Abstract
Background: Tongue squamous cell carcinoma (TSCC) is one of the most common oral cancers. Immune activity is significantly related to the initiation and progression of TSCC. Systemic analysis of the immunogenomic landscape and identification of crucial immune-related genes (IRGs) would help understanding of TSCC. Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) provide multiple TSCC cases for use in an integrated immunogenomic study. Methods: Immune landscape of TSCC was depicted by expression microarray data from GSE13601 and GSE34105. Univariate Cox analysis, in combination with survival analysis, was applied to select candidate IRGs with significant survival value. Survival predicting models were constructed by multivariate Cox regression and logistic regression analysis. Unsupervised clustering analysis was used to construct an immune gene panel based on prognostic IRGs to distinguish TSCC subgroups with different prognostic outcomes. Finally, IHC staining was performed to validate the clinical value of this immune-gene panel. Results: Differentially expressed IRGs were identified in two TSCC microarray datasets. Functional enrichment analysis revealed that ontology terms associated with variations in T cell function, were highly enriched. Infiltration status of activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells, had great prognostic value for TSCC progression. Unsupervised clustering analysis was further performed to classify TSCC patients into three subgroups. CTSG, CXCL13, and VEGFA were finally combined together to form an immune-gene panel, todistinguish different TSCC subgroups. IHC staining of TSCC sections further validated the clinical efficiency of the immune-gene panel consisting of prognostic IRGs to distinguish TSCC patients. Conclusion: VEGFA, CXCL13, and CTSG, correlated with T cell infiltration and prognostic outcome. They were screened to form an immune-gene panel to identify TSCC subgroups with different prognostic outcomes. Clinical IHC further validated the efficacy of this immune-gene panel to evaluate aggressiveness of TSCC development.
Keywords: Oral squamous cell carcinoma, tongue squamous cell carcinoma, immune-related genes, CXCL13, VEGFA, CTSG
Introduction
Cancers in the lip and oral cavity are one of the most common types of carcinoma worldwide, with 354,864 new cases in 2018 [1,2]. Tongue squamous cell carcinoma (TSCC) is one of the most common malignancies in the oral cavity, with an increasing incidence as well as poor prognosis [3,4]. In 2019, about 17% of tongue cancer patients died in the United States [5]. Currently, surgical treatment is considered the primary therapeutic option for TSCC patients. However, occult metastasis is often associated with TSCC due to the tongue’s anatomic structure [3,6,7]. Thus, the 5-year overall survival rate of TSCC is low [8]. In the past decades, an abundance of prognostic biomarkers for TSCC was identified, including P53, Ki-67, P16, VEGFs, and cyclin D1, which might participate in tumorigenesis [9-11]. Also, the tumor-infiltratingimmune cells can affect malignant progression, so corresponding immunotherapies have been widely used for treatment [12,13].
Tumor microenvironment (TME) consists of immune cells, fibroblasts, endothelial cells and tumor stroma, which play an important role in malignant progression [14]. As the core component of TME, immune cells and immune-related genes (IRGs) are associated with the response to anti-tumor therapies and subsequent prognosis. It has been reported that anti-tumor immune activities were suppressed in TSCC and the immune microenvironment maintained these complicated alterations [15]. TSCC patients with higher CD4+ and CD8+ cell infiltration had improved clinical outcome [16]. In addition, immune checkpoint inhibitors (ICIs), such as PD-L1/PD-1 inhibitors could effectively facilitate a reversal of immunosuppression [17]. Thus, the investigation of an abnormal TME is important for improving chemotherapy of OSCC. However, the relationship between tumor-infiltrating immune cells (TICs) and clinical outcome of OSCC patients remains undetermined. Few TSCC prognostic biomarkers from an immune perspective have been observed and validated. Since tumor immune microenvironment is becoming more important for the progression of TSCC, elucidating the relationship between local immune activity and TSCC, as well as exploring prognostic immune biomarkers in TSCC, is necessary.
In our study, we aimed to reveal the immune landscape of TSCC compared to normal tongue tissues by single-sample gene set enrichment analysis (ssGSEA). We identified differentially expressed IRGs by analyzing the transcriptomic data of TSCC patients. Next, we integrated prognostic IRG expression profiles and investigated the correlation of hub IRGs and immune cell infiltration levels in TSCC samples. Furthermore, three clusters were identified by unsupervised clustering according to expression level of IRGs. In conclusion, our work depicted the immune landscape of TSCC and identified three IRGs with potential for predicting the aggressiveness of TSCC progression.
Material and methods
Data source and acquisition
Two gene expression datasets, GSE13601 and GSE34105, were downloaded from the Gene Expression Omnibus (GEO) platform with the help of R package GEOquery [18], to figure out differentially expressed genes in TSCC. The platform for these two datasets was GPL570. Transcriptomic profiles of expression data from tongue squamous cell carcinoma patients were acquired from The Cancer Genome Atlas (TCGA) database. TSCC is one of the most important subtypes of head and neck squamous cell carcinoma (HNSCC) based on anatomic site. Due to the anatomic properties of tumor samples, TSCC samples were filtered out from the TCGA-HNSCC dataset.
Immune cell infiltration analysis
The ssGSEA [19] method was used to quantify the relative levels of 28 types of different immune cells in tongue squamous cell carcinoma tissues as well as normal tissues. The featured gene panels according to type of immune cells were based on a prior publication [20]. In detail, differentially expressed genes from both GSE13601 and GSE34105 were applied to calculate infiltration scores of 28 types of immune cells with the help of R packages GSVA and Genefilter. Subsequently, enrichment scores obtained from ssGSEA were regarded as reliable indexes to measure immune cell infiltration status. Finally, heatmaps were pictured to depict immune infiltration status.
Identification of differential expression immune-related genes
R package limma [21] was selected to identify the differentially expressed genes in these two gene expression datasets from the GEO platform as well as RNA sequencing data from TCGA. P value <0.05 and |log2 Fc| >1 were set as the cutoff values for the identification of differentially expressed genes. A standard list of immune-related genes (IRGs) was derived from the Immunology Database and Analysis Portal (Import) [22]. By comparison between genes from the list of IRGs and differentially expressed genes, differentially expressed IRGs in TSCC based on RNA sequencing data from TCGA were finally identified.
Functional enrichment analysis
The web tool Metascape, which provides a biologist-oriented resource for gene functional analysis [23], was selected to identify these significantly enriched gene functional terms in two datasets respectively. Cross-link interactions between enriched terms were also depicted. R package Clusterprofiler was used to further identify significantly enriched GO and KEGG terms for the intersection of IRGs from the two datasets [24]. Terms with a P-value <0.01, a minimum count of 3, and an enrichment factor >1.5 (the enrichment factor is the ratio between the observed counts and the counts expected by chance) were collected and grouped into clusters based on their membership similarities. FDR cutoff p-value applied to our functional enrichment analysis was set at 0.05.
Protein-protein interaction analysis
The web tool Metascape discussed above was also selected to analyze the protein-protein interaction relationship of two datasets. The inserted function, MCODE, was applied to explore core protein-protein interaction modules. These core modules play central roles altogether in the whole regulating network, and proteins inside them might be crucial biomedical targets.
Survival analysis
Patients with integrated clinical information from the TCGA database were selected for survival analysis. Kaplan-Meier analysis was used for survival analysis. Identified IRGs above were submitted to univariate analysis using R package survival. Potential independent prognostic gene indicators were further analyzed using R package survmine to identify IRGs with significant prognostic value. Similarly, the prognostic values of immune cells were also figured out based on immune infiltration scores.
Relationship between immune infiltration and crucial IRGs
Correlations between immune cell infiltration and IRGs selected above were measured by specific gene expression levels and enrichment scores for certain types of immune cells. Positive correlations between gene expression and enrichment scores for anti-tumor immune cells indicated oncological impacts by blockade of anti-tumor immune activity. On the other hand, negative correlations between gene expression and enrichment scores for anti-tumor immune cells suggested protective roles by the promotion of the anti-tumor immune activity.
Construction of different TSCC phenotypes
Three kinds of TSCC phenotypes were established with the help of unsupervised cluster analysis using based on the infiltration status of immune cells. In detail, the Unsupervised clustering method (cutoff =1.0) is an important type of machine learning model aimed at clustering substantial unmarked data into multiple subgroups, in order to make a maximum difference among subgroups and minimum difference inside subgroups. In our study, 6 immune-related genes with prognostic values were firstly filtered out by univariable Cox regression analysis and survival analysis. Then expression levels of these 6 genes from TSCC patients were ln(x+1)-transformed for further analysis. Clustering of subgroups based on these IRGs was completed by calculating Euclidean distance and Ward (unsquared distances) linkage.
Collection of clinical TSCC samples
A total of 30 cases of tongue squamous cell carcinoma (TSCC) tissue samples as well as paired adjacent normal tissue samples were acquired from clinically diagnosed TSCC patients during surgical tumor resection operations, at the Union hospital of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China) from December 2019 to November 2020. All patients had signed proper consent for the use of their clinical samples. Collected fresh tissue samples were frozen in liquid nitrogen within 15 minutes after resection. Clinical information, including age, gender, and TNM staging, wererecorded.
Immunohistochemical and immunofluorescent staining
Immunohistochemical and immunofluorescent stainings were applied to test the clinical efficiency of the constructed immune gene panel. Collected fresh TSCC tumor tissue samples as well as paired adjacent normal tissue samples were applied for further IHC staining. 4% PFA fixed TSCC samples were sectioned and incubated with primary antibodies overnight. Primary antibodies included anti-CTSG (Affinity, AF5167), anti-CXCL13 (Proteintech, 10927-1-AP), anti-VEGFA (Proteintech, 19003-1-AP), anti-IL17 (Proteintech, 13082-1-AP), anti-CD4 (Proteintech, 67786-1-Ig), anti-CD8 (Proteintech, 66868-1-Ig). The secondary antibody was added subsequently for the formation of colored precipitation.
Validation of IRGs with clinical and pathologic value
These IRGs with great prognostic value and consisting of the immune gene panel were further analyzed to determine significant clinical or pathological value. Comparison of relative expression levels among different pathologic T stages was completed, and P<0.05 was considered a significant difference.
Si-RNA transfection
Specific si-RNA and non-specific control from GenePharma were transfected into Cal27 cell lines with the help of Lipo6000 (Beyotime). Co-culture of Lipo6000 and Cal27 cells for 6 h was enough for transfection, and real-time qPCR was carried out for identification.
CCK8 assay
Proliferation rates of Cal27 cells under different treatments were measured by the CCK8 kit (Beyotime). Cal27 cells were seeded into 96-well plates (2000-5000 cells per well) and cultured for 72 h. Co-culture of Cal27 cells and CCK8 reagents for 1.5 h was enough for detection of cell proliferation rate at OD 450 nm.
Transwell assay
The migration ability of Cal27 cells was evaluated by a transwell assay kit (Corning Life Science). Cal27 cells with proper density were seeded onto the upper face of the transwell chamber and cultured for 24 h subsequently. After removal of cells remaining on the upper surface, crystal violet staining was performed for measurement of cells that migrated through the chamber.
Statistical analysis
Graphpad Prism 7 was applied for statistical analysis. An Independent t-test was applied for comparison of continuous variables between two groups, and P-value <0.05 was considered significant. A Chi-square test was used to analyze the independence of different clinical factors, IRG expression levels, and immune cell infiltration degree. Univariate Cox regression analysis and Kaplan-Meier survival analysis were performed for validation of survival value. Logistic regression analysis and multivariate Cox regression analysis were performed to construct risk score models.
Results
Description of immune cell infiltration status in tongue carcinoma
To investigate the comprehensive immunophenotypic status in TSCC, single-sample Gene Set Enrichment Analysis (ssGSEA), a quantitative method using microarray data to depict local immune status, was applied in this article. Heatmaps composing relative abundance of 28 infiltrating immune cell populations in two datasets (GSE13601 and GSE34105) were described for visualization. Interestingly, the immune status in tumor samples was markedly activated, as immune scores of tumor samples for multiple types of cells (e.g., activated CD4+ T cells, MDSCs) were higher compared with normal tongue samples (Figure 1A, 1C). Especially, a statistical analysis comparing immune cell infiltration between normal and tumor samples indicated that multiple types of T cells, including anti-tumor T cells (e.g., activated CD4+ T cell, natural killer T cell) as well as pro-tumor T cells (e.g., regulatory T cell), all showed higher infiltration scores in tumor samples. Representative images of activated CD4+ T cells and activated CD8+ T cells are shown (Figure 1E, 1F). It could be speculated from this phenomenon that variations of T cell function might play a crucial role in TSCC progression. Pearson’s correlation analysis revealed that abundances of anti-tumor and pro-tumor immune cells were positively correlated in local tissue sites. As expected, the distribution of pro-tumor and anti-tumor immune cells among TSCC tumor samples were similar in different TSCC datasets, suggesting that tongue carcinoma immune infiltrate is complex (Figure 1B, 1D). Clinical information for datasets GSE13601 and GSE34105 were listed below (Tables 1, 2).
Figure 1.

Immunogenomic landscape of tongue squamous cell carcinoma. A. The immune cell infiltration status in GSE13601 depicted by heatmap. B. Correlation between infiltration of immune cells undertaking anti-tumor functions and those executing pro-tumor functions in GSE13601. C. Immune cell infiltration status in GSE34105 depicted by heatmap. D. Correlation between infiltration of immune cells undertaking anti-tumor functions and those executing pro-tumor functions in GSE34105. E, F. Representative images of activated CD4+ T cell and activated CD8+ T cell infiltration status between normal and tumor tissues in GSE13601 and GSE34105 respectively, depicted by boxplots.
Table 1.
Clinical information of patients from GSE13601
| subgroups | N |
|---|---|
| Tumour | 31 |
| Normal | 26 |
Table 2.
Clinical information of patients from GSE34105
| Subgroups | Variable | N | Total number of patients |
|---|---|---|---|
| Tumour | 62 | ||
| Age | |||
| ≥60 | 35 | ||
| <60 | 27 | ||
| Gender | |||
| Male | 34 | ||
| Female | 28 | ||
| Stage | |||
| T stage | |||
| T1 | 18 | ||
| T2 | 27 | ||
| T3 | 9 | ||
| T3-4 | 1 | ||
| T4 | 7 | ||
| N stage | |||
| N0 | 46 | ||
| N1 | 7 | ||
| N2 | 7 | ||
| N3 | 1 | ||
| Nx | 1 | ||
| M stage | |||
| M0 | 59 | ||
| M1 | 1 | ||
| Mx | 1 | ||
| Control | 16 | ||
| Age | |||
| ≥60 | 5 | ||
| <60 | 11 | ||
| Gender | |||
| Male | 8 | ||
| Female | 8 |
Identification of differentially expressed IRGs
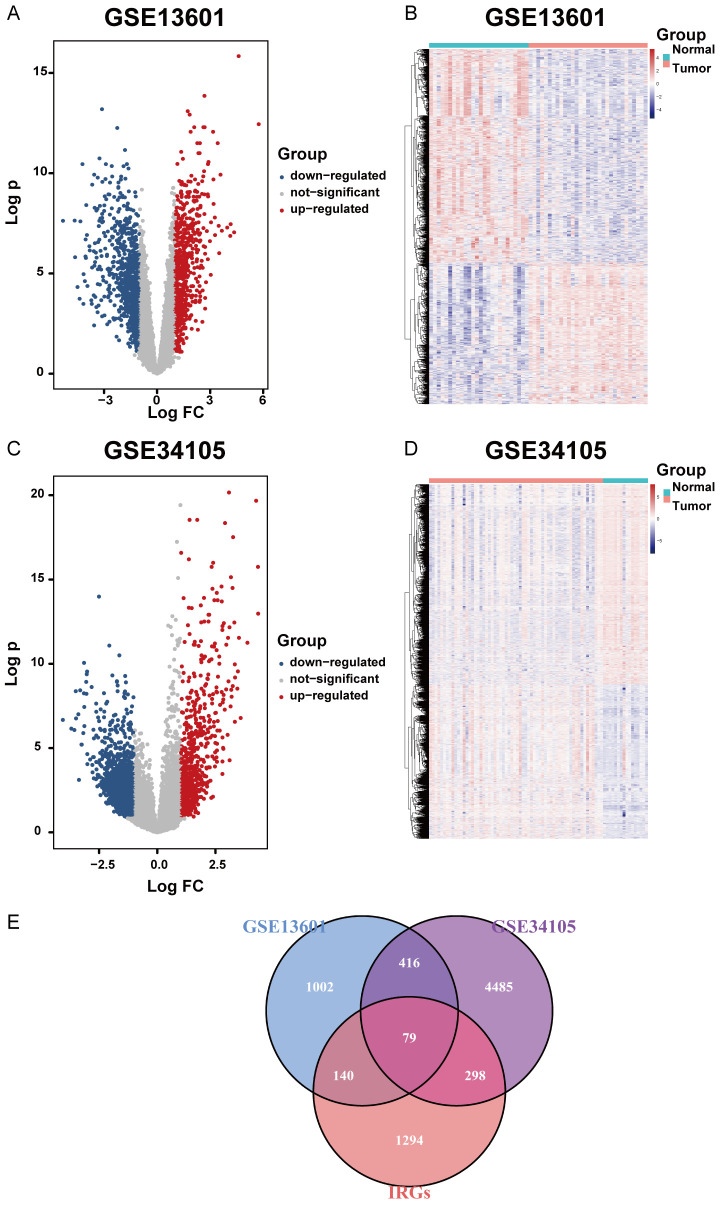
The R package Limma was used to classify differentially expressed immune-related genes among different datasets. In GSE13601, 1637 differentially expressed genes were identified, with 652 upregulated genes and 985 down-regulated ones (Figure 2A, 2B). In GSE34105, 5278 differentially expressed genes were found, with 2302 upregulated and 2976 downregulated (Figure 2C, 2D). An immune-related gene set (IRG) containing more than 2000 IRGs was implemented to further recognize 219 and 377 differentially expressed IRGs in GSE13601 and GSE34105 respectively, as well as the commonly shared 79 IRGs in both of the two datasets (Figure 2E). From this perspective, these intercept IRGs might be important factors in TSCC, and further analysis exploring the immune status of TSCC was dependent on them.
Figure 2.
Differentially expressed immune-related genes. A. Differentially upregulated and downregulated genes selected from GSE13601. P-value <0.05 and |log2 Fc| >1 were set as cutoff values for the selection of differentially expressed genes. B. Differentially expressed genes in GSE13601 depicted by heatmap. C. Differentially upregulated and downregulated genes selected from GSE34105. P value <0.05 and |log2 Fc| >1 were set as cutoff values for the selection of differentially expressed genes. D. Differentially expressed genes in GSE34105 depicted by heatmap. E. Venn diagram showing the relationship between differentially expressed genes and immune-related genes. The common part indicates IRGs differentially expressed by tongue squamous cell carcinoma.
Functional enrichment analysis of differentially expressed IRGs
Functional enrichment analysis revealed that immune-related terms, such as cytokine-related signaling pathway, cell chemotaxis, and defense response to other organisms, were the most highly-enriched terms in GSE13601 (Figure 3A), and these immune functions were broadly crosslinked altogether, indicating a complex immune regulating network in TSCC (Figure 3B). Similarly, In GSE34105, cytokine-mediated signaling pathway, chemotaxis as well as lymphocyte activation were the most enriched (Figure 3C, 3D). These results validated that differentially expressed IRGs might be broadly involved in multiple types of immune activities in the TSCC local microenvironment across datasets. As expected, GO analysis conducted on shared IRGs between these two datasets exhibited leukocyte migration, chemotaxis, response to the virus as the top three oncology processes. In KEGG analysis, immune-related pathways, such as cytokine-cytokine interaction, chemokine signaling pathway and natural killer cell mediated cytotoxicity, also exhibited high abundance (Figure 3E, 3F). These results further confirmed that IRGs among different datasets shared some immune activities and pathways as their common biologic functions.
Figure 3.
Gene functional enrichment analysis of differentially expressed immune-related genes. A. Terms of significantly enriched gene ontology and pathways in GSE13601. B. A network plot depicting interactions among those enriched ontological functions in GSE13601. C. Terms of significantly enriched gene ontology and pathways in GSE34105. D. A network plot depicting interactions among those enriched ontological functions in GSE34105. E. Enriched GO terms by shared immune-related genes between GSE13601 and GSE34105. F. Enriched KEGG terms by shared immune-related genes between GSE13601 and GSE34105.
PPI network analysis of differentially expressed IRGs
To figure out interaction among differential IRGs, protein-protein interaction (PPI) network analysis was conducted by STRING web tool. As expected, these IRGs involved in immune-related biologic processes, showed highly interacting patterns with complex protein-protein crosslinks in both datasets (Figure 4A, 4B). Broad interactions among IRGs were noted, which might play a significant role in regulation of immune activity in TSCC progression. To further explore core interaction networks playing central roles in immune activities, MCODE function was carried out to find out core protein modules. Six MCODE modules were figured out in GSE13601, the biological functions of which were peptide ligand-binding receptors, PI3K-Akt signaling pathway, anti-inflammatory, antigen presentation, GPCR ligand binding, and neutrophil chemotaxis respectively (Figure 4C). In GSE34105, ten interacting modules were figured out, and their biologic functions are all immune-related, such as antigen procession and presentation, interleukin-2 family signaling and immune response-regulating signaling pathway (Figure 4D). In addition, proteins in these MCOED modules, CXCL12, EGFR and TNF for example, exhibited the probability of being hub proteins in immune regulation of TSCC.
Figure 4.
Protein-protein interaction (PPI) analysis. A. PPI network constructed by differentially expressed immune-related genes in GSE13601. B. PPI network constructed by differentially expressed immune-related genes in GSE34105. C. Core PPI modules selected by MCODE function in GSE13601. D. Core PPI modules selected by MCODE function in GSE34105.
Identification of survival-related IRGs
Prediction of TSCC prognostic outcome is very necessary, as it could help in the selection of clinical treatments of surgical resection versus conservative drug treatment. However, precise and TSCC-targeted clinical biomarkers applied in predicting clinical outcomes of TSCC patients are still inadequate. Molecular biomarkers were needed that could serve as accurate immune-related prognostic indicators. Univariate Cox regression analysis was first conducted based on TCGA-HNSCC tongue squamous cell carcinoma samples to identify possible survival-associated IRGs, and eleven candidate molecules were figured out, and a univariate Cox proportional hazard model was established to show their prognostic values in a forest plot (Figure 5A). Accordingly, clinical information of patients in TCGA dataset were described in Table 3. Subsequently, survival analysis was performed on these candidate biomarkers to identify eight molecules with significant prognostic survival values by the P-value cut-off equal to 0.05 (Figure 5B-I). Among them, BPIL2, CTSG, and CXCL13 served as good survival indexes and the other five as bad survival predictors.
Figure 5.

Survival analysis of differentially expressed immune-related genes. A. Hazard ratio univariate Cox regression model constructed by immune-related genes with potential survival value. B-I. Kaplan-Meier curves depicting survival analysis of immune-related genes with significant prognostic value (BPIL2, CTSG, CXCL13, VEGFA, ADIPOQ, DKK1, LEFTY1, NPY).
Table 3.
Clinical information of patients from TCGA dataset
| Variable | Number |
|---|---|
| Age (years) | 116 |
| ≥60 | 65 |
| <60 | 51 |
| Gender | 116 |
| Male | 75 |
| Female | 41 |
| Clinical stage | 116 |
| Stage I | 10 |
| Stage II | 32 |
| Stage III | 29 |
| Stage IVA | 40 |
| Stage IVB | 1 |
| #N/A | 4 |
| Alcohol history | 116 |
| Yes | 72 |
| No | 41 |
| #N/A | 3 |
| History of neoadjuvant treatment | 116 |
| Yes | 2 |
| No | 114 |
| Lymphnode neck dissection | 116 |
| Yes | 107 |
| No | 9 |
| Neoplasm histologic grade | 116 |
| G1 | 16 |
| G2 | 78 |
| G3 | 22 |
| Pathologic stage | 116 |
| Stage I | 15 |
| Stage II | 20 |
| Stage III | 31 |
| Stage IVA | 49 |
| Stage IVB | 1 |
Identification of survival-related immune cells
The prognostic value of immune cells in TSCC was then explored using survival analysis. With a P-value cut-off of 0.05, a total of 3 types of infiltrating immune cells, activated CD8+ T cell, central memory CD4+ T cell, and type 17 T helper cell, were identified as key immune cells with significant prognostic value in TSCC (Figure 6A-C).
Figure 6.

Survival analysis of cancer-related infiltrating immune cells. A-C. Kaplan-Meier curves depicting survival analysis of three types of infiltrating immune cells (activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells) with high prognostic value.
Correlations between prognostic immune cells and genes
To explain the role of hub IRGs in the regulation of immune activities during TSCC, correlation analysis between the above eight IRGs and three immune cells was performed. Results indicated that six out of the eight IRGs, including LEFTY1, VEGFA, DKK1, BPIL2, CXCL13, and CTSG, showed significant correlations with at least one type of immune cell. Among them, a poor survival biomarker, VEGFA, had a negative correlation with infiltration of the above three types of prognostic immune cells, activated CD8+ T cells, central memory CD4+ T cell, and type 17 T helper cell (Figure 7A-C). In contrast, CXCL13, a favorable survival predictor, exhibited a significant positive correlation with infiltration of activated CD8+ T cells, central memory CD4+ T cell, and type 17 T helper cell (Figure 7D-F). As expected, CTSG, another favorable survival biomarker, also showed a significant positive correlation with infiltration of the above three types of anti-tumor immune cells (Figure 7G-I). The above correlations partly validate and explain the roles of IRGs in TSCC from an immune perspective. Especially, VEGFA, CXCL13, and CTSG, all of which were significantly correlated with both prognostic immune cell infiltration status and clinical outcomes, might be used as excellent predictors of TSCC tumor immune status and clinical outcome.
Figure 7.

Description of relationship between IRG expression and immune cell infiltration. A-C. Negative correlations between VEGFA expression level and enrichment scores for activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells. D-F. Positive correlations between CXCL13 expression level and enrichment scores for activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells. G-I. Positive correlations between VEGFA expression level and enrichment scores for activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells. The significance of all these correlation analysis depicted above were set by the cutoff P-value at 0.05.
Construction of predictive models with prognostic IRGs
ROC curves were first depicted based on expression levels of VEGFA, CXCL13, and CTSG. Results showed that other than VEGFA, CXCL13 and CTSG exhibited extremely low efficiency for prognostic prediction with their AUC values less than 0.5 (Figure 8A-C). While VEGFA might be used for 1-year prognostic prediction, its long-term predictive performance might not be sufficient. Given the above disadvantages for a single IRG to be used as a prognostic biomarker in TSCC, construction of predictive models with all 3 IRGs was necessary. Thus, logistic regression, as well as multivariate Cox regression analysis was performed to construct two prognostic predictive models (Figure 8D, 8E and Tables 4, 5). ROC curves for two of these models indicated ideal prognostic values for 1-year, 3-year, and 5-year survival of TSCC patients (Figure 8F, 8G). The above results suggested that the application of a combination of VEGFA, CXCL13, and CTSG might be an effective method for prediction of TSCC clinical outcomes.
Figure 8.

Construction of prognosis-predicting models using selected IRGs with significant prognostic value. A-C. ROC curves depicting the test efficiency of expression levels of VEGFA, CXCL13 and CTSG respectively. The measures of area under curve were defined as AUC value, which indicated their clinical outcome-predicting efficiency precisely. D, E. Information of prognostic-predicting models constructed by logistic regression analysis and multivariate Cox regression analysis. F, G. ROC curves depicting the test efficiency of logistic regression model and multivariate Cox regression model respectively. The measures of area under curve were defined as AUC value, which indicated their clinical outcome-predicting efficiency precisely.
Table 4.
Multivariate Cox regression analysis between IRGs and clinical factors
| Variable | N | chisp | P value |
|---|---|---|---|
| Age (years) | 116 | 0.02310 | 0.879 |
| ≥60 | 65 | ||
| <60 | 51 | ||
| Gender | 116 | 1.57591 | 0.209 |
| Male | 75 | ||
| Female | 41 | ||
| Clinical stage | 116 | 7.78277 | 0.051 |
| Stage I | 10 | ||
| Stage II | 32 | ||
| Stage III | 29 | ||
| Stage IVA | 40 | ||
| Stage IVB | 1 | ||
| #N/A | 4 | ||
| Alcohol history | 116 | 2.38329 | 0.123 |
| Yes | 72 | ||
| No | 41 | ||
| #N/A | 3 | ||
| History of neoadjuvant treatment | 116 | 1.50296 | 0.220 |
| Yes | 2 | ||
| No | 114 | ||
| Lymphnode neck dissection | 116 | 1.61382 | 0.204 |
| Yes | 107 | ||
| No | 9 | ||
| Neoplasm histologic grade | 116 | 5.89476 | 0.052 |
| G1 | 16 | ||
| G2 | 78 | ||
| G3 | 22 | ||
| Pathologic stage | 116 | 6.34098 | 0.175 |
| Stage I | 15 | ||
| Stage II | 20 | ||
| Stage III | 31 | ||
| Stage IVA | 49 | ||
| Stage IVB | 1 | ||
| Scores for logistic model | 116 | ||
| Scores for multivariate Cox model | 116 |
Table 5.
Multivariate Cox regression analysis for identified IRGs
| Variable | chisq | P value |
|---|---|---|
| Neck lymphnode dissection | 3.327 | 0.0682 |
| Tumor histological grade | 7.633 | 0.0220 |
| gender | 0.709 | 0.3999 |
| Tumor clinical stage | 6.352 | 0.0957 |
| Alcohol history | 1.643 | 0.1999 |
| CXCL13 | 4.124 | 0.0423 |
| VEGFA | 3.750 | 0.0500 |
| CTSG | 3.126 | 0.0889 |
| Score for logistic model | 3.837 | 0.0401 |
| Score for multivariate Cox model | 3.844 | 0.0499 |
Identification of specific TSCC subgroups with different prognosis
Heterogeneity of tumors is the main obstacle for accurate individual diagnosis and therapeutic strategies. Thus, a proper classification strategy was necessary to identify TSCC subgroups with different prognostic outcomes. Unsupervised clustering analysis, one of the most efficient machine learning methods for the classification of unmarked data, was then applied based on the relative expression levels of prognostic IRGs selected above, and three different TSCC subgroups were subsequently defined. It was subsequently observed that the above three subtypes of T cells with prognostic value that exhibited a similar infiltration tendency among different TSCC clusters (Figure 9A). Results of survival analysis indicated that cluster A showed a worse survival outcome compared to cluster B and cluster C (Figure 9B, 9C). In contrast, there was no difference in clinical survival between cluster B and cluster C (Figure 9D). Thus, it was concluded that TSCC patients belonging to cluster A might have poor clinical outcome, whereas patients in cluster B and cluster C tended to have better survival (Tables 6, 7 and 8). Finally, efforts were made to validate the immune infiltration status among different TSCC clusters statistically. As expected, the above observed three types of prognostic immune cells, including activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells, showed a significant increase of infiltration status among the three clusters, in which cluster A had the poorest anti-tumor immune infiltration (Figure 9E-G). This indicated that the poor prognosis of TSCC patients belonging to cluster A might be partly explained by a weak anti-tumor immune infiltration status, showing the importance of T cell immune function in TSCC prognosis.
Figure 9.

Exploration of an immune-gene panel to distinguish different TSCC clusters. A. Three types of TSCC subgroups, named cluster A, cluster B and cluster C, were constructed by unsupervised cluster analysis based on prognostic IRGs selected above. B-D. Prognostic analysis between different TSCC clusters. E-G. Comparison of activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells infiltration status among different TSCC clusters.
Table 6.
Association between TSCC subgroup classification and T stage
| TSCC subgroups | T1 | T2 | T3 | T4a | T4b | Total number of patients |
|---|---|---|---|---|---|---|
| cluster A | 24% | 36% | 20% | 16% | 4% | 25 |
| cluster B | 18% | 34% | 28% | 20% | 0% | 50 |
| cluster C | 17.07% | 39.02% | 34.15% | 9.76% | 0% | 41 |
Table 7.
Association between TSCC subgroup classification and N stage
| TSCC subgroups | N0 | N1 | N2 | N2a | N2b | N2c | Nx | Total number of patients |
|---|---|---|---|---|---|---|---|---|
| cluster A | 52% | 12% | 0% | 0% | 28% | 4% | 4% | 25 |
| cluster B | 46% | 8% | 4% | 2% | 28% | 10% | 2% | 50 |
| cluster C | 36.59% | 26.83% | 2.44% | 2.44% | 21.95% | 4.88% | 4.88% | 41 |
Table 8.
Association between TSCC subgroup classification and M stage
| TSCC subgroups | M0 | Mx | Total number of patients |
|---|---|---|---|
| cluster A | 76.92% | 23.08% | 13 |
| cluster B | 76.92% | 23.08% | 26 |
| cluster C | 86.67% | 13.33% | 15 |
Validation of immune-gene panel to distinguish TSCC subgroups
Efforts were then made to establishing a clinical detection method to better classify TSCC patients into corresponding subgroups. Validation was then made on the above prognostic 3-gene panel to see whether it could be used for TSCC subgroup classification. Statistical analysis showed that expression levels of CXCL13 and CTSG exhibited a decreasing tendency from cluster A to cluster C, while VEGFA showed an opposite trend (Figure 10A-C). These results coincided with their prognostic value, which suggests that a combination of these 3 IRGs can distinguish TSCC subgroups.
Figure 10.

Identification of TSCC immune subgroups by immune-gene panel. A-C. Relative expression levels of VEGFA, CXCL13 and CTSG in tongue squamous cell carcinoma samples among different TSCC subgroups in TCGA dataset. D-F. Positive and negative rate of VEGFA, CXCL13 and CTSG among different TSCC subgroups in TCGA dataset. Expression data of TSCC samples from TCGA dataset were homogenized for calculation of positive rate corresponding to each IRG.
From a clinical perspective, RNA expression levels of IRGs might not be rapidly and conveniently applied to clinical diagnosis. Efforts were further made to establish a better method in the clinical scenario. After homogenization of transcript expression levels of these differentially expressed IRGs in three TSCC subgroups, samples with gene expression levels greater than 0 were regarded as positive individuals, with the others as negative ones. This may help in establishing a mimetic diagnostic model using RNA sequencing data. Positive rates were then calculated to detect their possibility for distinguishing different TSCC subgroups. Results showed that CXCL13 and CTSG exhibited significantly lower positive rates in cluster A, the subgroup with better clinical outcome, with higher positive rates in the poor survival ones, cluster B and cluster C. In contrast, VEGFA showed an extremely high positive rate in cluster A, but a low positive rate in cluster B and cluster C (Figure 10D-F). Thus, in order to avoid the bias of diagnosis caused by a single biomarker, a combination of these IRGs might be helpful in distinguishing different TSCC subgroups.
Clinical validation of selected immune gene panel
To validate the efficiency of the selected immune gene panel during clinical diagnosis, IHC staining was employed on human TSCC sections to evaluate the efficiency of an immune-gene panel in distinguishing different TSCC sub-populations. However, since clinical TSCC samples in this study were freshly obtained within 2 years, survival information of these collected samples was not adequate for long-term survival analysis. As pathological T stage and N stage are golden clinical standards for evaluation of TSCC aggressiveness, efforts were then made to find a relationship between TSCC pathologic T stage, N stage, and immune-gene panel. Bioinformatic analysis was performed at first to compare the expression levels of prognostic immune-gene panel among TCGA TSCC samples at different pathologic T and N stages. Results suggested that patients at higher pathologic T stage showed a higher level of VEGFA and lower levels of CTSG and CXCL13, while patients at higher N stage showed a similar tendency but without statistical significance (Figure 11A, 11B). Clinical validation was performed to evaluate the intensity of IHC staining among TSCC sections at different T stages and N stages. As expected, TSCC patients at higher pathologic T stage showed stronger levels of VEGFA while weaker levels of CTSG and CXCL13 (Figure 11C). Similarly, TSCC samples at higher pathological N stage exhibited a relatively higher level of VEGFA, and lower levels of CXCL13 and CTSG (Figure 11D). It is known that TSCC survival is negatively correlated with cancer pathological T stage to a great extent, since substantial blood and lymph vessels are present around as well as inside the tongue, and local tumor invasion and infiltration may easily result in metastasis of TSCC. Therefore, the ability of the immune-gene panel constructed by VEGFA, CXCL13, and CTSG to distinguish TSCC at different T stages might explain its ability to evaluate TSCC survival outcome. Further studies are necessary to directly analyze the relationship between the immune-gene panel and TSCC clinical outcome with complete long-term patient survival. The above bioinformatic analysis revealed that the behaviors of Th17 cells, activated CD8+ T cells and central memory CD4+ T cells are closely correlated with clinical outcome of TSCC patients. To confirm this phenomenon, immunofluorescent staining was carried out on clinical TSCC samples. Results showed that IL-17+ cells were broadly distributed in TSCC samples at high pathological grade, which meant that Th17 cells might be largely increased in TSCC that is well differentiated (Figure 11E). Accordingly, CD4+ T cells and CD8+ T cells both showed higher abundance in well-differentiated TSCC than poorly differentiated TSCC (Figure 11F, 11G). In summary, these three types of T cells were identified to suggest better prognostic outcome of TSCC patients and might play anti-tumor roles during TSCC progression.
Figure 11.
Clinical validation for the predicting efficiency of immune-gene panel. A. Relative expression levels of CTSG, CXCL13 and VEGFA in tongue squamous cell carcinoma samples among different T stages in TCGA dataset. B. Relative expression levels of CTSG, CXCL13 and VEGFA in tongue squamous cell carcinoma samples among different N stages in TCGA dataset. C. Clinical validation of VEGFA, CXCL13 and CTSG expression levels in tumor samples at pathologic T1, T3 and T4 stage with the help of IHC staining images. D. Clinical validation of VEGFA, CXCL13, and CTSG expression levels in tumor samples at pathological N0 and N2 stages with the help of IHC staining images. E-G. Clinical validation for accumulation of Th17 cells, CD4+ T cells and CD8+ T cells in TSCC by immunofluorescent staining.
Role of BPIL2, CXCL13, and CTSG in TSCC development and metastasis
Considering that BPIL2, CXCL13, and CTSG have been confirmed to indicate better clinical outcome with bioinformatic analysis, efforts were then made to validate their function in TSCC. Si-RNA technique was carried out to knock down BPIL2, CTSG, and CXCR5, a classical membrane receptor for CXCL13. As expected, knockdown of all of the three genes showed an increased cell proliferation rate compared to control groups detected by the CCK8 assay. Specifically, downregulation of CTSG contributed the most to cancer proliferation and TSCC development among the three candidates (Figure 12A-C). Accordingly, transwell experiments and subsequent crystal violet staining were carried out on TSCC cell lines treated by the si-RNA technique. Results showed that loss of CXCR4, receptor of CXCL13, as well as loss of CTSG, contributed most to the migration and invasion of TSCC cell lines, while knockdown of BPIL2 exhibited little effect on migration and invasion. Results suggest that CTSG and CXCL13 are closely correlated to TSCC metastasis (Figure 12D-F). In conclusion, all of the three genes could play an anti-tumor effect in TSCC, and CXCL13, as well as CTSG, contributed more compared to BPIL2. Further exploration was still necessary to figure out detailed roles and subsequent mechanisms for these three genes.
Figure 12.

Validation for the role of BPIL2, CXCL13 and CTSG in TSCC development and metastasis. A-C. Role of BPIL2, CXCL13 and CTSG in TSCC malignant development validated by Cell Counting Kit (CCK8) assay. D-F. Role of BPIL2, CXCL13 and CTSG in TSCC metastasis confirmed by transwell assay. Si-RNA technique was implemented for efficient knockdown of BPIL2, CXCR5 and CTSG, and two target sites for each gene were applied in our experiment.
Discussion
With the development of tumor immunotherapy, tumor immunity has become a research hotspot. Growing evidence showed that an abundance of immune cells and immune-specific antigens have been detected in oral cancer and function in tumor immunity [25-28]. These findings provide immunotherapeutic targets for TSCC. Although the immune genomic differences and the prognostic signature genes have been investigated in many types of cancers, the overall immune landscape and differential prognostic IRGs have not been elucidated in TSCC [29,30]. Moreover, variant immune cells are associated with tumor promotion or inhibition, and immunotherapy seems a helpful strategy for tongue cancer patients [31]. Therefore, we estimated the relative 28 immune cell types based on the transcriptomic data from GEO datasets via ssGSEA methods, and the immunological microenvironment constitution was found to be heterogeneous in TSCC. Analysis results showed that multiple types of immune cells showed higher infiltration scores in TSCC tissues, which indicated that there were complicated immune responses during TSCC progression, revealing the complex and diverse immune microenvironment of TSCC. In addition, we conducted the correlation analysis of the immune scores in GEO datasets, and the immune scores of most normal tongue tissues exhibited an anti-tumor activity, while tongue cancer tissues exhibited both high anti-tumor scores and high pro-tumor scores, which were consistent with the above established ssGSEA immune model. The diversity of local immune status in TSCC patients might be explained by the immune infiltration changed along with different clinical and pathologic stages of tumor [32]. That is to say, the early TME in TSCC might exhibit an anti-tumor status and that of advanced TSCC might exhibit a pro-tumor immunity. However, the specific immune status at different tumor stages could not be demonstrated based on our results. As reported by other studies, the immune system plays a vital role in carcinoma progression and patients with higher anti-tumor immune infiltration have a better immunotherapeutic response [33-35]. Thus, estimation of immune infiltration predicts the immune status and efficiency of response for TSCC patients.
Next, differentially expressed IRGs were identified by comparing the expression profiles of TSCC samples with adjacent normal tissues. According to the functional enrichment analysis, differentially expressed IRGs derived from two independent cohorts were enriched in immune-related biological processes and signaling pathways, such as leukocyte migration, cytokine-cytokine receptor interaction, and chemokine signaling pathway, which were consistent with the results of ssGSEA analysis. It is believed that the most enriched modules, for leukocyte migration, and cytokine-cytokine receptor interaction, may be essential to this process. We used TSCC cohorts from the TCGA database to validate whether these genes were associated with the prognosis of TSCC. Based on the univariate Cox regression analysis and prognostic analysis, CXCL13, S100A3, ADIPOQ, DKK1, LEFTY1, NDP, NPY, SH3BP2, BPIL2, CTSG, and VEGFA were significantly correlated with the clinical outcomes of TSCC patients. TSCC patients with high BPIL2, CTSG, and CXCL13 expression had a better prognosis while the person with high the other five signatures had worse clinical outcomes. Previous studies have certificated that these genes played important roles in immune responses. CTSG could regulate the surface MHC class I molecule of immune and tumor cells [36], CXCL13 induced the lymphocyte infiltration within TME [37], and VEGFA could limit anti-tumor immune responses [38]. Apart from some conventional classical biomarkers applied in the diagnosis of TSCC, such as Ki-67 and P53 [39], novel molecular biomarkers are necessary for more accurate TSCC diagnosis. Considering the importance of immune response in TSCC progression, these observed prognostic IRGs in TSCC might have diagnostic and therapeutic significance.
In our findings, multiple types of T cells, including pro-tumor and anti-tumor T cells, all showed higher levels of infiltration status in tumor samples compared to adjacent normal tissues. Accumulation of anti-tumor T cells, including CD4+ T cell and CD8+ T cell in TSCC tissues, could act for the protection of tumor development and metastasis, especially in some TSCC patients at an early stage. Activation of the STING-IFN-γ signaling pathway could result in the recruitment of anti-tumor T cells and subsequent anti-tumor immune reaction [40]. On the other hand, increased expression of CXCL14 has been demonstrated to result in high-level infiltration of anti-tumor T lymphocytes, as a way to resist the growth and development of OSCC [41].
From another perspective, many studies have revealed an increase in pro-tumor T cells in TSCC. Overexpression of TIGIT/CD155 contributes to the immunosuppressive status of HNSCC [42]. Also, TIM3-related upregulation of regulatory T cell pro-tumor function was correlated with immunosuppression in HNSCC [43].
In conclusion, abnormal gene variations may contribute to the increase and activation of pro-tumor types of T cells, thus promoting the malignant progression of TSCC. Along with tumor development, the host might upregulate expression levels of tumor suppressor genes, leading to subsequent recruitment of anti-tumor T cells. Medical interventions have also been confirmed to influence the ratio of anti-tumor to pro-tumor T cells in TSCC. Studies showed that chemotherapies and immunotherapies augment the numbers of tumor-infiltrating T cells through increased expression of adhesive molecules and chemokines [44]. That is to say, due to the individual difference from molecular variations and medical treatment among TSCC patients, enhanced infiltration of pro-tumor and anti-tumor T cells could be reasonably explained, and this phenomenon also needs further exploration.
We also found that the amounts of activated CD8+ T cells, central memory CD4+ T cells, and type17 T helper cells were significantly associated with good clinical outcome of TSCC patients. As widely acknowledged, the three kinds of immune cells could execute a crucial anti-tumor effect in tumor progression [45-47]. Thus, they may have a vital anti-tumor role in TSCC progression. However, which type of immune cell is the major anti-tumor cell in TSCC still requires further research. Through correlation analysis of these prognostic IRGs and immune cells, we found a significant correlation between VEGFA, CTSG, CXCL13, DKK1, BPIL2, LEFTY1 with at least one of these three prognostic immune cell types. Among them, CTSG and CXCL13 were significantly positively correlated with these three prognostic immune cells, while VEGFA was negatively correlated with them. These results demonstrated that CTSG and CXCL13 are essential in anti-tumor immune infiltration and VEGFA was critical to pro-tumor immunity in TSCC patients. Since immune response is recognized as important for tumor progression, elucidating the molecular regulating network of immune response is necessary. However, correlation analysis completed in our research only initially explored the regulating mechanisms of immune cells in TSCC, which still need deep investigation. On the other hand, the correlation of prognostic immune cells and prognostic IRGs could validate the clinical significance mutually. Thus, these immune gene signatures and immune cell subtypes may be novel predictors for immune infiltration assessment and prognostic biomarkers for TSCC patients.
To further validate the importance of the immune landscape in TSCC, TSCC patients were classified into three subgroups according to mimetic scores of prognostic immune cells. Prognostic analysis further revealed a significant difference in the survival probability of TSCC patients among three clusters. A significant difference in survival outcomes of cluster A compared to cluster B and C validated the clinical value of our subgroup definition strategy. Also, immune infiltration statuses changed along with different clusters, validating that our strategy could distinguish TSCC patients with different immune statuses, along with different clinical outcome. It was apparent that significant heterogeneity of immune responses existed within different TSCC tissues, thus contributing to the heterogeneity of clinical diagnosis and therapeutic responses [48]. Accordingly, establishment of subgroup classification strategies is necessary for improving the accuracy of TSCC clinical diagnosis, as well as overcoming drug resistance related to tumor local immune activities. By calculating positive rates and expression levels of prognostic IRGs in different clusters, VEGFA, CTSG, and CXCL13 were selected and combined as an immune-gene panel applied for distinguishing different TSCC clusters. IHC staining on clinical TSCC sections further validated the efficiency of the immune gene panel in distinguishing patients at different pathologic T stages. The established three-gene signature could be used to distinguish TSCC patients with different immune responses and survivals. Clinical validation further confirmed its discriminability for TSCC patients at different T stages, indicating a clinical value of this immune gene panel.
However, there were still some limitations to this research. First, the transcriptional data could not represent the global alterations in TSCC patients, thus proteomics should also be considered in further studies. Secondly, sample volume ought to be expanded as a way to eliminate bias. Selected molecules would be more reliable with further in vitro and in vivo validation, and much more effort is required to explore these underlying mechanisms for immune responses in TSCC. In addition, limited to our survival information of TSCC patients, clinical validation of survival outcome-distinguishing efficiency was substituted by validation of pathologic T stage-distinguishing efficiency. The collection of detailed survival information might contribute to further validation in the future.
In conclusion, an integrated bioinformatic analysis revealed the overall immune landscape of TSCC patients, indicating the complexity of the TSCC immune microenvironment. Patients could be significantly classified into three immune clusters based on selected prognostic IRGs, and this subgroup-classification strategy was further validated by bioinformatic analysis and clinical sections to have great clinical significance. These findings might provide a basis for diagnosis and immunotherapy in TSCC.
Acknowledgements
This work was funded by Young Elite Scientist Sponsorship Program by CAST (2018QNRC001, to QT), and Key Supporting Program by Health Commission of Hubei Province (WJ2019C001, to LC).
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Mannelli G, Arcuri F, Agostini T, Innocenti M, Raffaini M, Spinelli G. Classification of tongue cancer resection and treatment algorithm. J Surg Oncol. 2018;117:1092–1099. doi: 10.1002/jso.24991. [DOI] [PubMed] [Google Scholar]
- 3.Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26:645–662. doi: 10.1007/s10555-007-9082-y. [DOI] [PubMed] [Google Scholar]
- 4.Gupta B, Johnson NW, Kumar N. Global epidemiology of head and neck cancers: a continuing challenge. Oncology. 2016;91:13–23. doi: 10.1159/000446117. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.Ho CM, Lam KH, Wei WI, Lau SK, Lam LK. Occult lymph node metastasis in small oral tongue cancers. Head Neck. 1992;14:359–363. doi: 10.1002/hed.2880140504. [DOI] [PubMed] [Google Scholar]
- 7.Bello IO, Soini Y, Salo T. Prognostic evaluation of oral tongue cancer: means, markers and perspectives (I) Oral Oncol. 2010;46:630–635. doi: 10.1016/j.oraloncology.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 8.van Dijk BA, Brands MT, Geurts SM, Merkx MA, Roodenburg JL. Trends in oral cavity cancer incidence, mortality, survival and treatment in the Netherlands. Int J Cancer. 2016;139:574–583. doi: 10.1002/ijc.30107. [DOI] [PubMed] [Google Scholar]
- 9.Almangush A, Heikkinen I, Makitie AA, Coletta RD, Laara E, Leivo I, Salo T. Prognostic biomarkers for oral tongue squamous cell carcinoma: a systematic review and meta-analysis. Br J Cancer. 2017;117:856–866. doi: 10.1038/bjc.2017.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garnaes E, Frederiksen K, Kiss K, Andersen L, Therkildsen MH, Franzmann MB, Specht L, Andersen E, Norrild B, Kjaer SK, von Buchwald C. Double positivity for HPV DNA/p16 in tonsillar and base of tongue cancer improves prognostication: insights from a large population-based study. Int J Cancer. 2016;139:2598–2605. doi: 10.1002/ijc.30389. [DOI] [PubMed] [Google Scholar]
- 11.Cho JH, Kim HS, Park CS, Kim JK, Jung KY, Shin BK, Kim HK. Maspin expression in early oral tongue cancer and its relation to expression of mutant-type p53 and vascular endothelial growth factor (VEGF) Oral Oncol. 2007;43:272–277. doi: 10.1016/j.oraloncology.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Kirkwood JM, Butterfield LH, Tarhini AA, Zarour H, Kalinski P, Ferrone S. Immunotherapy of cancer in 2012. CA Cancer J Clin. 2012;62:309–335. doi: 10.3322/caac.20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobold S, Pantelyushin S, Rataj F, Vom Berg J. Rationale for combining bispecific T cell activating antibodies with checkpoint blockade for cancer therapy. Front Oncol. 2018;8:285. doi: 10.3389/fonc.2018.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Zhang J, Li X, Huang H, Liu Y, Yu M, Zhang Y, Wang H. Rescue of iCIKs transfer from PD-1/PD-L1 immune inhibition in patients with resectable tongue squamous cell carcinoma (TSCC) Int Immunopharmacol. 2018;59:127–133. doi: 10.1016/j.intimp.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Nordfors C, Grun N, Tertipis N, Ahrlund-Richter A, Haeggblom L, Sivars L, Du J, Nyberg T, Marklund L, Munck-Wikland E, Nasman A, Ramqvist T, Dalianis T. CD8+ and CD4+ tumour infiltrating lymphocytes in relation to human papillomavirus status and clinical outcome in tonsillar and base of tongue squamous cell carcinoma. Eur J Cancer. 2013;49:2522–2530. doi: 10.1016/j.ejca.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu K, Iyoda T, Okada M, Yamasaki S, Fujii SI. Immune suppression and reversal of the suppressive tumor microenvironment. Int Immunol. 2018;30:445–454. doi: 10.1093/intimm/dxy042. [DOI] [PubMed] [Google Scholar]
- 18.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 19.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, Thomson E, Monteiro R, Guimaraes P, Walters B, Wiser J, Butte AJ. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JJ, Kao KC, Chiu YL, Jung CJ, Liu CJ, Cheng SJ, Chang YL, Ko JY, Chia JS. Enrichment of human CCR6(+) regulatory t cells with superior suppressive activity in oral cancer. J Immunol. 2017;199:467–476. doi: 10.4049/jimmunol.1601815. [DOI] [PubMed] [Google Scholar]
- 26.Lei Y, Xie Y, Tan YS, Prince ME, Moyer JS, Nor J, Wolf GT. Telltale tumor infiltrating lymphocytes (TIL) in oral, head & neck cancer. Oral Oncol. 2016;61:159–165. doi: 10.1016/j.oraloncology.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao Y, Li H, Mao L, Yang QC, Fu LQ, Wu CC, Liu B, Sun ZJ. CD103(+) T and dendritic cells indicate a favorable prognosis in oral cancer. J Dent Res. 2019;98:1480–1487. doi: 10.1177/0022034519882618. [DOI] [PubMed] [Google Scholar]
- 28.Koike K, Dehari H, Shimizu S, Nishiyama K, Sonoda T, Ogi K, Kobayashi J, Sasaki T, Sasaya T, Tsuchihashi K, Tsukahara T, Hasegawa T, Torigoe T, Hiratsuka H, Miyazaki A. Prognostic value of HLA class I expression in patients with oral squamous cell carcinoma. Cancer Sci. 2020;111:1491–1499. doi: 10.1111/cas.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Y, Qin X. Profiles of immune cell infiltration and their clinical significance in head and neck squamous cell carcinoma. Int Immunopharmacol. 2020;82:106364. doi: 10.1016/j.intimp.2020.106364. [DOI] [PubMed] [Google Scholar]
- 30.Feng B, Shen Y, Pastor X, Bieg M, Plath M, Ishaque N, Eils R, Freier K, Weichert W, Zaoui K, Hess J. Integrative analysis of multi-omics data identified EGFR and PTGS2 as key nodes in a gene regulatory network related to immune phenotypes in head and neck cancer. Clin Cancer Res. 2020;26:3616–3628. doi: 10.1158/1078-0432.CCR-19-3997. [DOI] [PubMed] [Google Scholar]
- 31.Luo JJ, Young CD, Zhou HM, Wang XJ. Mouse models for studying oral cancer: impact in the era of cancer immunotherapy. J Dent Res. 2018;97:683–690. doi: 10.1177/0022034518767635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh H, Horiuchi Y, Nagasaki T, Sakonju I, Kakuta T, Fukushima U, Uchide T, Yamashita M, Kuwabara M, Yusa S, Takase K. Evaluation of immunological status in tumor-bearing dogs. Vet Immunol Immunopathol. 2009;132:85–90. doi: 10.1016/j.vetimm.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Karn T, Jiang T, Hatzis C, Sanger N, El-Balat A, Rody A, Holtrich U, Becker S, Bianchini G, Pusztai L. Association between genomic metrics and immune infiltration in triple-negative breast cancer. JAMA Oncol. 2017;3:1707–1711. doi: 10.1001/jamaoncol.2017.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shalapour S, Karin M. Pas de deux: control of anti-tumor immunity by cancer-associated inflammation. Immunity. 2019;51:15–26. doi: 10.1016/j.immuni.2019.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collignon E, Canale A, Al Wardi C, Bizet M, Calonne E, Dedeurwaerder S, Garaud S, Naveaux C, Barham W, Wilson A, Bouchat S, Hubert P, Van Lint C, Yull F, Sotiriou C, Willard-Gallo K, Noel A, Fuks F. Immunity drives TET1 regulation in cancer through NF-kappaB. Sci Adv. 2018;4:eaap7309. doi: 10.1126/sciadv.aap7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giese M, Turiello N, Molenda N, Palesch D, Meid A, Schroeder R, Basilico P, Benarafa C, Halatsch ME, Zimecki M, Westhoff MA, Wirtz CR, Burster T. Exogenous cathepsin G upregulates cell surface MHC class I molecules on immune and glioblastoma cells. Oncotarget. 2016;7:74602–74611. doi: 10.18632/oncotarget.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazanietz MG, Durando M, Cooke M. CXCL13 and its receptor CXCR5 in cancer: inflammation, immune response, and beyond. Front Endocrinol (Lausanne) 2019;10:471. doi: 10.3389/fendo.2019.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmittnaegel M, Rigamonti N, Kadioglu E, Cassara A, Wyser Rmili C, Kiialainen A, Kienast Y, Mueller HJ, Ooi CH, Laoui D, De Palma M. Dual angiopoietin-2 and VEGFA inhibition elicits antitumor immunity that is enhanced by PD-1 checkpoint blockade. Sci Transl Med. 2017;9:eaak9670. doi: 10.1126/scitranslmed.aak9670. [DOI] [PubMed] [Google Scholar]
- 39.Motta Rda R, Zettler CG, Cambruzzi E, Jotz GP, Berni RB. Ki-67 and p53 correlation prognostic value in squamous cell carcinomas of the oral cavity and tongue. Braz J Otorhinolaryngol. 2009;75:544–549. doi: 10.1016/S1808-8694(15)30494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu N, Palmer DC, Robeson AC, Shou P, Bommiasamy H, Laurie SJ, Willis C, Dotti G, Vincent BG, Restifo NP, Serody JS. STING agonist promotes CAR T cell trafficking and persistence in breast cancer. J Exp Med. 2021;218:e20200844. doi: 10.1084/jem.20200844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parikh A, Shin J, Faquin W, Lin DT, Tirosh I, Sunwoo JB, Puram SV. Malignant cell-specific CXCL14 promotes tumor lymphocyte infiltration in oral cavity squamous cell carcinoma. J Immunother Cancer. 2020;8:e001048. doi: 10.1136/jitc-2020-001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Mao L, Liu JF, Chen L, Yu GT, Yang LL, Wu H, Bu LL, Kulkarni AB, Zhang WF, Sun ZJ. Blockade of TIGIT/CD155 signaling reverses T-cell exhaustion and enhances antitumor capability in head and neck squamous cell carcinoma. Cancer Immunol Res. 2019;7:1700–1713. doi: 10.1158/2326-6066.CIR-18-0725. [DOI] [PubMed] [Google Scholar]
- 43.Mei Z, Huang J, Qiao B, Lam AK. Immune checkpoint pathways in immunotherapy for head and neck squamous cell carcinoma. Int J Oral Sci. 2020;12:16. doi: 10.1038/s41368-020-0084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyauchi S, Kim SS, Pang J, Gold KA, Gutkind JS, Califano JA, Mell LK, Cohen EEW, Sharabi AB. Immune modulation of head and neck squamous cell carcinoma and the tumor microenvironment by conventional therapeutics. Clin Cancer Res. 2019;25:4211–4223. doi: 10.1158/1078-0432.CCR-18-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu C, Jiang A. Dendritic cells and CD8 T cell immunity in tumor microenvironment. Front Immunol. 2018;9:3059. doi: 10.3389/fimmu.2018.03059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res. 2014;2:91–98. doi: 10.1158/2326-6066.CIR-13-0216. [DOI] [PubMed] [Google Scholar]
- 47.Asadzadeh Z, Mohammadi H, Safarzadeh E, Hemmatzadeh M, Mahdian-Shakib A, Jadidi-Niaragh F, Azizi G, Baradaran B. The paradox of Th17 cell functions in tumor immunity. Cell Immunol. 2017;322:15–25. doi: 10.1016/j.cellimm.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 48.Andrews MC, Reuben A, Gopalakrishnan V, Wargo JA. Concepts collide: genomic, immune, and microbial influences on the tumor microenvironment and response to cancer therapy. Front Immunol. 2018;9:946. doi: 10.3389/fimmu.2018.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]