal infection due to enterohemorrhagic E. coli (008.04), or intestinal infection due to E. coli, unspecified (008.00). We also determined the number of patients whose discharge record listed another intestinal infectious diseases diagnosis code (ICD-9-CM 001-008.6) or a nonspecific gastroenteritis code. Based on previous studies [7–10, 12], we considered nonspecific gastroenteritis codes to be ICD-9-CM diagnosis codes 008.8, intestinal infection due to other organism, not elsewhere classified, 009, ill-defined infectious gastroenteritis, 558.9, other and unspecified noninfectious gastroenteritis and colitis; and 787.9, other symptoms involving digestive system: diarrhoea. We examined the frequency with which these intestinal infectious diseases codes and nonspecific gastroenteritis codes were assigned as the first diagnosis, one of the first three diagnoses and any diagnosis. The first three diagnoses have been used in US studies estimating the burden of foodborne disease as a compromise between limiting the analysis to hospitalisations for which gastroenteritis was listed as the primary cause and including hospitalisations for gastroenteritis may have been a manifestation of another illness [7, 10]. We assumed that the first three codes contained most main reason(s) for hospitalisation.
Stool sample submission
For each patient with a culture-confirmed Campylobacter, Salmonella, or E. coli O157 infection, we documented their status as an outpatient or inpatient at the time of specimen submission and calculated the number of days the specimen was submitted for culture before hospital discharge. We assumed that it takes an average of 2 days (range:1–3 day) for a positive result to be obtained [13]. We examined whether these factors were associated with having the pathogen-specific diagnosis listed.
In a separate analysis, we queried electronic medical records from 1 January 2004–31 December 2014 to determine how frequently hospitalised patients ⩾18 years of age with any nonspecific gastroenteritis code listed on their discharge record submitted a stool sample for bacterial culture during hospitalization or ⩽7 days before their hospital admission date.
We used SAS version 9.3 (SAS Institute, Cary, NC, USA) for these analyses. The chi-square test was used to test statistical significance between categorical variables. This study was approved by the Marshfield Clinic Institutional Review Board.
Results
We identified 138 hospitalised patients with culture-confirmed Campylobacter (n = 88), Salmonella (n = 39), or E. coli O157 (n = 11) infections. Most diagnoses were made by stool culture (97% of Campylobacter patients; 67% Salmonella and 100% E. coli O157) (Table 1). The median length of hospital stay was 3 days for patients with Campylobacter and E. coli O157 infection and 4 days for patients with Salmonella infection. Patient demographic characteristics are shown in Table 1.
Table 1.
Characteristics of hospitalised patients ⩾18 years of age with a culture-confirmed Campylobacter, Salmonella, or E. coli O157 infection, Marshfield Clinic, Wisconsin, 1 January 2004–31 December 2014 (1 January 2004–31 December 2013 for E. coli O157)
| Characteristic | Patients with a culture-confirmed infection | |||||
|---|---|---|---|---|---|---|
| Campylobacter (n = 88) | Salmonella (n = 39) | E. coli O157 (n = 11) | ||||
| N | % | N | % | N | % | |
| Specimen source | ||||||
| Stool | 85 | 97 | 26 | 67 | 11 | 100 |
| Blood | 3 | 3 | 8 | 21 | – | – |
| Urine | – | – | 3 | 8 | – | – |
| Othera | – | – | 2 | 3 | – | – |
| Duration of hospital stay (days) | ||||||
| 1 | 11 | 13 | 2 | 5 | 0 | 0 |
| 2 | 25 | 28 | 6 | 15 | 3 | 27 |
| 3 | 14 | 16 | 5 | 13 | 3 | 27 |
| 4 | 18 | 20 | 8 | 21 | 3 | 27 |
| 5–14 | 15 | 17 | 15 | 38 | 2 | 18 |
| >14 | 5 | 6 | 3 | 8 | 0 | 0 |
| Median (IQR) | 3 (2, 4) | 4 (3, 7) | 3 (2, 4) | |||
| Hospitalisation month | ||||||
| March–May | 14 | 16 | 8 | 21 | 1 | 9 |
| June–August | 34 | 39 | 16 | 41 | 6 | 55 |
| September–November | 29 | 33 | 9 | 23 | 4 | 36 |
| December–February | 11 | 13 | 6 | 15 | 0 | 0 |
| Male | 46 | 52 | 16 | 41 | 5 | 45 |
| Age (years) | ||||||
| 18–29 | 14 | 16 | 5 | 13 | 4 | 36 |
| 30–49 | 26 | 30 | 8 | 21 | 1 | 9 |
| 50–69 | 26 | 30 | 11 | 28 | 1 | 9 |
| ⩾70 | 22 | 25 | 15 | 38 | 5 | 45 |
| Median (IQR) | 52 (35, 70) | 62 (46, 75) | 54 (26, 74) | |||
IQR, inter quartile range.
Other specimen sources for Salmonella were abscess (1) and incision site (1).
Use of pathogen-specific intestinal infectious diseases diagnosis codes
Overall, 46% (64/138) of patients with culture-confirmed Campylobacter (43%; 38/88), Salmonella (56%; 22/39), or E. coli O157 (35%; 4/11) infection had that specific diagnosis as a code on the discharge record. Of the 22 patients with Salmonella listed, 13 were listed as Salmonella gastroenteritis, six as Salmonella septicemia, four as nonspecific Salmonella infection, two as Salmonella with other localised infection and one as typhoid fever (four patients were assigned two separate codes for Salmonella) Of the four patients with E. coli O157 listed, two were coded as intestinal infection due to enterohemorrhagic E. coli and two as intestinal infection due to unspecified E. coli.
Most (91%; 58/64) had the diagnosis listed as one of the first three codes [Campylobacter (95%; 36/38), Salmonella (82%; 18/22) and E. coli O157 (100%; 4/4)]. About two-thirds of patients (64%; 41/64) had the diagnosis listed as the first code [Campylobacter (66%; 25/38), Salmonella (59%; 13/22) and E. coli O157 (75%; 3/4)].
Three Salmonella patients also had Clostridium difficile listed on the discharge record (Table 2, Supplementary Appendix Table A). Among patients without their Campylobacter, Salmonella, or E. coli O157 infection listed as a diagnosis, one Campylobacter patient had a Salmonella infection listed on the discharge record and one had intestinal infection due to E. coli, unspecified listed; two patients had Clostridium difficile listed (one Campylobacter patient and one Salmonella patient) (Table 2, Supplementary Appendix Table B).
Table 2.
Frequency of ICD-9-CM diagnosis codes for intestinal infectious diseasea or nonspecific gastroenteritisb assigned to 138 patients with culture-confirmed Campylobacter, Salmonella and E. coli O157 infection, Marshfield Clinic, Wisconsin, 1 January 2004–31 December 2014 (1 January 2004–31 December 2013 for E. coli O157)
| ICD-9 diagnosis code | Culture-confirmed diagnosis listed | Culture-confirmed diagnosis not listed | Total | |||
|---|---|---|---|---|---|---|
| N = 64 | % | N = 74 | % | N = 138 | % | |
| Intestinal infectious disease | ||||||
| Typhoid fever (002) | 1 | 0 | 0 | 0 | 1 | 1 |
| Salmonella infections (003) | 21 | 33 | 1 | 1 | 22 | 16 |
| Intestinal infection due to E. coli, unspecified (008.00) | 2 | 3 | 1 | 1 | 3 | 2 |
| Intestinal infection due to enterohemorrhagic E. coli (008.04) | 2 | 3 | 0 | 0 | 2 | 1 |
| Intestinal infection due to Campylobacter (008.43) | 38 | 59 | 0 | 0 | 38 | 28 |
| Intestinal Infection due to Clostridium difficile (008.45) | 3 | 5 | 2 | 3 | 5 | 4 |
| Patients assigned ⩾1 diagnosis code for intestinal infectious diseasec | 64 | 100 | 4 | 54 | 68 | 49 |
| Nonspecific gastroenteritis | ||||||
| Intestinal infection due to other organism, not elsewhere classified (008.8) | 0 | 0 | 12 | 16 | 12 | 9 |
| Ill-defined infectious gastroenteritis (009)d | 0 | 0 | 15 | 20 | 15 | 11 |
| Other and unspecified noninfectious gastroenteritis and colitis (558.9) | 3 | 5 | 15 | 20 | 18 | 13 |
| Other symptoms involving digestive system: Diarrhoea (787.91) | 4 | 7 | 8 | 11 | 12 | 9 |
| Patients assigned ⩾1 diagnosis code for nonspecific gastroenteritisc | 7 | 11 | 48 | 65 | 55 | 40 |
| Patients assigned ⩾1 diagnosis code for intestinal infectious disease or nonspecific gastroenteritisc | 64 | 100 | 51 | 69 | 115 | 83 |
Intestinal infectious diseases: ICD-9-CM diagnosis codes 001-008.6.
Nonspecific gastroenteritis code: ICD-9-CM diagnosis codes 008.8, intestinal infection due to other organism, not elsewhere classified, 009, ill-defined infectious gastroenteritis, 558.9, other and unspecified noninfectious gastroenteritis and colitis; and 787.91, other symptoms involving digestive system: diarrhoea.
Patients may have been assigned >1 diagnosis code for intestinal infection disease and/or nonspecific gastroenteritis.
Ill-defined infectious gastroenteritis (ICD-9-CM 009) codes included: Infectious colitis, enteritis and gastroenteritis (009.0) (n = 8); Colitis, enteritis and gastroenteritis of presumed infectious origin (009.1) (n = 2); Infectious diarrhoea (009.2) (n = 3); Diarrhoea of presumed infectious origin (009.3).
Use of nonspecific gastroenteritis diagnosis codes
At least one nonspecific gastroenteritis code was assigned to only 65% (48/74) of patients whose Campylobacter, Salmonella, or E. coli O157 infection was not listed as a diagnosis (Table 2, Supplementary Appendix Table B). These diagnosis codes were: ill-defined infectious gastroenteritis (20%), other and unspecified noninfectious gastroenteritis and colitis (20%), intestinal infection due to other organism, not elsewhere classified (16%) and other symptoms involving digestive system: diarrhoea (11%). Among 17 Salmonella patients without their specific diagnosis listed, 73% (11/15) of those diagnosed by stool culture had a nonspecific gastroenteritis code listed compared with none of the two diagnosed by culture of another specimen. About half (53%; 39/74) of patients had a nonspecific code listed as one of the first three codes, including 29 (39%) for whom it was listed as the first diagnosis (Table 2). Of the 64 patients with Campylobacter, Salmonella, or E. coli O157 listed as a specific diagnosis, 11% also had a nonspecific gastroenteritis code assigned: other symptoms involving digestive system: diarrhoea (7%) and other, unspecified noninfectious gastroenteritis and colitis (5%) (Table 2, Supplementary Appendix Table A).
Use of pathogen-specific and nonspecific gastroenteritis diagnosis codes
Overall, among the 138 patients with Campylobacter, Salmonella, or E. coli O157 infection, 115 (83%) had a diagnosis code for Campylobacter, Salmonella, or E. coli or a nonspecific gastroenteritis code on the discharge record (Table 2); 75% (103) had a diagnosis code for Campylobacter, Salmonella, or E. coli or a nonspecific gastroenteritis code listed as one of the first three diagnosis (Supplementary Appendix Table C).
Stool sample submission
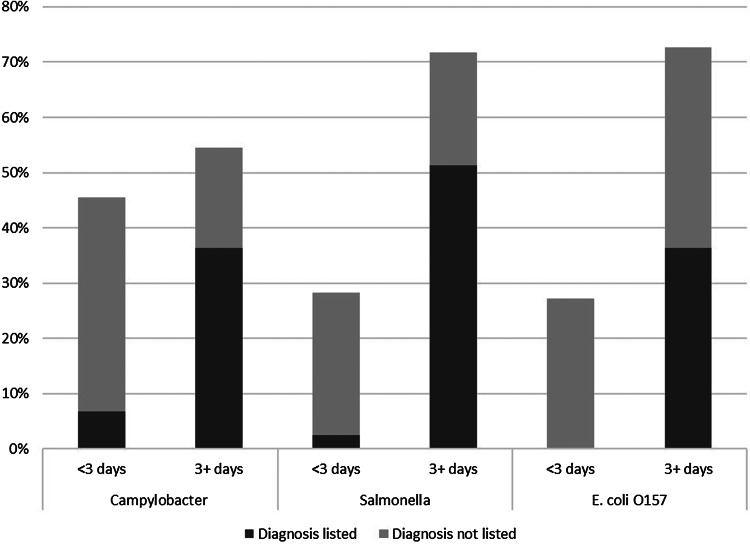
Of the 138 patients with culture-confirmed infection, 8% (11) submitted a specimen for culture before admission, 54% (74) on the day of admission and 29% (40) on the second day of hospitalisation. Only 9% (13) submitted a specimen ⩾3 days after admission. Having a pathogen-specific diagnosis for Campylobacter, Salmonella, or E. coli listed on the discharge record was a statistically significant association between the number of days the specimen was submitted for bacterial culture before discharge from hospital (P < 0.05 for each pathogen) (Fig. 1). Among patients with a specimen submitted for culture ⩾3 before discharge, 67% (56/84) had a pathogen-specific diagnosis code listed compared with 13% (7/54) whose specimen was submitted later. This finding was consistent across pathogens (Fig. 1).
Fig. 1.
Percentage of hospitalised patients ⩾18 years of age with a culture-confirmed Campylobacter, Salmonella, or E. coli O157 infection who submitted a specimen for bacterial culture <3 days and 3+ days before discharge from hospital and the proportion of these patients who had the culture-confirmed diagnosis listed as an ICD-9-CM diagnosis code on the hospital discharge record, Marshfield Clinic, Wisconsin, 1 January 2004–31 December 2014 (1 January 2004–31 December 2013 for E. coli O157).
In a separate analysis, we determined that 74% of 7862 hospitalised adults with a discharge code for infectious intestinal disease and/or nonspecific gastroenteritis submitted a stool sample for bacterial culture (Table 3). The percentage submitting a stool sample increased with age from 65% (294/449) among patients 18–29 years of age to 70% (851/1209) among those 30–49 years and 75% among those 50–69 year (1884/2499) and 70+ years of age (2888/3839) (P < 0.05). Among the patients with a nonspecific diagnosis code for gastroenteritis, 69% submitted a stool sample (Table 3).
Table 3.
Percentage of adults ⩾18 years of age who submitted a stool sample, by selected ICD-9-CM diagnosis codes, Marshfield Clinic, Wisconsin, 1 January 2004–31 December 2014
| ICD-9-CM diagnosis codesa | Stool sample submitted | |
|---|---|---|
| N | % | |
| Intestinal infectious disease | 1554/1681 | 92 |
| Nonspecific gastroenteritis | 4250/6181 | 69 |
| Other symptoms involving digestive system: Diarrhoea (787.91) | 2787/3855 | 72 |
| Other and unspecified noninfectious gastroenteritis and colitis (558.9) | 840/1372 | 61 |
| Intestinal infection due to other organism, not elsewhere classified (008.8) | 604/937 | 64 |
| Ill-defined infectious gastroenteritis (009)b | 132/151 | 87 |
| Total (intestinal infectious disease and nonspecific gastroenteritis) | 5804/7862 | 74 |
Intestinal infectious diseases: ICD-9-CM diagnosis codes 001-008.6; Nonspecific gastroenteritis code: ICD-9-CM diagnosis codes 008.8, intestinal infection due to other organism, not elsewhere classified, 009, ill-defined infectious gastroenteritis, 558.9, other and unspecified noninfectious gastroenteritis and colitis; and 787.91, other symptoms involving digestive system: diarrhoea; patients may have been assigned>1 diagnosis code for inflectional intestinal disease or nonspecific gastroenteritis.
Percentage of patients submitting a stool sample in subcategories of ill-defined infectious gastroenteritis ICD-9-CM 009): Infectious colitis, enteritis, and gastroenteritis (009.0) (89%); Colitis, enteritis & gastroenteritis of presumed infectious origin (009.1) (79%); Infectious diarrhoea (009.2) (87%); Diarrhoea of presumed infectious origin (009.3) (100%).
Discussion
Using linked laboratory and hospital discharge data, this study showed that hospital discharge data underreport enteric illnesses even when the infections are culture-confirmed. These data also give a sense of the proportion of culture-confirmed infections that are not captured with pathogen-specific diagnostic codes and provide some insight on the diagnostic codes used for these patients. In addition, this study suggests that data on the proportion of hospitalised patients with a nonspecific gastroenteritis code who submit a stool sample for bacterial culture can be used to estimate the degree to which illnesses caused by Salmonella, Campylobacter and E. coli O157 are underdiagnosed.
Fewer than half of patients with a culture-confirmed Salmonella, Campylobacter, or E. coli O157 infection had that diagnosis listed on the hospital discharge record. The strong association between having the laboratory-diagnosed infection listed on the discharge record and a longer lag time between specimen submission and hospital discharge could reflect health care providers not learning about the laboratory diagnosis before the patient is discharged. However, even when the specimen was submitted ⩾3 days before discharge, almost one-third of records of patients with culture-confirmed illness were missing the pathogen-specific diagnostic code. Our analysis shows that imprecise coding is one reason.
Our results also imply that studies that use ICD-9-CM diagnosis codes to estimate hospitalisations likely underestimate the number of hospitalisations due to gastroenteritis. More than 15% of patients with a culture-confirmed infection did not have an intestinal infectious disease diagnosis code for Salmonella, Campylobacter, or E. coli or a nonspecific gastroenteritis code on their discharge record. Thus, these patients would not have been captured in a tally of gastroenteritis hospitalisations. This may reflect the inaccurate discharge coding of patients hospitalised for gastroenteritis. However, there are other possible explanations. Patients with a culture-confirmed Salmonella, Campylobacter, or E. coli O157 infection may have been hospitalised for complications of the infection after their gastrointestinal symptoms had resolved and some may not have had gastroenteritis. In our analysis, one-third of Salmonella patients had the pathogen isolated from a site other than stool, suggesting they had sepsis, meningitis, or another extra-intestinal infection; however, the majority of them had a gastroenteritis code listed. It is also possible that patients had a hospital admission unrelated to their culture-confirmed Salmonella, Campylobacter, or E. coli O157 infection.
Most patients had only one diagnosis code for an intestinal infectious disease or non-specific gastroenteritis listed on the discharge record. Non-specific gastroenteritis codes used for patients who did not have their Salmonella, Campylobacter, or E. coli O157 diagnosis listed on the discharge record were most commonly ‘ill-defined infectious gastroenteritis,’ ‘other, unspecified noninfectious gastroenteritis & colitis,’ ‘intestinal Infection due to other organism, not elsewhere classified,’ and ‘Other symptoms involving digestive system: Diarrhoea.’ It is notable that patients with an infectious illness frequently received the nonspecific gastroenteritis code for noninfectious gastroenteritis and colitis noninfectious. The use of this code for infectious illness has been reported elsewhere [14] and should be considered when estimating the number of hospitalisations due to infectious gastroenteritis. A number of patients were also coded as ‘intestinal infection due to other organism, not elsewhere classified.’ Our data do not indicate if infection with another pathogen was confirmed or merely suspected for these patients, but our study determined that over one-third of patients with this code did not submit a stool sample for bacterial culture. This suggests that ‘intestinal infection due to other organism, not elsewhere classified’ may frequently be assigned to patients without a laboratory-diagnosed infection.
Many patients with a diagnosis code for Salmonella, Campylobacter, or E. coli or nonspecific gastroenteritis did not have this listed first. Including the first three codes captured almost 40% more patients with a culture-confirmed infection than the first code alone although some cases were still missed. This may reflect coding practices, billing requirements, or other factors related to the hospital admission. For instance, the infection may have been diagnosed as part of another disease or medical condition. Patient with co-morbidities, including HIV and other immune-compromising conditions may have more severe illness resulting in hospitalisation [15, 16]. Foodborne illness estimates in the USA have extracted records in which gastroenteritis was listed as one of the first three diagnoses in an effort to compromise between limiting the analysis to hospitalisations for which gastroenteritis was listed as the primary cause and including hospitalisations for which signs or symptoms of gastroenteritis may have been a manifestation of another illness [10, 17, 18]. Conversely, there is likely more underdiagnosis that we cannot assess particularly in vulnerable populations where dehydration or electrolyte imbalance from a gastrointestinal illness may result in hospitalisation well after resolution of the gastrointestinal illness.
The percentage of patients with nonspecific gastroenteritis diagnosis codes who submitted a stool sample for bacterial culture was 69%, ranging from 61 to 100% for each of the individual nonspecific gastroenteritis codes. These data are similar to the range reported in cross-sectional and health utilisation surveys in Germany, Italy, the Netherlands, Poland and Sweden (range: 70–93%), but higher than the studies conducted in the UK (29%) [19]. These data can be used to create multipliers and credibility limits to adjust for underdiagnosis when estimating the total number of gastroenteritis hospitalisations due to pathogens transmitted commonly through food. Other factors that may also contribute to underdiagnosis and that should be taken into consideration include the frequency with which laboratories test for specific pathogens and the sensitivity of stool culture. For example, the underreporting multiplier for Salmonella in the 2011 US estimates of the burden of foodborne illness was two [7]. Taking estimates for laboratory testing and laboratory test sensitivity from the 2011 paper for Salmonella (100% and 70%, respectively) and the median estimate for stool sample submission from this analysis (69%), the underreporting multiplier for Salmonella hospitalisation would be 2.1. Ranges can be used to describe uncertainty with these estimates.
There are a number of limitations to this study. First, the study was conducted using data from a single hospital within one integrated healthcare organisation. It is not known how coding practices at St. Joseph's hospital or the laboratory methods at Marshfield Clinic compare with other hospitals nationwide. Second, there are important differences between adults and children in terms of the burden and diagnosis of enteric pathogens [20, 21]. This pilot study was restricted to adults and may not reflect the diagnosis codes or frequency of stool sample submission among children. Third, we looked at only three pathogens—Salmonella, Campylobacter and E. coli O157. This pilot study demonstrated the feasibility of this approach for better understanding coding practices and underreporting in hospital data. Future studies could address the limitations above by expanding to other hospitals, including a broad set of enteric pathogens and including all age groups.
By linking laboratory and hospital discharge data from a healthcare organisation, this study enhanced our understanding coding practices and the degree to which enteric pathogens are underreported in hospital discharge data and underdiagnosed in surveillance data. The findings of this study can be used in several ways to improve estimates of the burden of foodborne and other enteric illnesses. Investigators using hospital discharge data to estimate the burden of Salmonella, Campylobacter and E. coli O157 may use these data to adjust for underreporting. These data also inform the selection of ICD-9 diagnostic codes when estimating the burden of gastroenteritis hospitalizations and in adjusting for underreporting of gastroenteritis episodes. Finally, data on rates of stool sample submission can be used to adjust for underreporting of hospitalised cases reported to public health surveillance.
Acknowledgements
This article was 100% funded with federal funds from a federal program of $131 270. This article was supported by Cooperative Agreement # 5NU60OE000103 funded by the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of CDC or the Department of Health and Human Services.
Supplementary material
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268818000882.
click here to view supplementary material
Conflict of interest
None.
References
- 1.Havelaar AH, et al. (2015) World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Medicine 12, e1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirk MD, et al. (2015) World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Medicine 12, e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tauxe RV, et al. (2010) Evolving public health approaches to the global challenge of foodborne infections. International Journal of Food Microbiology 139(Suppl 1), S16–S28. [DOI] [PubMed] [Google Scholar]
- 4.Cummings PL, et al. (2016) Salmonellosis hospitalizations in the United States: associated chronic conditions, costs, and hospital outcomes, 2011, trends 2000–2011. Foodborne Pathogens and Disease 13, 40–48. [DOI] [PubMed] [Google Scholar]
- 5.Kirk M, et al. (2014) Foodborne illness, Australia, circa 2000 and circa 2010. Emerging Infectious Diseases 20, 1857–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flint JA, et al. (2005) Estimating the burden of acute gastroenteritis, foodborne disease, and pathogens commonly transmitted by food: an international review. Clinical Infectious Diseases: an Official Publication of the Infectious Diseases Society of America 41, 698–704. [DOI] [PubMed] [Google Scholar]
- 7.Scallan E, et al. (2011) Foodborne illness acquired in the United States – major pathogens. Emerging Infectious Diseases 17, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas MK, et al. (2013) Estimates of the burden of foodborne illness in Canada for 30 specified pathogens and unspecified agents, circa 2006. Foodborne Pathogens and Disease 10, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas MK, et al. (2015) Estimates of foodborne illness-related hospitalizations and deaths in Canada for 30 specified pathogens and unspecified agents. Foodborne Pathogens and Disease 12, 820–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scallan E, et al. (2011) Foodborne illness acquired in the United States – unspecified agents. Emerging Infectious Diseases 17, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieke AL, et al. (2015) Validation of health event capture in the marshfield epidemiologic study area. Clinical Medicine & Research 13, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall G, et al. (2005) Estimating foodborne gastroenteritis, Australia. Emerging Infectious Diseases 11, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Council to Improve Foodborne Outbreak Response C. Guidelines for Foodborne Disease Outbreak Response, 2nd edn., Atlanta: Council of State and Territorial. [Google Scholar]
- 14.Pinner RW, et al. (1996) Trends in infectious diseases mortality in the United States. JAMA 275, 189–193. [PubMed] [Google Scholar]
- 15.Helms M, et al. (2003) Short and long term mortality associated with foodborne bacterial gastrointestinal infections: registry based study. BMJ (Clinical research ed.) 326, 357. [PMC free article] [PubMed] [Google Scholar]
- 16.Helms M, Simonsen J and Mølbak K (2006) Foodborne bacterial infection and hospitalization: a registry-based study. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 42, 498–506. [DOI] [PubMed] [Google Scholar]
- 17.Mounts AW, et al. (1999) Trends in hospitalizations associated with gastroenteritis among adults in the United States, 1979–1995. Epidemiology and Infection 123, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangarosa RE, et al. (1992) Hospitalizations involving gastroenteritis in the United States, 1985: the special burden of the disease among the elderly. American Journal of Epidemiology 135, 281–290. [DOI] [PubMed] [Google Scholar]
- 19.Haagsma JA, et al. (2013) Community incidence of pathogen-specific gastroenteritis: reconstructing the surveillance pyramid for seven pathogens in seven European Union member states. Epidemiology and Infection 141, 1625–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scallan E, et al. (2013) Estimates of illnesses, hospitalizations and deaths caused by major bacterial enteric pathogens in young children in the United States. The Pediatric Infectious Disease Journal 32, 217–221. [DOI] [PubMed] [Google Scholar]
- 21.Jones TF, et al. (2007) A population-based estimate of the substantial burden of diarrhoeal disease in the United States; FoodNet, 1996–2003. Epidemiology and Infection 135, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268818000882.
click here to view supplementary material