Abstract
4-Hydroxyphenylacetate 3-hydroxylase (HpaB and HpaC) of Escherichia coli W has been reported as a two-component flavin adenine dinucleotide (FAD)-dependent monooxygenase that attacks a broad spectrum of phenolic compounds. However, the function of each component in catalysis is unclear. The large component (HpaB) was demonstrated here to be a reduced FAD (FADH2)-utilizing monooxygenase. When an E. coli flavin reductase (Fre) having no apparent homology with HpaC was used to generate FADH2 in vitro, HpaB was able to use FADH2 and O2 for the oxidation of 4-hydroxyphenylacetate. HpaB also used chemically produced FADH2 for 4-hydroxyphenylacetate oxidation, further demonstrating that HpaB is an FADH2-utilizing monooxygenase. FADH2 generated by Fre was rapidly oxidized by O2 to form H2O2 in the absence of HpaB. When HpaB was included in the reaction mixture without 4-hydroxyphenylacetate, HpaB bound FADH2 and transitorily protected it from rapid autoxidation by O2. When 4-hydroxyphenylacetate was also present, HpaB effectively competed with O2 for FADH2 utilization, leading to 4-hydroxyphenylacetate oxidation. With sufficient amounts of HpaB in the reaction mixture, FADH2 produced by Fre was mainly used by HpaB for the oxidation of 4-hydroxyphenylacetate. At low HpaB concentrations, most FADH2 was autoxidized by O2, causing uncoupling. However, the coupling of the two enzymes' activities was increased by lowering FAD concentrations in the reaction mixture. A database search revealed that HpaB had sequence similarities to several proteins and gene products involved in biosynthesis and biodegradation in both bacteria and archaea. This is the first report of an FADH2-utilizing monooxygenase that uses FADH2 as a substrate rather than as a cofactor.
Two major groups of flavin-dependent monooxygenases have been reported. The first group usually consists of enzymes of single polypeptides that have either flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN) as their prosthetic group and use either NADH or NADPH as their cosubstrate (8, 30). The second group utilizes reduced FMN (FMNH2) as a cosubstrate rather than a prosthetic group and requires a separate FMN reductase to supply FMNH2. Bacterial luciferase of Photobacterium fischeri is the first reported FMNH2-utilizing monooxygenase (32). Recently, pristinamycin IIA synthase of Streptomyces pristinaespiralis (24), nitrilotriacetate monooxygenase of Chelatobacter heintzii (34), EDTA monooxygenases of two EDTA-degrading bacteria (22, 33), and two monooxygenases involved in desulfurization from Rhodococcus sp. strain IGTS8 (12, 19) have been characterized and shown to be FMNH2-utilizing monooxygenases. These FMNH2-utilizing monooxygenases appear to attack carbon-nitrogen bonds, carbon-sulfur bonds, carbon-carbon double bonds, or aldehyde groups of nonaromatic compounds. FMNH2 is supplied by FMN reductases with NADH as a reductant.
Recently, several two-component FAD-dependent monooxygenases have been reported (5, 23, 35); however, the function of each component in catalysis is unknown. In our recent study we have found that the FMN reductase that supplies FMNH2 to nitrilotriacetate monooxygenase has significant sequence similarities to the small component (HpaC) of a two-component monooxygenase, 4-hydroxyphenylacetate 3-hydroxylase (HpaB and HpaC) of Escherichia coli strain W (34). It has been suggested that HpaC is a coupling factor that enhances NADH oxidation for HpaB (23). Because HpaB and HpaC together catalyze the oxidation of 4-hydroxyphenylacetate in the presence of NADH and FAD, we hypothesized that HpaC was a flavin reductase that could reduce FAD to FADH2 and that HpaB was an FADH2-utilizing monooxygenase. We present here the characterization of HpaB as a new type of FADH2-utilizing monooxygenase. The large components of several other two-component FAD-dependent monooxygenases have both catalytic and sequence similarities to HpaB and may also be FADH2-utilizing monooxygenases.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli NovaBlue was used for pET-30 LIC cloning, and strain BL21(DE3) was used for gene expression (Novagen, Madison, Wis.). E. coli strains were routinely grown at 37°C in Luria-Bertani (LB) medium or on LB agar (26), except for strain BL21(DE3) that was incubated at room temperature when used to produce functional enzymes. Kanamycin (Sigma, St. Louis, Mo.) was used at 30 μg per ml in culture media.
Gene cloning and expression.
To overproduce NAD(P)H:flavin oxidoreductase (Fre) (27) in E. coli, PCR primers were designed to clone fre into pET-30 LIC vector (Novagen). The forward primer (Fre5, 5′-ACA-GAG-AAA-GCA-TAT-GAC-AAC-CTT-3′) was at base positions of 1439 to 1462 of gene fre (GenBank M61182) (27), in which an NdeI site (underlined) was introduced by altering two bases. The reverse primer (Fre3, 5′-AAA-TGC-CAC-TGA-ATT-CCA-GTT-TAG-3′) was located at positions 2196 to 2219 with an introduced EcoRI site. The gene was amplified with the primers and genomic DNA from E. coli DH5α for 25 cycles of PCR with a thermal profile of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. The amplification yielded a DNA product of 781 bp. The PCR product was cut by NdeI and EcoRI and then ligated into the plasmid pET30-LIC (Novagen) that was previously digested by NdeI and EcoRI to produce plasmid pES1. For overproduction of HpaB, the forward primer (HpaB5, 5′-GTA-GAG-GTC-CAT-ATG-AAA-CCA-GAA-3′) was from base positions 1100 to 1123 (GenBank Z29081) (24) with an introduced NdeI site, and the reverse primer (HpaB3, 5′-TGC-ATC-TTA-AGC-TTC-TGC-TGC-GTT-3′) was from base positions 2696 to 2673 with an introduced HindIII site. The gene was amplified by PCR with plasmid pAJ224 (24) DNA as template and cloned into pET-30 LIC vector to produce pES2. Plasmids pES1 and pES2 were transformed into E. coli NovaBlue for plasmid identification and recovery. The recovered plasmids were later transformed into E. coli BL21(DE3) for protein production upon IPTG (isopropyl-β-d-thiogalactopyranoside) induction (Novagen).
Protein purification.
Protein Fre was purified according to a previously reported method (9). Because the protein was overproduced, not all of the steps reported before were needed to purify it to homogeneity. BL21(DE3) cells (0.4 g [wet weight]) carrying pES1 harvested from 200 ml of culture were suspended in 10 ml of 20 mM potassium phosphate (KPi) buffer (pH 7.0) with 2 mM EDTA. The cell suspension was passed through a French pressure cell once at 18,000 lb/in2. The sample was centrifuged at 18,000 × g for 15 min to remove cell debris. Solid ammonium sulfate was added to the supernatant to bring it to 70% saturation. The precipitate was sedimented by centrifugation, resuspended in 1 ml of 20 mM KPi buffer (pH 7.0), and dialyzed overnight in the same buffer. The dialyzed sample was injected onto a Bioscale Q column (7 by 52 mm; Bio-Rad, Hercules, Calif.), and proteins were eluted with a 20-ml gradient of 0 to 400 mM NaCl in 20 mM KPi buffer (pH 7.0). Fre was eluted off the column around 200 mM NaCl as a major peak. Fre was collected, concentrated to less than 0.8 ml, and injected onto a Superdex 75 column (10 by 300 mm; Pharmacia, Alameda, Calif.). Fre was eluted with the 20 mM KPi buffer containing 150 mM NaCl. Protein HpaB was purified by using a slightly modified procedure from that previously reported (24). E. coli cells (2.5 g) producing HpaB were harvested from 1-liter cultures, suspended in 20 ml of 20 mM KPi buffer (pH 7.0) with 2 mM EDTA and 1 mM dithiothreitol (DTT), and broken by passage through a French press cell three times at 1,800 lb/in2. The cell lysate was then centrifuged at 18,000 × g for 15 min to remove cell debris. Solid ammonium sulfate was added to the supernatant to 20% saturation with constant mixing. The sample was then centrifuged as described above to remove precipitated proteins. The supernatant containing less than 130 mg of protein was loaded onto a phenyl agarose column (16 by 120 mm) and eluted with 100 ml of a linear gradient of ammonium sulfate (20 to 0% saturation) in the 20 mM KPi buffer. When the gradient finished, HpaB was eluted off the column by another 40 ml of the 20 mM KPi buffer. HpaB was concentrated to about 2 ml, and trace amounts of ammonium sulfate was removed by dialysis in the KPi buffer with 1 mM DTT for several hours. The sample was injected onto a 2-ml Bioscale Q column and eluted with a 20-ml gradient of NaCl from 0 to 200 mM in the KPi buffer with 1 mM DTT. HpaB was eluted off the column around 100 mM NaCl.
Enzyme assays.
4-Hydroxyphenylacetate 3-hydroxylase activity was measured by analysis of the conversion of 4-hydroxyphenylacetate to 3,4-dihydroxyphenylacetate with a high-pressure liquid chromatography (HPLC) method as previously reported (18). The reaction was normally performed with 20 mM KPi buffer (pH 7.0) containing 10 μM FAD, 1 mM 4-hydroxyphenylacetate, 2 mM NADH, 90 U of catalase (Sigma) per ml, and various amounts of Fre and HpaB at 24°C. One unit was defined as the amount of HpaB required to catalyze the consumption of 1 nmol of 4-hydroxyphenylacetate per min when HpaB was the limiting factor in the assays. Flavin reductase activity was determined by monitoring the consumption of NADH in 20 mM KPi buffer (pH 7.0) containing 200 μM NADH and 10 μM FAD or FMN at 24°C. One unit of Fre was defined as the amount required to catalyze the consumption of 1 nmol of NADH per min (9). FAD concentration changes during FAD reduction were monitored at 450 nm. The 4-hydroxyphenylacetate 3-hydroxylase activity of HpaB was also tested with chemically produced FADH2 by using a reaction mixture containing 1 mM 4-hydroxyphenylacetate, 10 μM HpaB, and 100 μM FAD in 20 mM KPi buffer (pH 7.0). The mixture was transferred into an anaerobic chamber, and FAD was reduced to FADH2 with 3 mM titanium citrate (36). The mixture was then removed from the anaerobic chamber, and O2 from air was permitted to diffuse into the reaction mixture to complete the reaction. After 30 min, the amount of 3,4-dihydroxyphenylacetate produced was analyzed by HPLC.
Analytical methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done by the method of Laemmli (17). Gels were stained for proteins with GelCode Blue (Pierce, Rockford, Ill.). Protein concentrations were determined with a protein dye reagent (6) with bovine serum albumin as the standard.
Oxygen consumption.
Oxygen consumption was determined in a closed reaction vessel (0.6 ml, total volume) fitted with a Clark-type oxygen electrode (Instech, Plymouth Meeting, Pa.). The electrode was calibrated with a chemical method by using N-methylphynazonium methosulfate and NADH to quantitatively consume O2 (25). Two sets of experiments were carried out with 10 μM FAD in the reaction mixture. The first set contained 53 U of Fre only. The second set contained 53 U of Fre and 10 μM HpaB. In both cases, NADH was added in a 2.5-μl volume to a final concentration of 84 μM to initiate O2 consumption. When O2 consumption stopped, 90 U of catalase (Sigma) was added to release O2 from H2O2. After the reactions were completed, the 3,4-dihydroxyphenylacetate produced was quantified by HPLC.
RESULTS
Protein production.
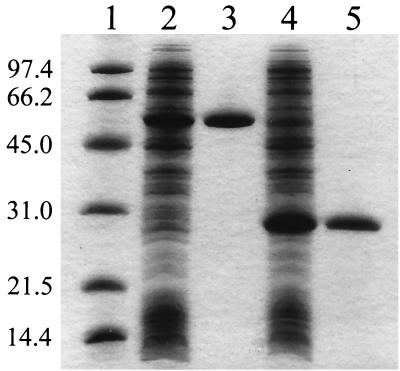
For ease of purification, the genes fre and hpaB were cloned into the pET-30 LIC vector to yield plasmids pES1 and pES2, respectively. Strain BL21(DE3) carrying pES1 or pES2 produced large quantities of soluble and active Fre or HpaB when growing at room temperature (Fig. 1). When growing at 37°C, the cells produced these proteins mainly in inclusion bodies. Neither Fre nor HpaB were fusion proteins. They were both purified to apparent homogeneity (Fig. 1). For a typical purification, 2.6 mg of Fre was purified from 27 mg of total protein in the cell extract with a 32% recovery of Fre activity, and 22 mg of HpaB was purified from 130 mg of the cell extract with a 55% recovery of HpaB activity. The purified Fre had a specific activity of 39,899 U mg−1 when reducing FMN or 33,515 U mg−1 when reducing FAD. All other Fre activity was reported as for FAD reduction. The purified HpaB had a specific activity of 231 U mg−1 for 4-hydroxyphenylacetate oxidation.
FIG. 1.
SDS-PAGE analysis of HpaB and Fre. Lane 1, molecular mass standards in kilodaltons (Bio-Rad); lane 2, 45 μg of the extract of E. coli cells overproducing HpaB; lane 3, 6.5 μg of HpaB; lane 4, 45 μg of the extract of E. coli cells overproducing Fre; and lane 5, 6.5 μg of Fre.
4-Hydroxyphenylacetate oxidation by HpaB.
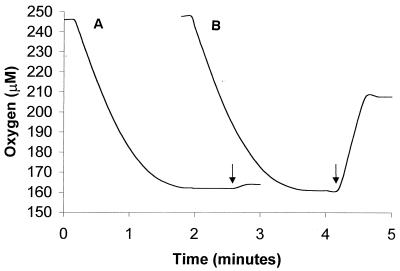
In order to demonstrate that HpaB was an FADH2-utilizing monooxygenase, a flavin reductase (Fre) was used to supply FADH2. When both HpaB and Fre were added to a reaction mixture containing FAD, 4-hydroxyphenylacetate was oxidized to 3,4-dihydroxyphenylacetate at the expense of NADH and O2. With 120 U of HpaB and 55 U of Fre in the reaction mixture, 50 nmol of O2 was consumed from 50 nmol of NADH added. Upon addition of catalase at the end of O2 consumption, about 1.2 ± 0.2 (standard deviation of three samples) nmol of O2 was released from H2O2 (Fig. 2). HPLC analysis showed the production of 46 ± 2 (standard deviation of three samples) nmol of 3,4-dihydroxyphenylacetate. When HpaB alone was added to the reaction mixture, there was no consumption of NADH, 4-hydroxyphenylacetate, or O2. In contrast, Fre alone led to the quantitative consumption of 50 nmol of NADH and 50 nmol of O2 without the oxidation of 4-hydroxyphenylacetate (Fig. 2). In this case, O2 was converted to H2O2, from which 25 ± 1 (standard deviation of three samples) nmol of O2 was released by catalase (Fig. 2). Therefore, in the presence of both proteins FADH2 generated by Fre was mainly oxidized by HpaB for the conversion of 4-hydroxyphenylacetate to 3,4-dihydroxyphenylacetate, whereas with Fre alone FAD was reduced to FADH2 and then autoxidized by O2 to generate H2O2. Since the oxygen consumption rates were almost the same in the presence or the absence of HpaB (Fig. 2), it is likely that the reduction of FAD was rate limiting; that is, FADH2 oxidation by either HpaB or free O2 was much faster than FAD reduction. HpaB also used chemically produced FADH2 for 4-hydroxyphenylacetate oxidation. In a mixture containing 100 nmol of chemically reduced FADH2 with an excess of titanium citrate, 128 nmol of 3,4-dihydroxyphenylacetate was produced. Since the oxidized FAD could be continuously reduced by titanium until all of the reducing agent was consumed, more than 100 nmol of end product was formed. Due to the extreme reactivity of both FADH2 and titanium with O2, it is difficult to quantitatively supply chemically produced FADH2 to HpaB for 4-hydroxyphenylacetate oxidation. Controls without HpaB or FAD did not produce 3,4-dihydroxyphenylacetate.
FIG. 2.
Oxygen consumption by HpaB and Fre (A) and by Fre only (B). At the end of each run, catalase was added as indicated by the arrows. Data presented in the text were calculated by using a volume of 0.6 ml.
FAD reduction and FADH2 oxidation.
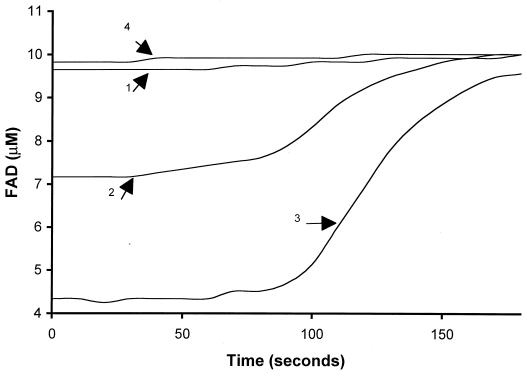
When the concentration of FAD was followed at 450 nm during FAD reduction in 1 ml of reaction mixture by 176 U of Fre, FAD (9.6 μM) and FADH2 (0.40 ± 0.05 μM) (standard deviation of three samples) concentrations were found to remain in a constant ratio until NADH was almost completely consumed at about 100 s (Fig. 3, curve 1). The lack of FADH2 accumulation during FAD reduction again indicates rapid autoxidation of FADH2. When HpaB was also added to the reaction mixture without 4-hydroxyphenylacetate, the concentration of FADH2 increased proportionally to the amount of HpaB added (Fig. 3, curves 2 and 3). With 5 μM HpaB, 3.1 ± 0.3 μM FADH2 (standard deviation of three samples) was stable for about 80 s. FADH2 was slowly oxidized at a maximal rate of 0.053 μM per s after most of the NADH was consumed (Fig. 3, curve 2). Since FADH2 was rapidly autoxidized by O2, the increased FADH2 should be due to the presence of HpaB, which bound and protected FADH2 from rapid oxidation by O2. When 10 μM HpaB was added, 6.3 ± 0.6 μM FADH2 (standard deviation of three samples) was detected and remained at that level for about 80 s. After most of the NADH was consumed, FADH2 was oxidized relatively slowly at a maximal rate of 0.088 μM per second (Fig. 3, curve 3). Even in the presence of 5 and 10 μM HpaB, FAD reduction as measured by NADH consumption remained the same as with Fre alone since the remaining FAD concentrations were still much higher than Fre's Km for FAD (0.8 μM) (9). When the NADH was completely consumed (by approximately 100 s), all FADH2 was slowly oxidized to FAD.
FIG. 3.
The FAD concentrations during FAD reduction. All reactions contained 200 μM NADH, 176 U of Fre, and various amount of HpaB in 1 ml of 20 mM KPi buffer (pH 7.0). The FAD concentration dynamics was monitored during FAD (10 μM) reduction by Fre only (arrow 1), by Fre with 5 μM HpaB (arrow 2), by Fre with 10 μM HpaB (arrow 3), and by Fre with 10 μM HpaB and 1 mM 4-hydroxyphenylacetate (arrow 4). NADH consumption rates were very similar for all of the reactions, and NADH was completely consumed within 100 s (data not shown).
Coupling the activities of HpaB and Fre.
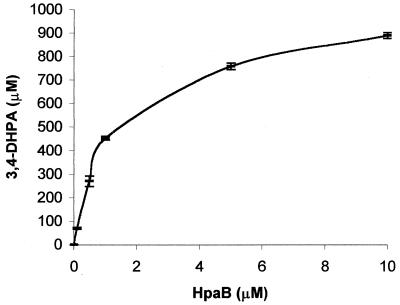
When 4-hydroxyphenylacetate, a substrate of HpaB, was also included in the reaction mixture, only 0.24 ± 0.06 μM FADH2 (standard deviation of three samples) was detected in the initial 30 s (Fig. 3, curve 4). In this case, FADH2 was oxidized by HpaB. 4-Hydroxyphenylacetate oxidation was detected by HPLC, and 186 ± 3 nmol (standard deviation of three samples) of 3,4-dihydroxyphenylacetate was produced from 200 nmol of NADH consumed. The coupling of Fre and HpaB activities was good in this case. When the amount of HpaB was reduced, the coupling decreased as the final production of 3,4-dihydroxyphenylacetate decreased (Fig. 4). This uncoupling was alleviated by reducing the amount of FAD in the reaction mixture. When FAD concentrations were around Fre's Km (0.8 μM), the final production of 3,4-dihydroxyphenylacetate clearly increased (Fig. 5). The uncoupling was also alleviated by reducing the amount of Fre instead of FAD in the reaction mixture (data not shown). Since autoxidation of FADH2 by O2 to H2O2 is well documented (11, 13) and was also demonstrated in Fig. 2, the H2O2 produced was not measured in these experiments. Fre also reduces riboflavin and FMN at the expense of NADH (9), but HpaB did not use the reduced riboflavin nor FMNH2 for 4-hydroxyphenylacetate oxidation. The reduced riboflavin and FMNH2 were immediately autoxidized by O2. In addition, HpaB did not protect FMNH2 and reduced riboflavin from rapid autoxidation by O2.
FIG. 4.
Relationship between HpaB concentrations and final 3,4-dihydroxyphenylacetate (3,4-DHPA) production. The reactions contained 176 U of Fre, 1 mM NADH, 1 mM 4-hydroxyphenylacetate, 10 μM FAD, and various amounts of HpaB in 1 ml of 20 mM KPi buffer. The 10 μM HpaB in 1 ml had 134 U of activity. NADH consumption was completed within 10 min, but samples were incubated for 2 h to ensure the completion of reactions.
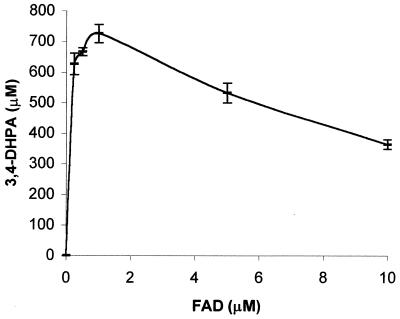
FIG. 5.
Relationship between FAD concentrations and final 3,4-dihydroxyphenylacetate (3,4-DHPA) production. The reactions contained 176 U of Fre, 1 mM NADH, 1 mM 4-hydroxyphenylacetate, 1 μM HpaB, and various amounts of FAD. The mixtures were incubated for 2 h. NADH in all reactions were completely consumed after 2 h of incubation as detected by the HPLC method used to detect 3,4-dihydroxyphenylacetate.
DISCUSSION
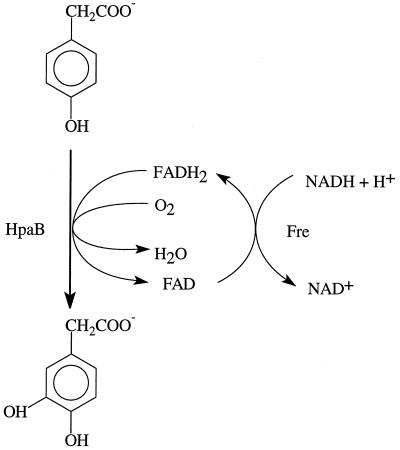
Functional analysis demonstrated that HpaB is an FADH2-utilizing monooxygenase that catalyzes the reaction as shown in Fig. 6. Our results show that only HpaB is responsible for 4-hydroxyphenylacetate oxidation. Since HpaB used either chemically generated FADH2 or enzymatically generated FADH2 for 4-hydroxyphenylacetate oxidation, HpaC was not required for the function of HpaB. A continuous supply of FADH2 was easily achieved by Fre reduction of FAD with NADH as a reductant. Fre has no apparent sequence similarities to HpaC. Fre reduces FAD, FMN, and riboflavin at the expense of NADH in the presence or absence of HpaB. Only FADH2 was used by HpaB because FAD could not be substituted by either FMN or riboflavin in the reaction mixture containing Fre. HpaC was not characterized in this report, but a recently reported NADPH:FAD oxidoreductase that supplies FMNH2 to an FMNH2-utilizing monooxygenase from Streptomyces viridifaciens (21) has significant sequence homology with HpaC and uses either NADPH or NADH to reduce either FAD or FMN. HpaC may possess similar catalytic properties.
FIG. 6.
Coupled reactions catalyzed by Fre and HpaB.
Although FADH2 is rapidly autoxidized by O2, HpaB competed effectively with O2 to use FADH2 for 4-hydroxyphenylacetate oxidation. In the absence of HpaB, FADH2 was quantitatively autoxidized to H2O2 (Fig. 2). In contrast, under the conditions of Fig. 2 only 5% of FADH2 was autoxidized by O2 to H2O2 during 4-hydroxyphenylacetate oxidation by HpaB. Comparison of FADH2 concentrations also indicates that FADH2 utilization by HpaB is slightly faster than the autoxidation process because the FADH2 concentration was lower during 4-hydroxyphenylacetate oxidation (Fig. 3, curve 4) than during FADH2 autoxidation (Fig. 3, curve 1). When HpaB concentrations were low, only a small fraction of FADH2 was used by HpaB for the oxidation of 4-hydroxyphenylacetate and a significant amount of FADH2 was autoxidized (Fig. 4 and 5). In the extreme case without HpaB, all FADH2 was autoxidized to H2O2 (Fig. 2) (11, 13). However, lowering the FAD concentration in the reaction mixture can slow down FAD reduction and increase the FADH2 utilization by HpaB (Fig. 5). If the coupling of the activities of an FAD reductase and an FADH2-utilizing monooxygenase is important, FAD concentrations should be lowered to near the Km of the reductase to ensure coupling. On the other hand, HpaB's activities did not affect Fre's activities because NADH consumption rates (data not shown) were very similar during FADH2 oxidation either by O2 or by HpaB. In addition, the oxygen consumption rates were also similar during FADH2 oxidation either by O2 to FAD and H2O2 or by HpaB for the oxidation of 4-hydroxyphenylacetate (Fig. 2). These data indicate that FADH2 oxidation by free O2 is faster than FAD reduction and HpaB can compete with free O2 for FADH2 utilization.
Figure 3 also provides evidence that HpaB binds FADH2. In the absence of 4-hydroxyphenylacetate, HpaB bound FADH2 and protected it from rapid autoxidation by O2. This protection was proportional to the amount of HpaB in the reaction mixture and was only transitory (Fig. 3). It is unclear whether FADH2 is slowly dissociated from HpaB and the free FADH2 then gets rapidly autoxidized, whether FADH2 is slowly oxidized by O2 while bound to HpaB and then FAD is rapidly released, or whether both processes occur simultaneously. No matter how FADH2 is oxidized by O2 in the presence of HpaB, it is clear that HpaB can bind FADH2 in the absence of 4-hydroxyphenylacetate.
The main difference between an FADH2-utilizing monooxygenase and FMNH2-utilizing monooxygenases is the cosubstrate (FADH2 or FMNH2) used. Among FMNH2-utilizing monooxygenases only isobutylamine N-hydroxylase involved in valanimycin synthesis has the ability to use either FMNH2 or FADH2, but it prefers to use FMNH2 (20). The second difference is the substrates. All the FMNH2-utilizing monooxygenases reported so far oxidize nonaromatic compounds (19, 22, 31–34), while the FADH2-utilizing monooxygenase reported here hydroxylates aromatic compounds. Structurally, HpaB does not have any apparent sequence similarities to any FMNH2-utilizing monooxygenases.
HpaB shows high homology with TftD (the large component of chlorophenol 4-monooxygenase) from Burkholderia cepacia AC1100, HadA (the large component of chlorophenol-4-hydroxylase) from B. pickettii DTP0602, and HpaA (the large component of 4-hydroxyphenylacetate 3-hydroxylase) from Klebsiella pneumoniae (14). On the basis of sequence similarity and the two component nature of these enzymes (10, 14, 29, 35), TftD (14, 35), HadA (29), and HpaA (10) may also belong to FADH2-utilizing monooxygenases that all use aromatic compounds as their substrates. We have recently demonstrated that TftD used FADH2 generated by Fre for the oxidation of chlorophenols (data not shown). Interestingly, there is a well-characterized two component 4-hydroxyphenylacetate 3-hydroxylase with the small component containing FAD and the large component serving as a coupling factor for 4-hydroxyphenylacetate oxidation (2, 3, 4). It seems that the large component of this enzyme is different from HpaC because it is only 38 kDa (2), whereas HpaC is about 58 kDa (23).
A BLAST search (1) with the amino acid sequence of HpaB revealed that HpaB also has sequence similarities to seven other proteins. These seven proteins include a phenol hydroxylase (PheA) that oxidizes phenol to catechol from Bacillus thermoleovorans (7). This is not surprising since HpaB also oxidizes phenol to catechol (23). Another protein is PvcC, whose gene is part of a gene cluster involved in siderophore pyoverdine biosynthesis but whose function has not been reported (28). PvcC may catalyze the oxidation of l-tyrosine to l-dopa, the first step in the biosynthesis pathway, because both HpaB (18) and HpaA (10) can oxidize l-tyrosine to l-dopa. All the other five proteins are deduced from genome projects on the basis of sequence similarities to HpaB, one from Photorhabdus luminescens (GenBank AF021838), one from Bacillus subtilis (16), and three from Archaeoglobus fulgidus (15). It is unclear why a strict anaerobic archaeon A. fulgidus has genes encoding three monooxygenases. If all these enzymes are FADH2-utilizing monooxygenases, they are a new class of enzymes widespread in the microbial world involved in both biodegradation and biosynthesis.
ACKNOWLEDGMENTS
This research was supported by NSF grant MCB-9722970.
We thank Jose L. Garcia for providing plasmid pAJ224.
REFERENCES
- 1.Altschul S F, Lipman D J. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arunachalam U, Massey V, Vaidyanathan C S. p-Hydroxyphenylacetate-3-hydroxylase: a two-protein component enzyme. J Biol Chem. 1992;267:25848–25855. [PubMed] [Google Scholar]
- 3.Arunachalam U, Massey V, Miller S M. Mechanism of p-Hydroxyphenylacetate-3-hydroxylase: a two-protein enzyme. J Biol Chem. 1994;269:150–155. [PubMed] [Google Scholar]
- 4.Arunachalam U, Massey V. Studies on the oxidative half-reaction of p-Hydroxyphenylacetate-3-hydroxylase. J Biol Chem. 1994;269:11795–11801. [PubMed] [Google Scholar]
- 5.Becker D, Schrader T, Andreesen J R. Two-component flavin-dependent pyrrole-2-carboxylate monooxygenase from Rhodococcus sp. Eur J Biochem. 1997;249:739–747. doi: 10.1111/j.1432-1033.1997.t01-1-00739.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Duffner F M, Mueller R. A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovorans strain A2: nucleotide sequence and analysis of the genes. FEMS Microbiol Lett. 1998;161:37–45. doi: 10.1111/j.1574-6968.1998.tb12926.x. [DOI] [PubMed] [Google Scholar]
- 8.Flashner M S, Massey V. Flavoprotein oxygenases. In: Hayaishi O, editor. Molecular mechanisms of oxygen activation. New York, N.Y: Academic Press, Inc.; 1974. pp. 245–283. [Google Scholar]
- 9.Fontecave M, Eliasson R, Reichard P. NAD(P)H:flavin oxidoreductase of Escherichia coli. J Biol Chem. 1987;262:12325–12331. [PubMed] [Google Scholar]
- 10.Gibello A, Ferrer E, Sanz L, Ramos J L. Polymer production by Klebsiella pneumoniae 4-hydroxyphenylacetic acid hydroxylase genes cloned in Escherichia coli. Appl Environ Microbiol. 1995;61:4167–4171. doi: 10.1128/aem.61.12.4167-4171.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson Q H, Hastings J W. The oxidation of reduced flavin mononucleotide by molecular oxygen. Biochem J. 1962;83:368–377. doi: 10.1042/bj0830368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray K A, Pogrebinsky O S, Mrachko G T, Xi L, Monticello D J, Squires C H. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat Biotechnol. 1996;14:1705–1709. doi: 10.1038/nbt1296-1705. [DOI] [PubMed] [Google Scholar]
- 13.Gutfreund H. The reactions of reduced flavine nucleotides with oxygen. Biochem J. 1960;74:17. p. [Google Scholar]
- 14.Hubner A, Danganan C E, Xun L, Chakrabarty A M, Hendrickson W. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia AC1100: characterization of the tftC and tftD genes and locations of the tft operons on multiple replicons. Appl Environ Microbiol. 1998;64:2086–2093. doi: 10.1128/aem.64.6.2086-2093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klenk H P, et al. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 16.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee J-Y, Xun L. Novel biological process for l-dopa production from l-tyrosine by p-hydroxyphenylacetate 3-hydroxylase. Biotechnol Lett. 1998;20:479–482. [Google Scholar]
- 19.Lei B, Tu S-C. Gene overexpression, purification, and identification of a desulfurization enzyme from Rhodococcus sp. strain IGTS8 as a sulfide/sulfoxide monooxygenase. J Bacteriol. 1996;178:5699–5705. doi: 10.1128/jb.178.19.5699-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parry R J, Li W. Purification and characterization of isobutylamine N-hydroxylase from the valanimycin producer Streptomyces viridifaciens MG456-HF10. Arch Bioch Biophys. 1997;339:47–54. doi: 10.1006/abbi.1996.9857. [DOI] [PubMed] [Google Scholar]
- 21.Parry R J, Li W. An NADPH:FAD oxidoreductase from the valanimycin producer Streptomyces viridifaciens. J Biol Chem. 1997;272:23303–23311. doi: 10.1074/jbc.272.37.23303. [DOI] [PubMed] [Google Scholar]
- 22.Payne J W, Bolton H, Campbell J A, Xun L. Purification and characterization of EDTA monooxygenase from the EDTA-degrading bacterium BNC1. J Bacteriol. 1998;180:3823–3827. doi: 10.1128/jb.180.15.3823-3827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto M A, Perez-Aranda A, Garcia J L. Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J Bacteriol. 1993;175:2162–2167. doi: 10.1128/jb.175.7.2162-2167.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prieto M A, Garcia J L. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. J Biol Chem. 1994;269:22823–22829. [PubMed] [Google Scholar]
- 25.Robinson J, Cooper J M. Method of determining oxygen concentrations in biological media, suitable for calibration of oxygen electrode. Anal Biochem. 1970;33:390–399. doi: 10.1016/0003-2697(70)90310-6. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Spyrou G, Haggard-Ljungquist E, Krook M, Jornvall H, Nilsson E, Reichard P. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J Bacteriol. 1991;173:3673–3679. doi: 10.1128/jb.173.12.3673-3679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stintzi A, Cornelis P, Hohnadel D, Meyer J-M, Dean C, Poole K, Lourambas S, Krishnapillai V. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiol. 1996;142:1181–1190. doi: 10.1099/13500872-142-5-1181. [DOI] [PubMed] [Google Scholar]
- 29.Takizawa N, Yokoyama H, Yanagihara K, Hatta T, Kiyohara H. A locus of Pseudomonas pickettii DTP0602, had, that encodes 2,4,6-trichlorophenol-4-dechlorinase with hydroxylase activity, and hydroxylation of various chlorophenols by the enzyme. J Ferment Bioeng. 1995;80:318–326. [Google Scholar]
- 30.Testa B. The nature and functioning of cytochromes P450 and flavin-containing-monooxygenases. In: Testa B, Caldwell J, editors. The metabolism of drugs and other xenobiotics: biochemistry of redox reactions. San Diego, Calif: Academic Press, Inc.; 1995. pp. 70–121. [Google Scholar]
- 31.Thibaut D, Ratet N, Bisch D, Faucher D, Debussche L, Blanche F. Purification of the two enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J Bacteriol. 1995;177:5199–5205. doi: 10.1128/jb.177.18.5199-5205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu S-C, Mager H I X. Biochemistry of bacterial bioluminescence. Photochem Photobiol. 1995;62:615–624. doi: 10.1111/j.1751-1097.1995.tb08708.x. [DOI] [PubMed] [Google Scholar]
- 33.Witschel M, Nagel S, Egli T. Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103. J Bacteriol. 1997;179:6937–6943. doi: 10.1128/jb.179.22.6937-6943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Mortimer M W, Fisher T S, Kahn M L, Brockman F J, Xun L. Cloning, sequencing and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH:flavin mononucleotide oxidoreductase. J Bacteriol. 1997;179:1112–1116. doi: 10.1128/jb.179.4.1112-1116.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xun L. Purification and characterization of chlorophenol 4-monooxygenase from Pseudomonas cepacia AC1100. J Bacteriol. 1996;178:2645–2649. doi: 10.1128/jb.178.9.2645-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zehnder A J B, Wuhrmann K. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science. 1976;194:1165–1166. doi: 10.1126/science.793008. [DOI] [PubMed] [Google Scholar]