Abstract
BACKGROUND
Obstructive sleep apnea (OSA)-hypopnea syndrome (OSAHS) has been recognized as a comorbidity of type 2 diabetes mellitus (T2DM); more than half of T2DM patients suffer from OSAHS. Intermittent hypoxia (IH) plays an important role in metabolic diseases, such as obesity and OSAHS, through various mechanisms, including altering the gut microecological composition and function. Therefore, it is important to study the role of gut microbiota in T2DM patients with OSAHS, which has a high incidence and is prone to several complications.
AIM
To assess whether IH is involved in altering the fecal microbiome in T2DM patients with OSAHS.
METHODS
Seventy-eight participants were enrolled from Henan Province People’s Hospital and divided into healthy control (HC, n = 26), T2DM (n = 25), and T2DM + OSA (n = 27) groups based on their conditions. The fecal bacterial DNA of the research participants was extracted and subjected to 16S ribosomal RNA sequencing. The clinical indices, such as insulin resistance index, homocysteine (HCY) concentration, and the concentrations of inflammatory factors in the peripheral blood, were assessed and recorded.
RESULTS
Group T2DM + OSA had the highest apnea-hypopnea index (AHI) (2.3 vs 3.7 vs 13.7), oxygen desaturation index (0.65 vs 2.2 vs 9.1), HCY concentration (9.6 μmol/L vs 10.3 μmol/L vs 13.81 μmol/L) and C-reactive protein (CRP) concentrations (0.3 mg/L vs 1.43 mg/L vs 2.11 mg/L), and lowest mean oxygen saturation (97.05% vs 96.6% vs 94.7%) among the three groups. Twelve and fifteen key differences in amplicon sequence variants were identified when comparing group T2DM + OSA with groups T2DM and HC, respectively. We found progressively decreased levels of Faecalibacterium, Eubacterium, and Lachnospiraceae, and an increase in the level of Actinomyces, which strongly correlated with the HCY, CRP, fasting plasma glucose, and hemoglobin A1c concentrations, AHI, mean oxygen saturation, and insulin resistance index in group T2DM + OSA (P < 0.05).
CONCLUSION
For T2DM patients with OSAHS, IH may be involved in selective alterations of the gut microbiota, which may affect the pathophysiological development of T2DM and DM-related complications.
Keywords: Gut microbiota, Obstructive sleep apnea-hypopnea syndrome, Type 2 diabetes mellitus, Intermittent hypoxia, Obstructive sleep apnea
Core Tip: Clinically, type 2 diabetes mellitus (T2DM) patients have a significantly higher prevalence of obstructive sleep apnea-hypopnea syndrome (OSAHS) than non-T2DM patients and are more prone to diabetes-related complications and metabolic syndrome, including obesity and hypertension. In recent years, the imbalance of gut microbiota has been found to be associated with various metabolic disorders. This study revealed that intermittent hypoxia was associated with changes in the gut microbiota in patients with T2DM complicated by OSAHS. These changes may be involved in the progression of metabolic disorders through increased proinflammatory factors and impaired intestinal barrier function.
INTRODUCTION
Obstructive sleep apnea (OSA)-hypopnea syndrome (OSAHS) is characterized by recurrent partial or complete pharyngeal obstruction during sleep[1], resulting in intermittent hypoxia (IH); cyclical adverse changes in heart rate, blood pressure (BP), and sympathetic activity; and disruption of sleep architecture[2]. Epidemiological studies have shown that OSAHS is a globally prevalent chronic sleep disorder, especially in adults with type 2 diabetes mellitus (T2DM), with rates varying from 23% to 87%[3,4]. Moderate to severe OSAHS has been found to be associated with an increase in the incidence of T2DM, and the incidence of OSAHS in patients with T2DM was > 50% higher than that in those without diabetes, independent of traditional risk factors, such as obesity and other confounders. IH, a hallmark of OSAHS, plays an important role not only in the pathogenesis of OSAHS but also in reducing glycemic control and insulin resistance in patients with T2DM and diabetes-related complications[5-7], probably by activating several systematic inflammatory mediators and enhancing the oxidative stress cascade and hypothalamic-pituitary-adrenal function, among others[8].
Commensal bacteria, termed microbiota, cover every surface of our body exposed to the external environment, with 70% of these residing in the gastrointestinal tract[9]. The microbiome is a vast and complex polymicrobial ecosystem that coexists with the human organism and has been identified as playing important roles in the development of host immunological phenotypes. Gut microbiota dysbiosis has been linked to a series of metabolic disorders, such as diabetes and hypertension[10]. In a study of IH mimicking OSAHS, in an Ldlr−/− mice fed a high-fat diet, the imbalance of gut microbiota was found to be associated with adverse cardiovascular events and metabolic disorders[11,12]. Similarly, sleep fragmentation induced by OSAHS alters feeding behavior and promotes obesity and metabolic abnormalities, while the host gut microbiome changes, leading to increased intestinal permeability and chronic inflammation of adipose tissues[11,12]. For T2DM patients with OSAHS, only a few studies have focused on the relationship between severe metabolic disorders caused by IH and gut microbial dysbiosis, independent of conventional risk factors, such as obesity and hypertension. Hence, our study aimed to investigate the gut microbiota changes in T2DM patients with OSAHS. Clinical indices, such as inflammatory factors and homocysteine (HCY), were compared for the groups as well.
MATERIALS AND METHODS
Participants
In total, 78 participants, including 25 hospitalized patients with T2DM (Group T2DM), 27 hospitalized patients with T2DM complicated by OSAHS (Group T2DM + OSA), and 26 healthy controls (HC) who were examined with a type IV portable monitor (PM, Sleep Fairy-A7, China) overnight (from 10 p.m. to 7 a.m.), were recruited from July 2019 to July 2020. Most of the sleep recording took place in a quiet and comfortable hospital ward at Henan Provincial People’s Hospital; the remainder was performed by participants sleeping at home after being systematically trained in using the PM. The patient-reported periods before sleep onset and after awakening in the morning were excluded before manual scoring[13]. We collected fasting blood and fecal samples the next morning. General questionnaires were used to collect information on demographic characteristics and health status.
OSAHS assessment
Pulse oximetry was used to assess oxyhemoglobin saturation, and respiratory effort was measured with a pneumatic sensor attached to an effort belt. Nasal airflow was recorded using a nasal cannula connected to a pressure transducer. The final data were automatically generated by the software system. The scoring rules were based on the 2007 American Academy of Sleep Medicine manual. The apnea-hypopnea index (AHI) was calculated as the total number of episodes of apnea (continuous cessation of airflow for at least 10 s) and hypopnea (reduction in airflow for ≥ 10 s with oxygen desaturation of ≥ 4%) divided by the total duration of sleep events, and OSAHS was defined as an AHI of 5 events/h.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (1) Age of 18-70 years; (2) Treatment-naïve type 2 diabetes based on the 1999 World Health Organization Criteria[14]; (3) 6.5% ≤ glycated hemoglobin A1c (HbA1c) ≤ 11%; (4) No antibiotic use within the 12 wk before enrollment and no probiotics and/or prebiotics; and (5) No glucose-lowering drugs other than insulin during the 12 wk before enrollment in patients with diabetes.
The exclusion criteria were as follows: (1) Body mass index (BMI) of > 28 kg/m2; (2) Diagnoses of chronic respiratory disease, central system sleep apnea syndrome, severe heart failure, BP of ≥ 140/90 mmHg, and severe organic diseases such as cancer, myocardial infarction, and stroke; (3) Diagnoses of other types of diabetes, for example, type 1 diabetes; (4) Diagnoses of inflammatory bowel disease, coagulation disorders, connective tissue diseases, or gastrointestinal surgery, except for appendicitis and hernia surgery; (5) Participation in other programs during the previous 3 mo; (6) Alcoholism (drinking more than five times in 1 wk: > 100 g of spirits, 250 g of rice wine, or 5 bottles of beer); (7) Pregnancy; and (8) Taking drugs that may affect respiratory function, for example, anxiolytics, hypnotics, or mood stabilizers.
Sample collection
Biological samples and anthropometric data were obtained without medical treatment. Blood samples were collected after overnight fasting and centrifuged using centrifuge (Multifuge X3R, Thermo Fisher Scientific, United States) at 3000 rpm for 20 min after standing at 24 °C for 30 min to obtain serum. Fresh fecal samples, morning urine, and serum samples were immediately frozen on dry ice after collection and stored at -80 °C until further analysis.
Clinical indices
Fasting plasma glucose (FPG) concentrations were measured with the Automatic Biochemical Analyzer (TBA-120 FR, Toshiba, Japan). Fasting insulin concentrations were measured using immunochemiluminometric assays (ADVIA Centaur, Siemens A.G., German). Plasma HbAlc concentrations were measured by high-performance liquid chromatography (Bio-Rad D-10, Bio-Rad Laboratories Co., Ltd., Germany). Routine blood tests were performed using the Swelab Alfa Cell analyzer (Boule Diagnostics AB, Sweden). The Homeostatic Model Assessment for Insulin Resistance (HOMA-IR) was calculated using the following formula: FPG*FIN/22.5, where FIN is fasting insulin.
Inflammatory factors and HCY
C-reactive protein (CRP) and HCY concentrations were measured with the Automatic Biochemical Analyzer. The determination of the plasma concentrations of the lipopolysaccharide-binding protein (LBP), interleukin (IL)-6, IL-17, and transforming growth factor (TGF)-β1 was performed using human ELISA kits according to the manufacturer's instructions. The ELISA kits for the assessment of the concentrations of human LBP, tumor necrosis factor (TNF)-α, IL-17, and TGF-β1 were all from Cusabio Biotech, Wuhan, China. The kits for high sensitivity IL-6 were purchased from Multi-Science, Hangzhou, China.
16S rRNA gene amplification sequencing
The DNA of the participants was extracted using the E.Z.N.A.® Stool DNA Kit (Omega Bio-tek, Inc., Norcross, GA, Unite States). The isolated DNA was used as the template for polymerase chain reaction (PCR) amplification of the V3-V4 region of 16S rRNA genes. The forward primer (341F) was 5'-CCTACGGGNGGCWGCAG-3', and the reverse primer (805R) was 5'-GACTACHVGGGTATCTAATCC-3.' PCR was performed with an EasyCycler 96 PCR system (Analytik Jena Corp., AG), and the products from different samples were indexed and mixed at equal ratios for sequencing by Shanghai Mobio Biomedical Technology Co. Ltd. using the Miseq platform (Illumina Inc., San Diego, CA, Unite States) according to the manufacturer’s instructions.
Sequencing data analysis
Amplicon sequence variants (ASVs) were identified with the DADA2 algorithm. The representative sequences for each ASV were annotated using the SILVA reference database (SSU138). The alpha diversity metrics (Shannon–Wiener diversity index and Simpson diversity index) were assessed by using Mothur v1.42.1. The non-parametric Mann–Whitney U test was used to test for significant differences between the two groups. The comparison of multiple groups was performed using a nonparametric Kruskal–Wallis test. Principal coordinates analysis (PCoA) based on Bray–Curtis and unweighted UniFrac distance was conducted using the R program (version 3.6.0, http://www.R-project.org/) to display microbiome space between samples. The key ASVs associated with T2DM and OSA were identified by random forest models, and those significantly associated with BMI selected by MaAsLin2 (https://github.com/biobakery/Maaslin2) were removed. A heatmap plot was drawn to indicate the distributions of the rest of the key ASVs. The linear discriminant analysis effect size (LEfSe) method (lefse 1.1, https://github.com/SegataLab/Lefse) was used to detect taxa with differential abundance. PICRUSt2 v2.4.1 (https://github.com/picrust/picrust2/wiki) was used to predict functional abundance based on 16S rRNA gene sequences.
Statistical analyses
Continuous variables are expressed as means ± SD or medians with interquartile ranges for normal distribution data or non-normal distribution data, respectively. Categorical variables are expressed as percentages. All the statistical analyses, including one-way analysis of variance, Kruskal–Wallis test, Mann-Whitney U test, and least significant difference t-test, were performed using SPSS version 26.0, with a two-sided P value of < 0.05 denoting statistical significance. Significant differences were adjusted by Bonferroni correction. The correlation coefficients for the concentrations of the gut microbiota, inflammatory factors, and HCY were evaluated using Spearman correlation.
RESULTS
Clinical parameters and inflammatory levels among three groups
Applying the strict inclusion and exclusion criteria described above, a total of 78 fecal samples were collected for analysis after PM assessment. We characterized the clinical indices and gut microbiomes of patients in groups HC, T2DM, and T2DM + OSA (Figure 1). There were no significant differences in sex and age among the three groups (P > 0.05). Compared with group T2DM, there were no significant differences in the insulin resistance index (HOMA-IR), BMI, waist-to-hip ratio, neck circumference, FPG, HbA1c, systolic BP, and diastolic BP in group T2DM + OSA; however, they were significantly increased compared with those in group HC (P < 0.05).
Figure 1.
Flowchart of the study.
The HCY level was the highest in group T2DM + OSA compared with the other two groups. Our PM data revealed that the AHI and oxygen desaturation index (ODI) increased significantly, accompanied by a decrease in the mean oxygen saturation (SpO2); group T2DM + OSA had lower SpO2 than group HC. Further details on the clinical parameters of the participants with diverse severities are shown in Table 1.
Table 1.
Participant characteristics and clinical parameters
|
|
HC (n = 26)
|
T2DM (n = 25)
|
T2DM + OSA (n =27)
|
P
value
|
| Sex, male/female | 17/9 | 17/8 | 21/6 | 0.584 |
| Age, yr | 45.58 ± 8.81 | 45.92 ± 13.89 | 47.59 ± 5.18 | 0.728 |
| BMI, kg/m2 | 24.63 ± 2.61 | 25.84 ± 3.45 | 27.03 ± 2.11b | 0.009 |
| WHR, cm | 0.85 ± 0.06 | 0.95 ± 0.07 | 0.98 ± 0.05b | < 0.001 |
| NC, cm | 32.88 ± 3.70 | 36.25 ± 4.75 | 38.37 ± 2.31b | < 0.001 |
| FPG, mmol/L | 5.30 (4.70, 5.43) | 6.90 (6.30, 8.05) | 8.00 (6.40, 9.25)b | < 0.001 |
| HOMA-IR | 1.50 (1.09, 2.03) | 2.10 (1.02, 3.53) | 2.38 (1.37, 3.22)a | 0.099 |
| HbA1c, % | 5.30 (5.10, 5.60) | 9.70 (7.30, 10.80) | 8.70 (7.70, 9.70)b | < 0.001 |
| SBP, mmHg | 117.50 (112.00, 123.25) | 130.00 (117.00, 143.00) | 131.00 (128.00, 144.00)b | < 0.001 |
| DBP, mmHg | 71.96 ± 5.44 | 81.13 ± 11.33 | 82.26 ± 8.58b | < 0.001 |
| HCY, μmol/L | 9.60 (8.30, 12.53) | 10.30 (8.05, 12.03) | 13.81 (10.73, 20.54)b,c | < 0.001 |
| AHI | 2.30 (1.48, 3.05) | 3.70 (2.00, 4.15) | 13.70 (9.80, 20.10)b,c | < 0.001 |
| Mean SpO2, % | 97.05 (96.50, 97.53) | 96.60 (96.05, 96.95) | 94.70 (93.80, 95.20)b,c | < 0.001 |
| Lowest SpO2, % | 85.50 (82.00, 90.25) | 85.00 (76.00, 87.50) | 81.00 (73.00, 84.00)b | 0.002 |
| ODI | 0.65 (0.40, 1.23) | 2.20 (1.10, 5.45) | 9.10 (5.90, 15.30)b,c | < 0.001 |
P < 0.05 vs HC.
P < 0.01 vs HC.
P < 0.01 vs T2DM.
P value: Comparison of three groups. Data are presented as the mean ± SD or median with interquartile range. The P value was based on Kruskal–Wallis test or one-way ANOVA. Mann–Whitney U test or least significant difference t-test was used to compare two groups. Significant differences were adjusted by Bonferroni correction. WHR: Waist-hip ratio; NC: Neck circumstance; FPG: Fasting plasma glucose; HOMA-IR: Insulin resistance index; HbA1c: Hemoglobin A1c; SBP: Systolic pressure; DBP: Diastolic pressure; HCY: Homocysteine; AHI: Apnea–hypopnea indices; ODI: Oxygen desaturation index; HC: Healthy control; T2DM: Type 2 diabetes mellitus; OSA: Obstructive sleep apnea.
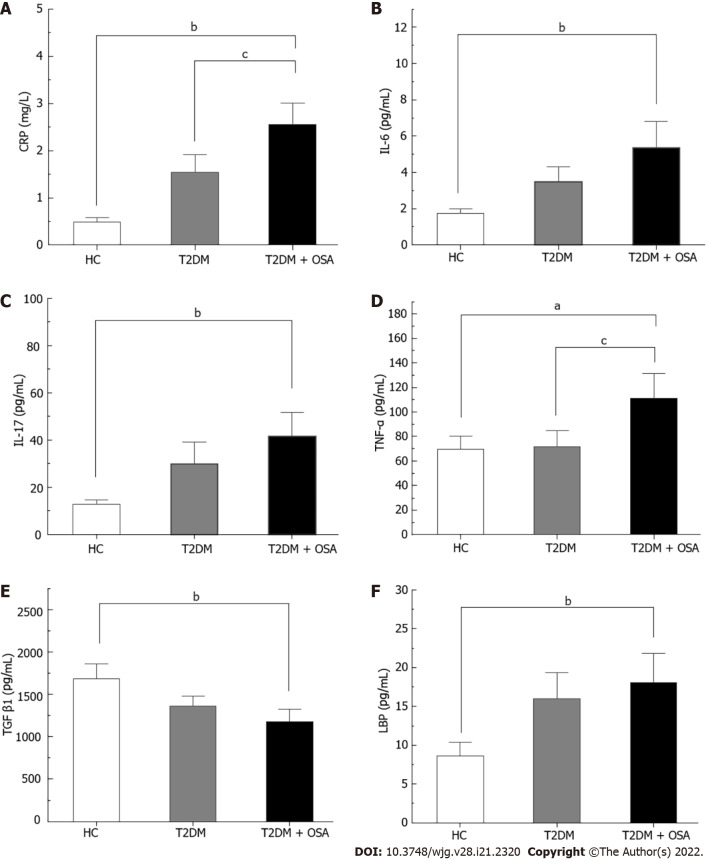
The concentration of CRP increased gradually from the HC group to the T2DM group and even more in the T2DM + OSA group. In addition, the levels of TNF-α, IL-17, IL-6, and LBP increased and the levels of TGF-β1 decreased in group T2DM + OSA compared with those in group HC, while the level of TNF-α increased compared with that in group T2DM (P < 0.05) (Figure 2 and Supplementary Table 1).
Figure 2.
The concentrations of inflammatory factors in groups healthy control, type 2 diabetes mellitus, and type 2 diabetes mellitus plus obstructive sleep apnea. A: CRP; B: IL-6; C: IL-17; D: TNF-α; E: TGF-β1; F: LBP. Bar charts show the means ± SE of the mean. aP < 0.05 vs healthy control (HC); bP < 0.01 vs HC; cP < 0.05 vs type 2 diabetes mellitus. The P value was based on Kruskal–Wallis test or one-way ANOVA. A significant difference was shown by the least significant difference t-test or Mann–Whitney U test. Significant differences were adjusted by Bonferroni correction. CRP: C-reaction protein; IL-6: Interleukin-6; IL-17: Interleukin-17; TNF-α: Tumor necrosis factor-α; TGF-β1: Transforming growth factor beta 1; LBP: Lipopolysaccharide-binding protein; HC: Healthy control; T2DM: Type 2 diabetes mellitus; OSA: Obstructive sleep apnea.
IH is related to differences in the gut microbiota of T2DM + OSA patients
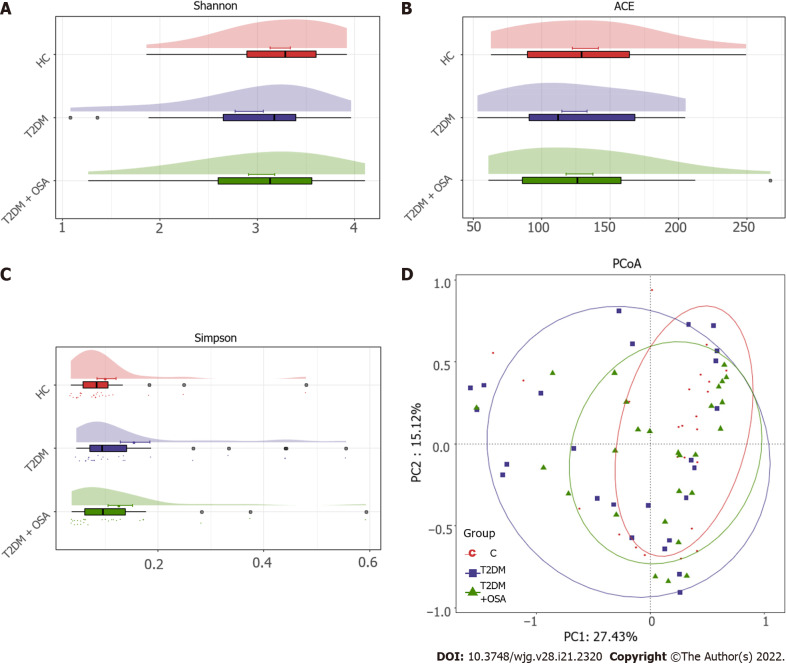
The resulting rarefaction curves showed that the microbial richness of the sampled guts was near saturation at the applied sequencing depth (Supplementary Figure 1), which was sufficient to identify most of the bacterial community members for each microbiome. The alpha diversity of the gut microbiota expressed by the Shannon estimator, ACE estimator, and Simpson index (Figure 3A-C and Supplementary Table 2) showed that there were no significant differences in groups HC, T2DM, and T2DM + OSA (P > 0.05). The overall structures of the gut microbiota in the three groups showed a minimal difference as revealed by the PCoA plot (Figure 3D); however, the difference was not significant (Adonis, P > 0.05).
Figure 3.
The gut microbial diversity and structures of the gut microbiota in groups healthy control, type 2 diabetes mellitus, and type 2 diabetes mellitus plus obstructive sleep apnea. A: Shannon index; B: ACE estimator; C: Simpson index; D: Principal coordinates analysis plot of the gut microbiota among groups healthy control, type 2 diabetes mellitus, and type 2 diabetes mellitus plus obstructive sleep apnea. HC: Healthy control; T2DM: Type 2 diabetes mellitus; OSA: Obstructive sleep apnea.
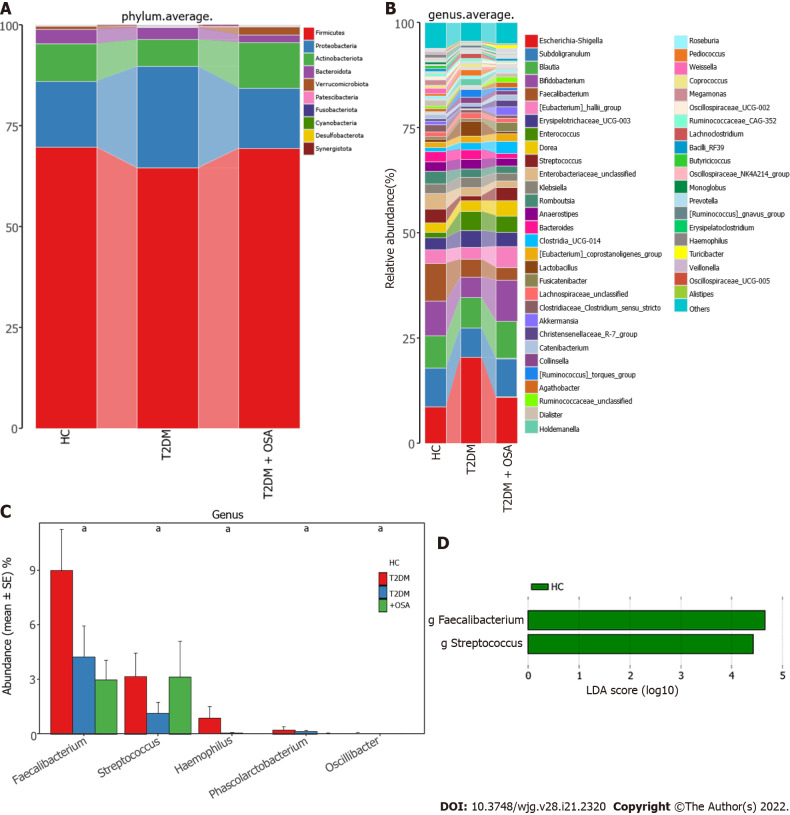
We further analyzed the taxonomic composition and alterations of the gut microbiome. The composition and abundance of the bacterial community in each sample at the phylum and genus levels are shown in Supplementary Figure 2. At the phylum level, Firmicutes and Proteobacteria were the dominant bacteria in groups HC, T2DM, and T2DM + OSA (Figure 4A). At the genus level, we found no significant differences in the relative abundance of Escherichia−Shigella, which had the highest abundance among the three groups (P > 0.05) (Figure 4B). The relative abundance of the following genera significantly differed among groups: Faecalibacterium (P = 0.0434), Streptococcus (P = 0.0393), Haemophilus (P = 0.0286), Phascolarctobacterium (P = 0.0242), and Oscillibacter (P = 0.0274) (Figure 4C). Among the above genera, the levels of Phascolarctobacterium decreased and the levels of Oscillibacter increased in group T2DM + OSA compared with group T2DM, and the levels of Faecalibacterium significantly decreased in group T2DM + OSA compared with group HC. A gradually decreasing trend of Faecalibacterium abundance from group HC to groups T2DM and T2DM + OSA was observed (Figure 4C). Through LEfSe, we also found a significant decrease in the level of Faecalibacterium in group T2DM + OSA (Figure 4D).
Figure 4.
Average compositions and relative abundance of the bacterial community among three groups at different levels. A: Average relative abundance histogram of dominant species composition stratified by group at the phylum levels; B: Average relative abundance histogram of dominant species composition stratified by group at the genus levels; C: Histograms for genera with significant differences. Bar chart showing the mean ± SE of each group; D: Linear discriminant analysis analysis column diagram. LDA: Linear discriminant analysis. aP < 0.05. Significant difference shown by the Kruskal–Wallis rank-sum test.
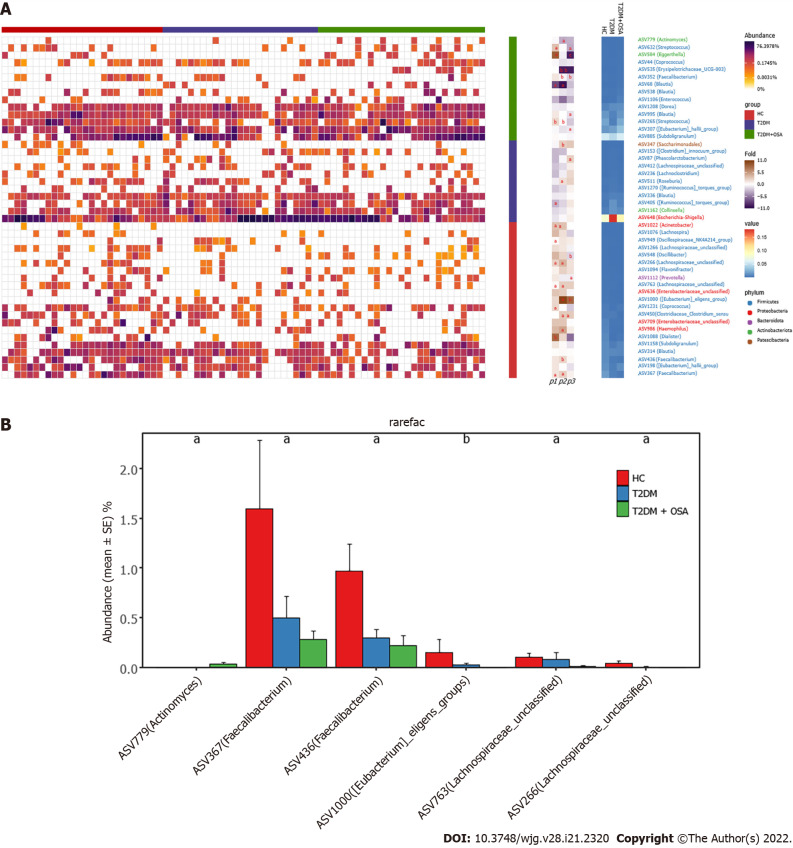
We found 12 ASVs associated with gut microbial dysbiosis in group T2DM + OSA compared with group T2DM, including ASV632 (Streptococcus), ASV450 (Clostridiaceae_Clostridium_sensu_stricto), ASV352 (Faecalibacterium), ASV511 (Roseburia), ASV307 ([Eubacterium]_hallii_group), ASV1000 ([Eubacterium]_eligens_group), ASV995 (Blautia), ASV584 (Eggerthella), ASV535 (Erysipelotrichaceae_UCG-003), ASV87 (Phascolarctobacterium), ASV1112 (Prevotella), and ASV548 (Oscillibacter). On comparing groups HC and T2DM + OSA, 15 ASVs were different: ASV450 (Clostridiaceae_Clostridium_sensu_stricto), ASV511 (Roseburia), ASV1000 ([Eubacterium]_eligens_group), ASV266 (Lachnospiraceae_unclassified), ASV763 (Lachnospiraceae_unclassified), ASV367 (Faecalibacterium), ASV779 (Actinomyces), ASV986 (Haemophilus), ASV352 (Faecalibacterium), ASV436 (Faecalibacterium), ASV265 (Streptococcus), ASV535 (Erysipelotrichaceae_UCG-003), ASV68 (Blautia), ASV347 (Saccharimonadales), and ASV1022 (Acinetobacter) (Figure 5A). Therein, the relative abundance of ASV1022 (Acinetobacter), ASV367 (Faecalibacterium), ASV436 (Faecalibacterium), ASV763 (Lachnospiraceae_unclassified), and ASV266 (Lachnospiraceae_unclassified) were progressively decreased, and the abundance of ASV779 (Actinomyces) increased from group HC to group T2DM and to group T2DM + OSA (Figure 5B).
Figure 5.
Amplicon sequence variants contributing to the changes in the gut microbiota in groups healthy control, type 2 diabetes mellitus, and type 2 diabetes mellitus plus obstructive sleep apnea. A: The heatmap shows the relative abundance of the 46 amplicon sequence variants (ASVs) significantly different in the three groups. The fold ratio (log2-transformed) indicated the relative abundance of the 46 ASVs between the two different groups [P1: Type 2 diabetes mellitus (T2DM) + obstructive sleep apnea (OSA) vs T2DM; P2: T2DM + OSA vs healthy control (HC); P3: HC vs T2DM]. aP < 0.05, bP < 0.01, cP < 0.001. Significant difference shown by the Mann–Whitney U test; B: With the increase in disease components, six key ASVs showed increasing or decreasing trends. aP < 0.05, bP < 0.01.
IH-related gut microbiota dysbiosis was associated with abnormal metabolic and inflammatory indicators of T2DM + OSA patients
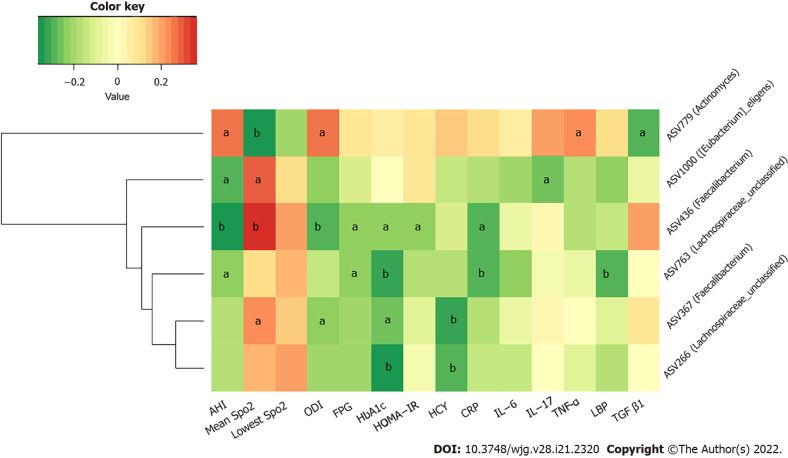
We further studied the correlations among metabolic indicators, inflammatory factors, and the above six key ASVs with ascending or decreasing trends. Spearman correlation analysis revealed that the decreased abundance of ASVs showed a significant negative correlation with indicators related to IH or respiratory disorders, such as the ODI and AHI; glucose metabolism indicators, such as HbA1c, FPG, and HOMA-IR; cardiovascular disease-related metabolic indicators, such as HCY; and inflammatory factors, such as CRP, TNF-α, and LBP. The ASV436 (Faecalibacterium) was negatively correlated with the AHI, ODI, FPG, HbA1c, CRP concentrations, and HOMA-IR, while the ASV763 (Lachnospiraceae_unclassified) was negatively correlated with the AHI and FPG, HbA1c, CRP, and LBP concentrations (P < 0.05). Furthermore, the ASV436 (Faecalibacterium), ASV1000 ([Eubacterium]_eligens_group), and ASV367 (Faecalibacterium) were positively correlated with the mean SpO2, another IH-related indicator. Among the three groups, the gradually increasing abundance of ASV779 (Actinomyces) showed an opposite relationship; it was significantly positively correlated with the AHI, ODI, and TNF-α concentration and negatively correlated with the mean SpO2 (P < 0.05) (Figure 6 and Supplementary Table 3).
Figure 6.
Heatmap of correlation among key amplicon sequence variants of gut microbiota and metabolic and inflammatory indicators. The color of the cells represents the Spearman’s correlation coefficient between each amplicon sequence variant and clinical parameter. aP < 0.05; bP < 0.01.
DISCUSSION
T2DM has been linked to gut dysbiosis and chronic inflammation in several clinical and animal experiments[15-17], which may be a consequence of the loss of or deficiency in a beneficial function, such as short-chain fatty acid (SCFA)-producing bacteria production from carbohydrate fermentation, in the gut ecosystem[16]. Diseases previously partially attributed to lifestyle, such as obesity and OSAHS, are now considered microbiota-related as well[12,18]. Although much epidemiological and clinical evidence has suggested that OSAHS is an independent risk factor for the development of T2DM[19], the underlying pathogenesis of altered glucose metabolism in T2DM patients with OSAHS remains to be elucidated. Meanwhile, a longitudinal cohort study over a period of 6 years found that the insulin resistance index (HOMA-IR) was a predictor of incident “witnessed apnea”, independent of obesity[20]. This showed that dysglycemia and insulin resistance may lead to the development of OSAHS. Together with the findings of the above-mentioned studies, this strongly suggests that the relationship between T2DM and OSAHS may be bidirectional[2]. Therefore, it is necessary to investigate whether the imbalance of the intestinal microbiota plays a key role in the pathophysiology underlying metabolic dysfunction of patients with T2DM complicated by OSAHS.
In our study, the sequencing analysis of the 16S rRNA gene-tags applied to fecal samples from T2DM patients complicated by OSAHS showed differences in the relative abundances of the predominant taxa of the genera levels. We found that the concentrations of various IH-related gut bacteria, including SCFA-producing bacteria such as Faecalibacterium and Lachnospiraceae, were significantly correlated with the concentrations of FPG HbA1c and HOMA-IR, as well as the concentration of HCY, a risk predictor of hypertension and arteriosclerosis[21,22]. IH can result in hypoxia/re-oxygenation cycling events within the gut microbiome and, as a result, the biological diversity of gut microorganisms may be modified[16]. Although the intestinal epithelium is significantly resistant to hypoxia, regulating the absorption and barrier function of the intestinal epithelium is sensitive to the oxygen level in the intestine[23]. Hypoxia/re-oxygenation can directly impair cellular function via an increase in permeability and bacterial translocation and a decrease in tight junction integrity[16]. In addition, studies have shown that after prolonged normoxic recovery after IH exposure, the gut microbiota and circulating endotoxemia remain negatively affected[8]. Our results do not show significant differences in α-diversity and β−diversity. However, the gradual decrease in the relative abundance of SCFA-producing bacteria (such as the ASVs of Faecalibacterium, Eubacterium, and Lachnospiraceae) associated with abnormal indicators of oxygen metabolism, as well as elevated levels of inflammatory indicators (including CRP, IL-17, and TNF-α), which are critically involved in the development of insulin resistance and pathogenesis of T2DM, was observed in T2DM + OSA patients[24]. Emerging evidence shows that SCFAs can modulate glycemic control, exhibit anti-inflammatory and antitumorigenic activity, and decrease oxidative stress[25-28]. Short chain fatty acids contribute to mucin synthesis, decrease bacterial translocation, maintain gut integrity, and mitigate inflammation in the intestine[29,30]. Thus, we speculated that SCFAs may be regarded as potential targets for recognizing metabolic comorbidities in patients with T2DM complicated by OSAHS.
There are several potential mechanisms by which IH mediates its effect on metabolic dysfunction. It induces macrophages to polarize toward the pro-inflammatory subtype of M1, leading to the production of more pro-inflammatory mediators in visceral adipose tissue, such as TNF-α, IL-6, and IL-8, resulting in subsequent impairment of insulin signaling pathways and insulin resistance[7]. Here, we found that the concentration of Lachnospiraceae was negatively correlated with that of LBP, which is one of the reference indicators of intestinal barrier disruption, suggesting that the induction of inflammatory processes may be due to the leakage of microbial metabolites into the circulation induced by IH. We found certain changes in the gut microbiota among the three groups over time; however, there were no significant differences between patients with T2DM with and without OSAHS. Nonetheless, the concentrations of inflammatory indicators, such as CRP, TNF-α, and IL-17, were significantly increased in T2DM patients with OSAHS, which indicates that the changes in gut microbiota may have been delayed relative to the chronic inflammatory changes in T2DM patients with OSAHS. On the other hand, some patients with relatively mild disease were included to prevent confounding by factors, including hyperglycemia and obesity, which affect the composition of intestinal microbiota[31]; as a result, the intestinal flora appeared not to have changed remarkably.
Various respiratory diseases have been associated with dysbiosis not only in the airway microbiota but also in the intestinal microbiota[32,33]. This evidence reinforced the existence of a gut-lung axis and the close relationship between intestinal and respiratory compartments; changes at one of the two sites could impact the other[34]. As a minor constituent of the airway microbiota, Actinomyces is related to anaerobic enzymes through GLUT1-dependent glucose elevation and MCT4-dependent lactate transport[35,36]. In our study, we observed that the increase in the relative abundance of Actinomyces was positively correlated with OSAHS severity indices and the concentration of TNF-α. We did not test for lung microbes; however, we speculate that they may be related to the gut-lung axis. Further experimental approaches to exploring causal links may be needed.
There are some limitations to our study. First, the sample size was relatively small. Second, the causal relationship between T2DM complicated by OSAHS and the gut microbiota was unclear. Large-scale clinical trials and gnotobiotic mice model validation may be required in the future.
CONCLUSION
T2DM patients with OSAHS may have a higher prevalence of gut microbial dysbiosis. IH may be involved in selective alterations of gut microbiota, which may be related to increased gut permeability and concurrent systemic inflammatory changes in patients with T2DM complicated by OSAHS. These findings provide foundations for further studies on the mechanisms and interventional approaches aimed at restoration of the gut microbiota to prevent or to palliate the adverse effects of T2DM patients with OSAHS.
ARTICLE HIGHLIGHTS
Research background
Obstructive sleep apnea (OSA)-hypopnea syndrome (OSAHS), as a chronic and treatable sleep disorder, has a high prevalence in type 2 diabetes mellitus (T2DM) patients. As a landmark feature of OSAHS, intermittent hypoxia (IH) plays an important role in the occurrence and development of related complications in T2DM patients. However, the pathological mechanisms are varied and unknown. Therefore, it is important to study the role of gut microbiota, a meaningful new target, in T2DM patients with OSAHS.
Research motivation
In recent years, it has been found that gut microbiota imbalance is related to metabolic diseases. However, most studies have not discussed the relationship between gut microbiota changes and T2DM patients with OSAHS.
Research objectives
In this study we focused on IH that might be involved in altering the gut dysbiosis in T2DM patients with OSAHS. Meanwhile, we further assessed the changes of clinical indicators and inflammatory factors related to dysbiosis, aiming to provide new targets and perspectives for the pathogenesis and prevention strategies of T2DM patients complicated with OSAHS.
Research methods
A case-control study was conducted to select subjects who were divided into T2DM + OSA group, T2DM group, and healthy control group. They were examined with a type IV portable monitor overnight. The clinical indexes, respiratory parameters, inflammatory indexes, and gut microbial community of the three groups were measured.
Research results
Among the three groups, T2DM + OSA group showed the most severe changes in sleep apnea parameters and increased systemic inflammatory factors. We found the decreased levels of short-chain fatty acid-related Faecalibacterium, Eubacterium, and Lachnospiraceae and the increased levels of Actinomyces at the amplicon sequence variant level. The changes in these gut microbiotas were closely related to clinical indicators as well.
Research conclusions
IH may be involved in the selective changes of intestinal microbiota, which may be related to the increased intestinal permeability and systemic inflammation response in T2DM patients with OSAHS.
Research perspectives
This study shows that IH may change the state of gut microbiota and systemic inflammation, which participate in the occurrence and development of T2MD complicated with OSAHS. In the future, large-scale clinical randomized controlled prospective trials and animal trials may be needed to further explore the corresponding causality.
ACKNOWLEDGEMENTS
We would like to thank the participants involved in this study.
Footnotes
Institutional review board statement: This study was reviewed and approved by the Ethics Committee of Henan Provincial People’s Hospital (IRB No. 2019-62).
Informed consent statement: All study participants provided informed written consent before the study enrollment.
Conflict-of-interest statement: All authors declare no conflicts of interest.
STROBE statement: The authors have read the STROBE Statement—checklist of items, and the manuscript was prepared and revised according to the STROBE Statement—checklist of items.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: January 8, 2022
First decision: March 9, 2022
Article in press: April 22, 2022
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Papazafiropoulou A, Greece; Trujillo X, Mexico S-Editor: Fan JR L-Editor: A P-Editor: Yu HG
Contributor Information
Sha-Sha Tang, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Cheng-Hong Liang, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Ya-Lei Liu, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Wei Wei, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Xin-Ru Deng, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Xiao-Yang Shi, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Li-Min Wang, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Li-Jun Zhang, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China.
Hui-Juan Yuan, Department of Endocrinology, Henan Provincial Key Medicine Laboratory of Intestinal Microecology and Diabetes, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, Henan Province, China. hjyuan@zzu.edu.cn.
Data sharing statement
Raw sequencing data and associated metadata were deposited in the NCBI short-read archive with BioProject accession number PRJNA788301.
References
- 1.Xu H, Wang H, Guan J, Yi H, Qian Y, Zou J, Xia Y, Fu Y, Li X, Jiao X, Huang H, Dong P, Yu Z, Yang J, Xiang M, Li J, Chen Y, Wang P, Sun Y, Li Y, Zheng X, Jia W, Yin S. Effects of continuous positive airway pressure on neurocognitive architecture and function in patients with obstructive sleep apnoea: Study protocol for a multicentre randomised controlled trial. BMJ Open. 2017;7:e014932. doi: 10.1136/bmjopen-2016-014932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subramanian A, Adderley NJ, Tracy A, Taverner T, Hanif W, Toulis KA, Thomas GN, Tahrani AA, Nirantharakumar K. Risk of Incident Obstructive Sleep Apnea Among Patients With Type 2 Diabetes. Diabetes Care. 2019;42:954–963. doi: 10.2337/dc18-2004. [DOI] [PubMed] [Google Scholar]
- 3.Foster GD, Sanders MH, Millman R, Zammit G, Borradaile KE, Newman AB, Wadden TA, Kelley D, Wing RR, Sunyer FX, Darcey V, Kuna ST Sleep AHEAD Research Group. Obstructive sleep apnea among obese patients with type 2 diabetes. Diabetes Care. 2009;32:1017–1019. doi: 10.2337/dc08-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West SD, Prudon B, Hughes J, Gupta R, Mohammed SB, Gerry S, Stradling JR ROSA trial investigators. Continuous positive airway pressure effect on visual acuity in patients with type 2 diabetes and obstructive sleep apnoea: a multicentre randomised controlled trial. Eur Respir J. 2018;52 doi: 10.1183/13993003.01177-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahrani AA. Obstructive sleep apnoea in diabetes: Does it matter? Diab Vasc Dis Res. 2017;14:454–462. doi: 10.1177/1479164117714397. [DOI] [PubMed] [Google Scholar]
- 6.Leong WB, Jadhakhan F, Taheri S, Chen YF, Adab P, Thomas GN. Effect of obstructive sleep apnoea on diabetic retinopathy and maculopathy: a systematic review and meta-analysis. Diabet Med. 2016;33:158–168. doi: 10.1111/dme.12817. [DOI] [PubMed] [Google Scholar]
- 7.Ryan S. Adipose tissue inflammation by intermittent hypoxia: mechanistic link between obstructive sleep apnoea and metabolic dysfunction. J Physiol. 2017;595:2423–2430. doi: 10.1113/JP273312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-Indias I, Torres M, Sanchez-Alcoholado L, Cardona F, Almendros I, Gozal D, Montserrat JM, Queipo-Ortuño MI, Farré R. Normoxic Recovery Mimicking Treatment of Sleep Apnea Does Not Reverse Intermittent Hypoxia-Induced Bacterial Dysbiosis and Low-Grade Endotoxemia in Mice. Sleep. 2016;39:1891–1897. doi: 10.5665/sleep.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schippa S, Conte MP. Dysbiotic events in gut microbiota: impact on human health. Nutrients. 2014;6:5786–5805. doi: 10.3390/nu6125786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okubo H, Nakatsu Y, Kushiyama A, Yamamotoya T, Matsunaga Y, Inoue MK, Fujishiro M, Sakoda H, Ohno H, Yoneda M, Ono H, Asano T. Gut Microbiota as a Therapeutic Target for Metabolic Disorders. Curr Med Chem. 2018;25:984–1001. doi: 10.2174/0929867324666171009121702. [DOI] [PubMed] [Google Scholar]
- 11.Tripathi A, Melnik AV, Xue J, Poulsen O, Meehan MJ, Humphrey G, Jiang L, Ackermann G, McDonald D, Zhou D, Knight R, Dorrestein PC, Haddad GG. Intermittent Hypoxia and Hypercapnia, a Hallmark of Obstructive Sleep Apnea, Alters the Gut Microbiome and Metabolome. mSystems. 2018;3 doi: 10.1128/mSystems.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko CY, Liu QQ, Su HZ, Zhang HP, Fan JM, Yang JH, Hu AK, Liu YQ, Chou D, Zeng YM. Gut microbiota in obstructive sleep apnea-hypopnea syndrome: disease-related dysbiosis and metabolic comorbidities. Clin Sci (Lond) 2019;133:905–917. doi: 10.1042/CS20180891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjorvatn B, Lehmann S, Gulati S, Aurlien H, Pallesen S, Saxvig IW. Prevalence of excessive sleepiness is higher whereas insomnia is lower with greater severity of obstructive sleep apnea. Sleep Breath. 2015;19:1387–1393. doi: 10.1007/s11325-015-1155-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Fang Z, Zhang C, Xia H, Jie Z, Han X, Chen Y, Ji L. Effects of Acarbose on the Gut Microbiota of Prediabetic Patients: A Randomized, Double-blind, Controlled Crossover Trial. Diabetes Ther. 2017;8:293–307. doi: 10.1007/s13300-017-0226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019;15:261–273. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 16.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, Yu L, Xu C, Ren Z, Xu Y, Xu S, Shen H, Zhu X, Shi Y, Shen Q, Dong W, Liu R, Ling Y, Zeng Y, Zhang Q, Wang J, Wang L, Wu Y, Zeng B, Wei H, Zhang M, Peng Y, Zhang C. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 17.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3:279–288. doi: 10.4161/gmic.19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labarca G, Reyes T, Jorquera J, Dreyse J, Drake L. CPAP in patients with obstructive sleep apnea and type 2 diabetes mellitus: Systematic review and meta-analysis. Clin Respir J. 2018;12:2361–2368. doi: 10.1111/crj.12915. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Bi Y, Zhang Q, Pan F. Obstructive sleep apnoea and the risk of type 2 diabetes: a meta-analysis of prospective cohort studies. Respirology. 2013;18:140–146. doi: 10.1111/j.1440-1843.2012.02267.x. [DOI] [PubMed] [Google Scholar]
- 20.Balkau B, Vol S, Loko S, Andriamboavonjy T, Lantieri O, Gusto G, Meslier N, Racineux JL, Tichet J Epidemiologic Study on the Insulin Resistance Syndrome Study Group. High baseline insulin levels associated with 6-year incident observed sleep apnea. Diabetes Care. 2010;33:1044–1049. doi: 10.2337/dc09-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monneret D, Tamisier R, Ducros V, Garrel C, Levy P, Baguet JP, Faure P, Pépin JL. The impact of obstructive sleep apnea on homocysteine and carotid remodeling in metabolic syndrome. Respir Physiol Neurobiol. 2012;180:298–304. doi: 10.1016/j.resp.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, Xu Y, Wang G. Homocysteine Levels are Associated with Endothelial Function in Newly Diagnosed Type 2 Diabetes Mellitus Patients. Metab Syndr Relat Disord. 2019;17:323–327. doi: 10.1089/met.2019.0011. [DOI] [PubMed] [Google Scholar]
- 23.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdel-Moneim A, Bakery HH, Allam G. The potential pathogenic role of IL-17/Th17 cells in both type 1 and type 2 diabetes mellitus. Biomed Pharmacother. 2018;101:287–292. doi: 10.1016/j.biopha.2018.02.103. [DOI] [PubMed] [Google Scholar]
- 25.Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI, Mehrabian M, Denu JM, Bäckhed F, Lusis AJ, Rey FE. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol. 2018;3:1461–1471. doi: 10.1038/s41564-018-0272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Liang R, Zhang W, Tian K, Li J, Chen X, Yu T, Chen Q. Faecalibacterium prausnitzii-derived microbial anti-inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. J Diabetes. 2020;12:224–236. doi: 10.1111/1753-0407.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang JD, Myers CJ, Harris SC, Kakiyama G, Lee IK, Yun BS, Matsuzaki K, Furukawa M, Min HK, Bajaj JS, Zhou H, Hylemon PB. Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem Biol. 2019;26:27–34.e4. doi: 10.1016/j.chembiol.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boesmans L, Valles-Colomer M, Wang J, Eeckhaut V, Falony G, Ducatelle R, Van Immerseel F, Raes J, Verbeke K. Butyrate Producers as Potential Next-Generation Probiotics: Safety Assessment of the Administration of Butyricicoccus pullicaecorum to Healthy Volunteers. mSystems. 2018;3 doi: 10.1128/mSystems.00094-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox AJ, West NP, Cripps AW. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015;3:207–215. doi: 10.1016/S2213-8587(14)70134-2. [DOI] [PubMed] [Google Scholar]
- 30.Moustafa A, Li W, Anderson EL, Wong EHM, Dulai PS, Sandborn WJ, Biggs W, Yooseph S, Jones MB, Venter JC, Nelson KE, Chang JT, Telenti A, Boland BS. Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin Transl Gastroenterol. 2018;9:e132. doi: 10.1038/ctg.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He FF, Li YM. Role of gut microbiota in the development of insulin resistance and the mechanism underlying polycystic ovary syndrome: a review. J Ovarian Res. 2020;13:73. doi: 10.1186/s13048-020-00670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruzzese E, Callegari ML, Raia V, Viscovo S, Scotto R, Ferrari S, Morelli L, Buccigrossi V, Lo Vecchio A, Ruberto E, Guarino A. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS One. 2014;9:e87796. doi: 10.1371/journal.pone.0087796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, Delhaes L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front Cell Infect Microbiol. 2020;10:9. doi: 10.3389/fcimb.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsland BJ, Trompette A, Gollwitzer ES. The Gut-Lung Axis in Respiratory Disease. Ann Am Thorac Soc. 2015;12 Suppl 2:S150–S156. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- 35.Huang D, Su X, Yuan M, Zhang S, He J, Deng Q, Qiu W, Dong H, Cai S. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am J Cancer Res. 2019;9:2047–2063. [PMC free article] [PubMed] [Google Scholar]
- 36.Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, Zhang Y, Surendra A, Gong Y, Elizabeth Tullis D, Yau YC, Waters VJ, Hwang DM, Guttman DS. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. 2015;5:10241. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing data and associated metadata were deposited in the NCBI short-read archive with BioProject accession number PRJNA788301.